Abstract
MINT‐7968768, MINT‐7968779: Tudor‐SN (uniprotkb:Q7KZF4) physically interacts (MI:0915) with G3BP (uniprotkb:Q13283) by anti bait coimmunoprecipitation (MI:0006) MINT‐7968800: Tudor‐SN (uniprotkb:Q7KZF4) and TIA‐1 (uniprotkb:P31483) colocalize (MI:0403) by fluorescence microscopy (MI:0416) MINT‐7968789: Tudor‐SN (uniprotkb:Q7KZF4) and G3BP (uniprotkb:Q13283) colocalize (MI:0403) by fluorescence microscopy (MI:0416)
Keywords: Tudor-SN, G3BP, Stress granule, Tudor domain containing protein
1. Introduction
Stress granules (SGs) are dynamic dense structures that are rapidly formed in the cytosol in response to a variety of environmental stress stimuli. Stress conditions induce extensive reprogramming in mRNA metabolism including induction of transcription and translation of specific genes to repair stress‐induced damage and adapt to changed conditions. As a consequence, many other genes are silenced via the recruitment of mRNA into SG that stalled with translation pre‐initiation complexes [1]. Once the stress condition is released, the SGs are disassembled, and mRNAs are repacked into translationally competent mRNPs and proteins are synthesized.
Several components of SGs have been identified, but their composition is still only partially known. SGs are composed of mRNAs in conjunction with a subset of translation initiation factors, including eIF2, eIF2B, eIF4E, the 40S ribosomal subunit, as well as RNA binding proteins. Notable RNA‐binding proteins in SGs include TIA‐1 [2], and G3BP [3], all of which have self‐interaction domains that can contribute to SGs formation. In addition to these core components, SGs contain an eclectic number of proteins, for example deacetylase [4], RNA helicases [5], hnRNP [6], and vary depending on the cell type [7] or duration of the stress signal [8].
Tudor‐SN protein was first identified as a coactivator of EBNA2 (Epstein‐Barr virus nuclear protein 2) [9], and subsequently discovered as coregulator of pim‐1 [10], STAT6 transcription factor in IL‐4 mediated gene regulation [11, 12], and STAT5 in prolactin (PRL) signaling [13]. It was also copurified with U5 snRNP complex and promote spliceosome assembly in vitro [14]. These studies suggest that Tudor‐SN protein participates in several biological responses and may play distinct roles in various cellular events. Interestingly, Tudor‐SN is an integral part of RISC (RNA‐induced silencing complex) [15], and could recognize hyper‐edited double‐stranded RNAs (I‐dsRNAs) [16], while I‐dsRNA molecules specifically binds a complex which comprises many SG components, including G3BP, TIA‐1 [17]. Very recently, Tudor‐SN was identified as an essential protein for RNA stability and stress tolerance in plants [18]. In our previous study, we identified G3BP as an interaction protein of Tudor‐SN in the GST‐pull down assay and MOLDI‐TOF analysis, which encouraged us to investigate whether Tudor‐SN is directly involved in SGs.
2. Materials and methods
2.1. Cells and plasmids
COS‐7 cells and HeLa cells were cultured as described previously [12]. COS‐7 cells were transfected by electroporation at 220 V/950 mF with a Bio‐Rad gene pulser. The transfection of HeLa cells were performed using FuGENE transfection reagent (Roche, Indianapolis) according to the manufacturers’ procedures.
Plasmids encoding GFP epitope‐tagged G3BP (GFP‐G3BP) was kindly provided by Dr. Jamal Tazi. The pSG5 expression plasmids containing full‐length Tudor‐SN tagged with Flag epitope (pSG5‐Tudor‐SN), the pGEXT‐4T‐1 plasmids containing SN domain (GST‐SN, 1‐639aa), TSN domain (GST‐TSN, 640–885aa) or Tudor domain (678–769aa, GST‐TD) of Tudor‐SN protein were generated as previously described [11, 12]. The full‐length Tudor‐SN (pRFP‐Tudor‐SN), SN (pRFP‐SN, 1–639aa) or TSN (pRFP‐TSN, 640–885aa) fragment was cloned and inserted into the EcoRI/XhoI sites of the vector pCherry‐C1which was kindly provided by Dr. Johan Peranen. All PCR products were sequenced.
2.2. GST‐pull down assay
GST (glutathione S‐transferase) pull down experiments were performed as previously described [12]. The beads‐bound GST fusion proteins were incubated with the total cell lysate of transfected COS7 cells or in vitro translated 35S‐labeled G3BP protein. After washing, the bound proteins were separated by SDS–PAGE and analyzed by immunoblotting with mouse monoclonal anti‐GFP antibody (Sigma, St. Louis, MO, USA) or autoradiography.
The cell‐free in vitro translation of full‐length G3BP was carried out in a nuclease‐treated rabbit reticulocyte lysate (RRL) system (Promega BioSciences, CA, USA) according to the manufacturer's recommendations. The proteins were labeled with L‐[35S]‐methionine (Amersham Biosciences, USA).
2.3. Co‐immunoprecipitation
The total cell lysates of HeLa cells without stress stimuli were collected with Nonidet P‐40 lysis buffer (50 mM Tris–HCl, pH 7.6, 300 mM NaCl, 0.1 mM EDTA, 0.5% Nonidet P‐40, 20% glycerol, 0.1 mM sodium orthovanadate, 1 mM sodium butyrate), and then incubated with mouse monoclonal anti‐Tudor‐SN or anti‐G3BP (Abcam, Cambridge, UK), as well as rabbit polyclonal IgG (Santa Cruz biotechnology) as control, followed by incubation with protein‐G/A‐Sepharose (Amersham Pharmacia Biotech). The bound proteins were analyzed by SDS–PAGE and blotted with anti‐Tudor‐SN or anti‐G3BP antibody. The mouse monoclonal anti‐Tudor‐SN antibody was generated against SN4 domain (amino acids 507–674) of Tudor‐SN in Dr. Silvennoinen's lab. The rabbit polyclonal anti‐Tudor‐SN antibody was generated against TSN domain (amino acids 640–885) of Tudor‐SN in our lab.
2.4. Immunofluorescence and confocal microscopy
Cells were grown on glass cover slips. Cellular stress was induced either by treatment with 0.5 mM sodium arsenite (Sigma–Aldrich, St. Louis, MO, USA), or by incubation at 45 °C for different time point (Heat shock). Control cells and treated cells were fixed and permeabilized, and then incubated with mouse monoclonal anti‐G3BP and rabbit polyclonal anti‐Tudor‐SN antibodies, or mouse monoclonal anti‐Tudor‐SN and rabbit polyclonal anti‐TIA‐1 (Santa Cruz biotechnology). After washing, cells were incubated with anti‐mouse Alexa fluor 488 (Invitrogen) and anti‐rabbit Texas‐red (Molecular probes, Eugene, Oregan USA) conjugated secondary antibodies. Confocal images were collected using LSM5 program and Zeiss confocal microscope, equipped with an Argon laser (488 nm) and HeNe laser (543 nm) and a ×63 objective. Green emission was detected using a 505‐nm low pass filter and red emission using a 630‐nm low pass filter [11]. Approximately 200 cells were scored per experiment independently by two different individuals.
For living cell imaging, HeLa cells were transfected with GFP‐tagged G3BP, and RFP‐Tudor‐SN, RFP‐SN or RFP‐TSN, respectively by using FuGENE transfection reagent according to the manufacturers’ procedures. As a control, HeLa cells were transfected with empty vector pEGFP‐C1 and p Cherry‐C1. After 24 h, the cells were seeded onto glass‐bottom dishes (Mat‐Tek, Ashland, MA) and cultured overnight. Before observation, the cells with 2 ml culture medium were maintained in a chamber system at 37 °C and 5% CO2. The images of timed series were acquired as described above.
HeLa cells were transfected with Tudor‐SN siRNA or scramble siRNA according to previously described [11]. After 72 h, the cells were seeded onto glass‐bottom dishes and cultured overnight. After heat shock at 45 °C for 60 min or treated with 0.5 mM sodium arsenite for 45 min, confocal images were obtained as described above.
2.5. Cell proliferation assay
Cell proliferation was measured with MTS assay. Briefly, cells were plated in 96‐well plates at a density of 2 × 103 per well and incubated for 24 h or 48 h, and then the cells were incubated with 20 μl of MTS solution (Promega) for 4 h at 37 °C. The absorbance was measured at 490 nm using ELISA microplate reader Multiskan (Thermo Labsystems).
3. Results
3.1. Tudor‐SN interacts with G3BP in vivo and in vitro
Tudor‐SN is a multi functional protein composed of four repeats of SN and a Tudor domain followed by a SN5 domain (Fig. 1 A). We initially performed GST‐pull down assay to verify the interaction of Tudor‐SN and G3BP. GST and different GST fusion proteins were bound to glutathione‐coupled beads (Fig. 1B) and incubated with total cell lysates of COS7 cells transfected with GFP‐G3BP. As shown in Fig. 1C, neither Tudor nor TSN domain associated with G3BP, but the SN domain readily precipitated the GFP‐G3BP protein. The beads‐bound different GST fusion proteins were also incubated with in vitro translated 35S‐labelled full‐length G3BP protein. As shown in Fig. 1D, in vitro translated G3BP was found to interact with the GST‐SN fusion protein, but not the others. These results indicate that the SN domain of Tudor‐SN interacts with G3BP.
Figure 1.

SN domain of Tudor‐SN protein interacts with G3BP protein in vitro. (A) The schematic structure of Tudor‐SN protein. (B) The loading control of GST, GST‐SN, GST‐TSN and GST‐TD fusion proteins for (C) and (D), are visualized by Coomassie blue. (C) COS‐7 cells were transfected with GFP‐G3BP. After 36 h, the total cell lysate (TCL) were collected and incubated with beads‐bound GST, or different GST fusion proteins. The interacted proteins were subjected to SDS–PAGE and analyzed by blotting with anti‐GFP antibody. Twenty percent of TCL was included as input. (D) G3BP was 35S‐labeled by in vitro translation and incubated with beads loaded with different GST fusion proteins or GST. The bound proteins were subjected to SDS–PAGE and visualized by autoradiography. Twenty percent of the in vitro‐translated protein was included as input.
To substantiate the in vivo interaction of Tudor‐SN and G3BP, the co‐immunoprecipitation experiment was performed with endogenous proteins of HeLa cells. As shown in Fig. 2 A, G3BP only precipitated with anti‐Tudor‐SN antibody, but not the control rabbit IgG antibody. Reciprocally (Fig. 2B), Tudor‐SN was found to co‐precipitate with G3BP, but not the control antibody. These data demonstrate that endogenous Tudor‐SN and G3BP proteins physically form the complex in vivo.
Figure 2.

Physical interactions between endogenous Tudor‐SN and G3BP in vivo. (A) The total cell lysate of HeLa cells was immunoprecipitated with mouse monoclonal anti‐Tudor‐SN antibody, or polyclonal rabbit IgG antibody as negative control. The precipitated proteins were separated by SDS–PAGE and blotted with anti‐Tudor‐SN antibody (upper panel). The same filter was stripped and re‐blotted with anti‐G3BP antibody (lower panel). (B) The total cell lysate of HeLa cells was immunoprecipitated with anti‐G3BP antibody, or polyclonal rabbit IgG antibody as negative control. The precipitated proteins were separated by SDS–PAGE and blotted with anti‐G3BP antibody (upper panel). The same filter was stripped and re‐blotted with anti‐Tudor‐SN antibody (lower panel). Twenty percent of TCL was included as input in (A) and (B).
3.2. Tudor‐SN and G3BP colocalize into SGs in response to stress stimuli
G3BP plays an essential role in SGs formation. To investigate whether Tudor‐SN is also involved in SGs, we examined the localization of endogenous Tudor‐SN and G3BP in response to heat shock for different time points. As shown in Fig. 3 A, in normal HeLa cells, G3BP was distributed in the cytoplasm (green, a), Tudor‐SN was mainly in cytoplasm and little in nucleus (res, b). The co‐localized area was merged in yellow (c). Upon heat shock for 10 min or 30 min, both G3BP (d, g) and Tudor‐SN (e, h) were visualized in the same cytoplasmic foci, and the merged picture showed the co‐localization of the two proteins (f, i). At 60 min, the SGs were characterized as large granule aggregates containing both Tudor‐SN (k) and G3BP (j), which formed around the nucleus (l). To confirm that the cytoplasmic foci are stress granules, we also detected the distribution of Tudor‐SN with another marker protein of stress granule, TIA‐1. As shown in Fig. 3B, after heat shock, both Tudor‐SN (green, d) and TIA‐1 (red, e) were found in the same cytoplasmic foci (yellow, f). These data verified that the Tudor‐SN protein is bona fide novel member of stress granules.
Figure 3.

Tudor‐SN distributes to the stress granules under stress condition. (A) The endogenous Tudor‐SN co‐localizes with G3BP in stress granules after heat shock treatment. HeLa cells were left untreated (a–c), or heat shocked by incubation at 45 °C for 10 min (d–f), 30 min (g–i) or 60 min (j–l). Cells were fixed and stained with rabbit polyclonal anti‐Tudor‐SN and mouse monoclonal anti‐G3BP antibodies, followed by Alexa 488 and Texas Red‐conjugated secondary antibodies. (B) The endogenous Tudor‐SN co‐localizes with TIA‐1 in stress granules after heat shock treatment at 45 °C for 45 min. Cells were fixed and stained with rabbit polyclonal anti‐TIA‐1 and mouse monoclonal anti‐Tudor‐SN antibodies, followed by Alexa 488 and Texas Red‐conjugated secondary antibodies. Confocal images were collected using LSM510 program and Zeiss confocal microscope with a ×63 objective. Scale bar, 10 μm.
Furthermore, we overexpressed GFP‐G3BP and RFP‐Tudor‐SN in HeLa cells, and then performed kinetic experiments to monitor the assembly of SGs in living cells treated with 0.5 mM sodium arsenite. The results in Fig. 4 showed the co‐ordinated recruitment of G3BP (green) and Tudor‐SN (red) to SGs during the assembly process. Without stimulation, GFP‐G3BP (green, a) and RFP‐Tudor‐SN (red, b) were distributed and co‐localized (merged yellow, c) mainly in the cytoplasm. As reported earlier [3], we also observed that overexpression of G3BP efficiently trigger the assembly of SGs even in the absence of stress stimuli (Fig. 4a, white arrows in the inset), and ectopically expressed Tudor‐SN could also recruit into the same foci (Fig. 4b, white arrows in the inset). With arsenite treatment for 5 min, G3BP (green, d) and Tudor‐SN (red, e) were gradually re‐distributed into some small stress granules which merged into yellow foci (f). At 10 min, lots of stress granules were formed which contained both Tudor‐SN (red, h) and G3BP (green, g). After 20 min, the small SGs fused into larger ones which showed double positive staining of G3BP (green, g, j) and Tudor‐SN (red, h, k) as indicated in the enlarged areas. In summary, in response to stress stimuli, the stress granules containing both Tudor‐SN and G3BP are aggregated first in small size dispersed in the cytoplasm, and then in large granules around the nucleus. These observations demonstrate that Tudor‐SN and G3BP are recruited simultaneously to the SGs in response to stress conditions.
Figure 4.
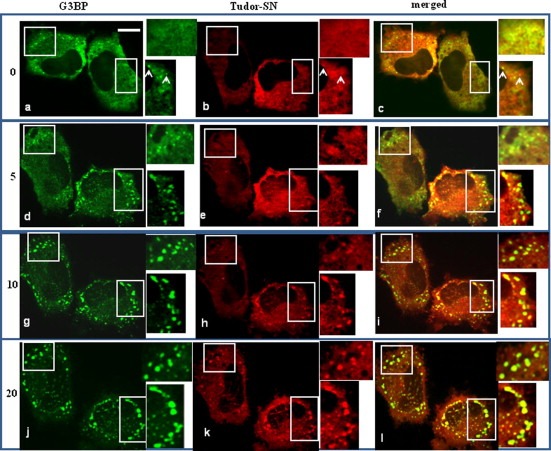
Kinetic experiments to monitor the assembly of SGs in living cells. HeLa cells were transfected with GFP‐G3BP and RFP‐Tudor‐SN, and cultured for 24 h. Sodium arsenite (0.5 mM) was then added to initiate the stress response. Cellular fluorescence was viewed and photographed for the same living cells at the indicated time points (0 min, 5 min, 10 min and 20 min) from the start of the treatment. The represented co‐localization of GFP‐G3BP and RFP‐Tudor‐SN are indicated by the write squares in each panel, and the enlarged insets were shown on the right side. Scale bar, 10 μm. The white arrows in a–c, indicated the overexpression of GFP‐G3BP induced the assembly of SGs in the absence of stress stimuli, and ectopically expressed RFP‐Tudor‐SN recruited into the same foci. The white arrows in the insets indicate the formation of SGs in the absent of stress stimuli.
3.3. SN is the functional domain in the re‐localization into SGs
Next we investigated whether the SN domain of Tudor‐SN participates in the SGs assembly. HeLa cells were transfected with GFP‐G3BP and RFP‐Tudor‐SN, RFP‐SN, or RFP‐TSN, respectively. After 24 h, the cells were seeded on glass cover slips, and then treated with either sodium arsenite or heat shock. The images were collected with confocal microscope. In normal cells, RFP‐Tudor‐SN (Fig. 5 A, b) and RFP‐SN were primarily distributed in the cytoplasm (Fig. 5B, b), RFP‐TSN was found to mainly localize in the nucleus (Fig. 5C, b). Under two different stress stimuli, both RFP‐Tudor‐SN (Fig. 5A, e and h) and RFP‐SN (Fig. 5B, e and h) were efficiently recruited into SGs with GFP‐G3BP (merged yellow, f, i). However, as shown in Fig. 5C, RFP‐TSN domain did not redistribute to the cytoplasmic foci (e, h) with GFP‐G3BP (d, g). Fig. 5C, (f and i) demonstrated the separate localization of RFP‐TSN and GFP‐G3BP. This data is consistent with the previous results showing the interaction between G3BP and SN domain, and the lack of interaction with TSN domain. These results demonstrate that the SN domain of Tudor‐SN protein is involved in the formation of SGs. To exclude the possibility that the co‐localization was caused by GFP and RFP, HeLa cells transfected with empty vector of pEGFP‐C1 and p Cherry‐C1 were also treated with heat shock or arsenite. As shown in Fig. 5D, the localization of RFP or GFP alone was not affected by the stress treatment (c, f, i), and the insets showed that no cross‐detection occurred between the green and red channels.
Figure 5.

SN domain of Tudor‐SN protein co‐localizes with G3BP in stress granules. HeLa cells were transfected with GFP‐G3BP with RFP‐Tudor‐SN (A), RFP‐SN (B), or RFP‐TSN (C), respectively. HeLa cells were also transfected with GFP and RFP vector as control (D). 24 h after transfection, the cells were either untreated, or treated with 0.5 mM sodium arsenite for 45 min, or heat shocked at 45° for 45 min. The localization of the GFP‐ or RFP fusion protein was analyzed by confocal microscopy. The enlarged insets were shown on the lower part. Scale bar, 10 μm. The white arrows in the insets indicate the formation of SGs in the absent of stress stimuli.
3.4. Knockdown of Tudor‐SN retards the aggregation of SGs
To investigate the significance of Tudor‐SN in the formation of stress granules, we performed knockdown experiments with siRNAs which directed against Tudor‐SN or scrambled siRNA as control. As shown in Fig. 6 A, transfection of Tudor‐SN siRNAs significantly reduced the expression of endogenous Tudor‐SN protein by about 80% (upper panel) comparing with the scrambled siRNA control, but has no effect on the abundance of G3BP (middle panel) (Fig. 6D) or GAPDH (lower panel). And the knockdown of Tudor‐SN protein inhibited the cell proliferation (Fig. 6B), but did not affect the cell viability. The transfected HeLa cells were seeded on glass cover slips, and incubated at 45 °C for 60 min or treated with 0.5 mM sodium arsenite for 45 min. Immunofluorescence experiments were performed using monoclonal anti‐G3BP antibody and polyclonal anti‐Tudor‐SN antibody. As shown in Fig. 6C, in control cells, both G3BP (a) and Tudor‐SN (b) were visualized in the cytoplasm of all the cells. Upon heat shock or arsenite treatment, G3BP and Tudor‐SN were efficiently recruited into the large SGs around the nucleus in all the HeLa cells (f and i). With knockdown of endogenous Tudor‐SN protein, the green staining of G3BP protein was clearly observed in all the cells, but the red staining of Tudor‐SN protein was not observed in about 70% percent of the cells. Photographs of three representative cells were selected for illustration in Fig. 6D. In the HeLa cells with knockdown of endogenous Tudor‐SN protein (e, h), the heat shock or arsenite treatment caused the formation of small SGs (G3BP in green, d, g) dispersed throughout the cytoplasm, which cannot aggregate into large foci as in the control cells (Fig. 6C, d and g). These results indicate that although Tudor‐SN may not be an essential factor to trigger the formation of SGs, it is likely to play important roles in the aggregation of the SGs.
Figure 6.

Tudor‐SN affects the aggregation of SGs. (A) Endogenous Tudor‐SN was sufficiently down‐regulated with RNA interference. The cell lysate of HeLa cells transfected with Tudor‐SN siRNA, or scrambled siRNA were loaded onto SDS–PAGE and then blotted with anti‐Tudor‐SN (upper panel), anti‐G3BP (middle panel) or anti‐GAPDH as control (lower panel). (B) Transfection of Tudor‐SN siRNA inhibits cell proliferation. HeLa cells and their transfectants were cultured in 96‐well plates at 2 × 103 per well for 24 and 48 h. The cell growth was assessed by MTS assay. Values are presented as the mean ± S.D. of three experiments with triple samples. (a) Compared with parental control. (b) Compared with scramble siRNA control. ∗, P < 0.05; ∗∗, P < 0.01; #, P > 0.05. After heat shock at 45 °C for 60 min or treated with sodium arsenite for 45 min, the scramble control cells (C) and Tudor‐SN knockdown HeLa cells (D) were fixed and stained with rabbit polyclonal anti‐Tudor‐SN and mouse monoclonal anti‐G3BP antibodies, followed by Alexa 488 and Texas Red‐conjugated secondary antibodies. Confocal images were collected using LSM510 program and Zeiss confocal microscope with ×63 objective. Scale bar, 10 μm.
4. Discussion
Tudor‐SN, also known as p100 or SND1, is a ubiquitously expressed protein and highly conserved in eukaryotes except Saccharomyces cerevisiae. Crystal structure indicates that the Tudor‐SN is composed of a tandem repeat of the SN domain which could capture double‐stranded nucleic acids, and Tudor region with an aromatic cage potentially capable of association with proteins with dimethylarginine‐modification [19, 20].
Consistent with the structure architecture and functional consequence, Tudor‐SN could recognize hyper‐edited double‐stranded RNAs (I‐dsRNAs) which are generated during stress, as a result, induces SG assembly [17]. Our present study provides direct evidence that Tudor‐SN is a bona fide novel component of SGs, which efficiently co‐localizes with G3BP in the SGs in response to various stress conditions. To monitor the coordination of Tudor‐SN and G3BP, the process of SG assembly was examined in living cells. It is revealed that SG formation begins with appearance of many small foci, which subsequently fuse into larger structures, and the two proteins assemble to SGs with similar kinetics, suggesting that they are recruited in a coordinate manner as a complex. Interestingly, knockdown of endogenous Tudor‐SN did not inhibit the formation of SGs, but retarded the aggregation of small SGs into large SGs, while a phosphomimetic mutant (S149E) of G3BP protein significantly inhibited the formation of SGs [3]. It supports the idea that although Tudor‐SN participates in the formation of SGs, it may not be an initiator as essential as G3BP to trigger the formation of SGs, but affects the aggregation efficiency of SGs.
In the present study, we clarified that the SN domain, but not the Tudor containing TSN domain of Tudor‐SN, is responsible for the recruitment to SGs. This is based on the observations that SN domain directly interacts with the 35S‐labled in vitro translated G3BP in the in vitro binding assay, and recruited to the SGs with G3BP. It is the first report that the SN domain is related to the SG formation. Recent evidence indicates that the SN domain of Tudor‐SN mediates the interaction with AT1R 3′‐UTR, and leads to both stabilization and enhanced translation of AT1R 3′‐UTR [21]. Thus Tudor‐SN may have potential functions in the regulation of mRNA stability under stress condition via the RNA binding ability of SN domain.
Accumulating evidences indicate that the formation of SGs may relate to diseases. For example, the SGs formed within the tumors in the hypoxic area which are thought to contribute to the radio‐resistance of the tumor vasculature [22]. Notably, Tudor‐SN is up‐regulated in colon cancer [23], breast cancer [24] and prostate cancer [25]. In addition, some viral infections transiently trigger stress granule formation [26], and may be part of the host defense response to limit virus infection. It was reported recently that Tudor‐SN interacts with 3′ end of transmissible gastroenteritis coronavirus (TGEV) genome [27] and the equine arteritis virus nsp1 [28]. Interestingly, long dsRNAs in cells often indicate the viral infection [29]. Moreover, dsRNAs can be modified to I‐dsRNAs which is proposed to induce SG formation, consequently facilitate cell survival during stress and trigger the antiviral activity in the host cells [17]. Alternatively, I‐dsRNAs could be recognized and cleaved by Tudor‐SN to switch off the I‐dsRNA‐induced silencing pathway [17]. Thus, Tudor‐SN may play important role in defense against viral infection via the formation of SGs. It is likely to generate new insights into future studies on the roles of Tudor‐SN in viral infections.
Stress could facilitate the cells to form SGs to protect RNAs from damaging condition. On the other hand, stress could also induce apoptosis. The consequence is dictated by the intensity of stress, as well as cell intrinsic pathways. However, the underline mechanisms remain unclear. Intriguingly, Caspase 3 could cleave Tudor‐SN between Tudor and SN5 domains during stress‐induced apoptosis, and this cleavage inhibits its ribonuclease activity [30]. Considering that SN domain of Tudor‐SN is responsible for the RNA interaction and SGs formation, we hypothesis that Tudor‐SN is likely to take part in both stress and apoptosis regulation and there is potential crosstalk between these two phenomena. Thus further work should be carried out to explore the Tudor‐SN function at the molecular and cellular levels which will shed light on the regulation of apoptosis and SGs pathways.
Acknowledgments
We thank Dr. Jamal Tazi for GFP‐G3BP plasmids, Dr. Johan Peranen for vector pCherry‐C1. This work was supported by grants from (2007AA02Z115), NSFC (90919032, 30970562, 30670441), 973 program (2009CB918903), Specialized Fund for the Doctoral Program of Higher Education (20091202110001), TSTC (08ZCGHHZ01900, 08JCYBJC07700), Tianjin Educational Committee Foundation (2008ZD01), Medical Research Council of Academy of Finland, Finnish Foundation for Cancer Research.
Gao Xingjie,Ge Lin,Shao Jie,Su Chao,Zhao Hong,Saarikettu Juha,Yao Xuyang,Yao Zhi,Silvennoinen Olli and Yang Jie(2010), Tudor-SN interacts with and co-localizes with G3BP in stress granules under stress conditions, FEBS Letters, 584, doi: 10.1016/j.febslet.2010.07.022
Contributor Information
Zhi Yao, Email: yaozhi@tmu.cn.
Jie Yang, Email: yangj@tijmu.edu.cn.
References
- 1. Kedersha N., Anderson P., Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans., 30, (2002), 963– 969. [DOI] [PubMed] [Google Scholar]
- 2. Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., Anderson P., Stress granules assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell, 15, (2004), 5383– 5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J.M., Bertrand E., Tazi J., The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol., 160, (2003), 823– 831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4. Kwon S., Zhang Y., Matthias P., The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev., 21, (2007), 3381– 3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chalupníková K., Lattmann S., Selak N., Iwamoto F., Fujiki Y., Nagamine Y., Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J. Biol. Chem., 283, (2008), 35186– 35198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guil S., Long J.C., Cáceres J.F., HnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol. Cell. Biol., 26, (2006), 5744– 5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Scheu S., Stetson D.B., Reinhardt R.L., Leber J.H., Mohrs M., Locksley R.M., Activation of the integrated stress response during T helper cell differentiation. Nat. Immunol., 7, (2006), 644– 651. [DOI] [PubMed] [Google Scholar]
- 8. Fujimura K., Katahira J., Kano F., Yoneda Y., Murata M., Microscopic dissection of the process of stress granule assembly. Biochim. Biophys. Acta, 1793, (2009), 1728– 1737. [DOI] [PubMed] [Google Scholar]
- 9. Tong X., Drapkin R., Yalamanchili R., Mosialos G., Kieff E., The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol. Cell. Biol., 15, (1995), 4735– 4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leverson J.D., Koskinen P.J., Orrico F.C., Rainio E.M., Jalkanen K.J., Dash A.B., Eisenman R.N., Ness S.A., Pim-1 kinase and p100 cooperate to enhance c-Myb activity. Mol. Cell, 2, (1998), 417– 425. [DOI] [PubMed] [Google Scholar]
- 11. Välineva T., Yang J., Palovuori R., Silvennoinen O., The transcriptional co-activator protein p100 recruits histone acetyltransferase activity to STAT6 and mediates interaction between the CREB-binding protein and STAT6. J. Biol. Chem., 280, (2005), 14989– 14996. [DOI] [PubMed] [Google Scholar]
- 12. Yang J., Aittomäki S., Pesu M., Carter K., Saarinen J., Kalkkinen N., Kieff E., Silvennoinen O., Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J., 21, (2002), 4950– 4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paukku K., Yang J., Silvennoinen O., Tudor and nuclease-like domains containing protein p100 function as coactivators for signal transducer and activator of transcription 5. Mol. Endocrinol., 17, (2003), 1805– 1814. [DOI] [PubMed] [Google Scholar]
- 14. Yang J., Välineva T., Hong J., Bu T., Yao Z., Jensen O.N., Frilander M.J., Silvennoinen O., Transcriptional co-activator protein p100 interacts with snRNP proteins and facilitates the assembly of the spliceosome. Nucleic Acids Res., 35, (2007), 4485– 4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Caudy A.A., Ketting R.F., Hammond S.M., Denli A.M., Bathoorn A.M., Tops B.B., Silva J.M., Myers M.M., Hannon G.J., Plasterk R.H., A micrococcal nuclease homologue in RNAi effector complexes. Nature, 425, (2003), 411– 414. [DOI] [PubMed] [Google Scholar]
- 16. Scadden A.D., The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol., 12, (2005), 489– 496. [DOI] [PubMed] [Google Scholar]
- 17. Scadden A.D.J., Inosine-containing dsRNA binds a stress-granule-like complex and downregulates gene expression in Trans. Mol. Cell, 28, (2007), 491– 500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dit Frey, N.F., Muller, P., Jammes, F., Kizis, D., Leung, J., Perrot-Rechenmann, C. and Bianchi, M.W. (2010) The RNA binding protein Tudor-SN is essential for stress tolerance and stabilizes levels of stress-responsive mRNAs encoding secreted proteins in Arabidopsis. Plant Cell 18, epub ahead of print. [DOI] [PMC free article] [PubMed]
- 19. Li C.L., Yang W.Z., Chen Y.P., Yuan H.S., Structural and functional insights into human Tudor-SN, a key component linking RNA interference and editing. Nucleic Acids Res., 36, (2008), 3579– 3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shaw N., Zhao M., Cheng C., Xu H., Saarikettu J., Li Y., Da Y., Yao Z., Silvennoinen O., Yang J., Liu Z.J., Wang B.C., Rao Z., The multifunctional human p100 protein ‘hooks’ methylated ligands. Nat. Struct. Mol. Biol., 14, (2007), 779– 784. [DOI] [PubMed] [Google Scholar]
- 21. Paukku K., Kalkkinen N., Silvennoinen O., Kontula K.K., Lehtonen J.Y., P100 increases AT1R expression through interaction with AT1R 3′-UTR. Nucleic Acids Res., 36, (2008), 4474– 4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moeller B.J., Cao Y., Li C.Y., Dewhirst M.W., Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell, 5, (2004), 429– 441. [DOI] [PubMed] [Google Scholar]
- 23. Tsuchiya N., Ochiai M., Nakashima K., Ubaqai T., Suqimura T., Nakagama H., SND1, a component of RNA-induced silencing complex, is up-regulated in human colon cancers and implicated in early stage colon carcinogenesis. Cancer Res., 67, (2007), 9568– 9576. [DOI] [PubMed] [Google Scholar]
- 24. Ho J., Kong J.W., Choong L.Y., Loh M.C., Toy W., Chong P.K., Wong C.H., Wong C.Y., Shah N., Lim Y.P., Novel breast cancer metastasis-associated proteins. J. Proteome Res., 8, (2009), 583– 594. [DOI] [PubMed] [Google Scholar]
- 25. Kuruma H., Kamata Y., Takahashi H., Igarashi K., Kimura T., Miki K., Miki J., Sasaki H., Hayashi N., Egawa S., Staphylococcal nuclease domain-containing protein 1 as a potential tissue marker for prostate cancer. Am. J. Pathol., 174, (2009), 2044– 2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Beckham C.J., Parker R., P bodies, stress granules, and viral life cycles (review). Cell Host Microbe, 3, (2008), 206– 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Galán C., Sola I., Nogales A., Thomas B., Akoulitchev A., Enjuanes L., Almazán F., Host cell proteins interacting with the 3′ end of TGEV coronavirus genome influence virus replication. Virology, 391, (2009), 304– 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tijms M.A., Snijder E.J., Equine arteritis virus non-structural protein 1, an essential factor for viral subgenomic mRNA synthesis, interacts with the cellular transcription co-factor p100. J. Gen. Virol., 84, (2003), 2317– 2322. [DOI] [PubMed] [Google Scholar]
- 29. Maquat L.E., Carmichael G.G., Quality control of mRNA function. Cell, 104, (2001), 173– 176. [DOI] [PubMed] [Google Scholar]
- 30. Sundström J.F., Vaculova A., Smertenko A.P., Savenkov E.I., Golovko A., Minina E., Tiwari B.S., Rodriguez-Nieto S., Zamyatnin A.A. Jr, Välineva T., Saarikettu J., Frilander M.J., Suarez M.F., Zavialov A., Ståhl U., Hussey P.J., Silvennoinen O., Sundberg E., Zhivotovsky B., Bozhkov P.V., Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat. Cell Biol., 11, (2009), 1347– 1354. [DOI] [PubMed] [Google Scholar]


