Abstract
Biological membranes with cubic morphology are a hallmark of stressed or diseased cellular conditions; both protein–protein interactions and lipid alterations appear to contribute to their biogenesis, yet their specific cellular functions are unknown. The occurrence of cubic membranes strikingly correlates with viral infections; notably, virus entry, proliferation, and release are processes closely linked to cellular cholesterol metabolism, and dys-regulation of cholesterol synthesis at the level of HMG-CoA reductase also induces cubic membrane formation, in the absence of viral infection. We propose that virus-induced cubic membranes could result from viral interference of cellular cholesterol homeostasis, generating a protective membrane environment to facilitate virus assembly and proliferation. Preventing cubic membrane formation might thus disrupt the ‘virus factory’ and offer new avenues to combat viral infections.
Virus infections induce major rearrangements of the host endomembrane system
Distinct steps of the viral replication cycle occur in close association with the endomembrane system of the host cell. The adaptation and modification of these membranes in the course of infection are either part of the cellular reprogramming process to facilitate viral replication or a specific infection-induced host defense response, or perhaps both 1, 2. Notably, many of the virus-induced membrane proliferations are of highly ordered three-dimensional nano-periodic structure, termed cubic membranes (Box 1 ). A thorough review of the literature demonstrates that basically all types of viruses (plus-, minus-single, or double-stranded RNA or DNA viruses) could potentially be associated with striking membrane modifications in the infected host cell (Table 1 ), which have even been considered to serve as ultrastructural markers of infection [3]. In particular, infections with plus-single-stranded RNA viruses [(+)ssRNA], which are important human pathogens, appear to be always associated with the induction of host membrane proliferations, many of which indeed represent cubic membranes or precursors thereof. These viruses are able to cross host barriers and move from animals to humans, resulting in serious zoonotic-human epidemics. (+)ssRNA families include Coronaviridae (SARS-CoV), Picornaviridae (poliovirus, hepatitis A virus), Togaviridae (rubella virus, Semliki Forest virus) and Flaviviridae (Dengue, West Nile virus) and are, therefore, of tremendous biomedical interest. Although their specific roles in the life cycle of a virus are unclear, the striking similarities in cubic membrane formation in the course of viral infection and that occurring under conditions of de-regulated sterol biosynthesis suggest a direct mechanistic link between virus-induced cubic membranes and cellular sterol metabolism and trafficking. Not surprisingly, virus entry, replication, and proliferation are strongly affected by drugs interfering with cellular cholesterol homeostasis 4, 5, 6, 7. Here, we advance the hypothesis that viral-induced cubic membranes are induced by the virus’ interference with the regulatory machinery controlling sterol biosynthesis. First we discuss the similarities in membrane proliferations triggered by virus infection and deregulated sterol biosynthesis, and outline cellular mechanisms that might lead to morphological membrane rearrangements; we point out potential regulatory links to cellular defense systems, such as the unfolded protein response and autophagy, and finally speculate how these data suggest novel ways of interfering with viral replication.
Box 1. Characteristics of cubic membranes.
In contrast to the typically rather flat and non-periodic bilayer structure of most biological membranes, cubic membranes represent highly curved, three-dimensional (3D) folded lipid bilayer structures; their nano-periodic appearance corresponds to mathematically well-defined triply periodic minimal surfaces 16, 17, 52, 53. Such membrane arrangements can be unequivocally identified and resolved by electron tomography (ET) 54, 55, 56, or by comparing the two-dimensional (2D) transmission electron microscopy (TEM) images to computer-simulated projections that are based on 3D membrane models 40, 52. Application of this ‘direct template matching’ (DTM) method indeed led to the characterization of the OSER membrane [18], which occurs owing to weak dimerization of over-expressed ER-resident proteins, as ‘cubic membranes’ [17]. The major advantage of the DTM method is that it can be applied also in retrospect to identify the appearance of cubic membranes in published TEM representations of ‘tubuloreticular structures’ (Figure 1).
Multiple electron microscopy observations document the occurrence of ‘highly convoluted membrane structures’ in host cells in the course of viral infection. Owing to their morphological and 3D complexity, 2D TEM images typically do not adequately represent the true nature of these membrane structures. Thus, such virus-induced membrane rearrangements have been described in the literature by approximately 15 different nicknames, including ‘tubulocrystalline inclusion’ in HCV-infected liver [57], ‘convoluted membranous mass’ in viral St. Louis encephalitis [58] and TRS in HIV [3] and in SARS-corona virus (SARS-CoV)-infected Vero cells [41]; in recent years, the TRS has become the most common denomination for such structurally organized membrane proliferations, which have even been suggested to serve as an ultrastructural marker [3] for HIV infection.
Many of the documented TEM micrographs of convoluted membrane structures that develop during viral infection or in stressed cells are indeed depictions of cubic membrane morphologies [40], and studies using electron tomography have confirmed the cubic architecture of virus-induced membrane modifications in Dengue-virus-infected cells [56]. Certain transition states between cubic morphologies and interconnected tubular structures, however, might be induced by the fixation procedure, a pitfall that therefore needs careful consideration [56]. Indeed, the existence of membranes with cubic morphology in living systems has been a matter of discussion [59]. However, the multitude of observations demonstrating cubic membranes in a large variety of cell systems and experimental conditions leaves little doubt as to the existence of such structures in living cells, and is now well accepted [16]. It is evident that formation of cubic membranes, or morphologically related membrane arrangements, such as TRS, is indeed a specific response of the cell, either induced by the virus to promote efficient replication and proliferation, or a cellular defense mechanism to cope with virus invasion.
Table 1.
Virus-induced host cubic membranes
| Virus | Genome | Envelope | Cell system | Pathology | Descriptive name | References |
|---|---|---|---|---|---|---|
| Poxviridae | ||||||
| Swinepox virus | dsDNA | Yes | Swine skin cells | Swinepox | Lamellar body | [70] |
| Poxvirus | dsDNA | Yes | Subcutaneous Yaba lesions of rhesus monkey (Macaca mulatta) | Smallpox | Elongated crystalloid with honeycombed structure | [71] |
| Tana poxvirus | dsDNA | Yes | Rhesus embryonic monkey kidney cells | Tana disease | Honeycomb crystal | [72] |
| Stomatitis papulosa virus | dsDNA | Yes | Bovine and ovine embryonic lung cell cultures | Stomatitis papulosa | Intranuclear tubules | [73] |
| Hepadnaviridae | ||||||
| Hepatitis B virus | dsDNA-RT | Yes | Chimpanzee hepatocytes | Hepatitis B | Paracrystalline | [57] |
| Herpesviridae | ||||||
| Herpes Simplex virus | dsDNA | Yes | Macrophage of rabbit neuron and astrocytes | Encephalitis | Crystalline array | [74] |
| Herpes Simplex virus | dsDNA | Yes | Endothelial cells and macrophages of New Zealand white rabbit | Encephalitis | Crystalline aggregate | [75] |
| Herpes Simplex virus | dsDNA | Yes | Human cerebral tissue, endothelial cells and macrophage | Encephalitis | Tubular structure | [76] |
| Baculoviridae | ||||||
| Baculoviruses | dsDNA | Yes | Pink shrimp cell | Penaeid shrimp disease | Membrane labyrinths | [77] |
| Papovaviridae | ||||||
| Simian virus 40 | dsDNA | No | African green monkey kidney cells (CV-1) | Sarcomas | Tubular membrane; virus-containing aggregates | [78] |
| Retroviridae | ||||||
| Rous Sarcoma virus | (+)ssRNA-RT | Yes | Canine tumor cells | Leptomeningeal sarcomas | Crystalline aggregates | [79] |
| Rous Sarcoma virus | (+)ssRNA-RT | Yes | New World monkey (Saquinus sp.) | RSV-induced tumor | Membrane complex with repeating pattern | [80] |
| Simian Immunodeficiency virus (SIV) | (+)ssRNA-RT | Yes | Macrophage and endothelial cells in rectal biopsies of Rhesus monkey | AIDS | TRS | [81] |
| HIV | (+)ssRNA-RT | Yes | Human KS tissues | AIDS-associated Kaposi's sarcoma | TRS | [3] |
| HIV-1 | (+)ssRNA-RT | Yes | Human monocyte-derived macrophage | AIDS | Sponge-like meshwork of interconnected membrane | [82] |
| HTLV-III | (+)ssRNA-RT | Yes | Human lymphocytes | AIDS | TRS | [83] |
| Coronaviridae | ||||||
| SARS-CoV | (+)ssRNA | Yes | African green monkey kidney epithelial (Vero) cells | Severe acute respiratory syndrome | TRS | [41] |
| Murine Hepatitis virus | (+)ssRNA | Yes | Mouse hepatocytes | Murine hepatitis | Peculiar tubular structure | [84] |
| Mouse Hepatitis virus (MHV-A59) | (+)ssRNA | Yes | HeLa-CEACAM1a cells | Murine hepatitis | Cubic membrane structures | [42] |
| Flaviviridae | ||||||
| Dengue virus | (+)ssRNA | Yes | Rhesus monkey kidney cells (LLC-MK2) | Dengue fever | Lamellar structures and lattice crystals | [85] |
| Dengue-1 virus | (+)ssRNA | Yes | Human leukemic leukocyte line (J-111) | Dengue fever | Lamellar and crystalloid structure | [86] |
| Wesselsbron virus | (+)ssRNA | Yes | Fetal lamb kidney cells | Wesselsbron disease | Crystalline-like inclusion; honeycomb pattern and reticulate array | [87] |
| Kunjin virus | (+)ssRNA | Yes | African green monkey kidney epithelial cell (Vero) | Meningitis or encephalitis | Paracrystals; convoluted membranes | [88] |
| Kunjin virus | (+)ssRNA | Yes | Vero cells | Meningitis or encephalitis | Hexagonal units of microtubule paracrystals; convoluted membranes | [89] |
| Kunjin virus | (+)ssRNA | Yes | Vero cells | Meningitis or encephalitis | Convoluted membranes | [90] |
| St. Louis encephalitis virus | (+)ssRNA | Yes | Mouse CNS tissue | St. Louis encephalitis | Crystalline arrays of tubules and convoluted masses of membranes (knots) | [58] |
| Ilheus virus | (+)ssRNA | Yes | human epidermoid cancer (Hep-2) cells | Encephalitis | Membrane complex, labyrinthine and membrane-lined tunnels | [91] |
| Hepatitis C virus | (+)ssRNA | Yes | Chimpanzee hepatocytes | Hepatitis C | Intracytoplasmic crystalloid inclusions | [57] |
| Picornoviridae | ||||||
| Hepatitis A virus | (+)ssRNA | No | HeLa cells; fetal rhesus monkey kidney (FRhK-4) cells | Hepatitis A | Crystalloid ER | [9] |
| Poliovirus | (+)ssRNA | No | Swine kidney cells | Polio-encephalomyelitis | Viral crystals | [92] |
| Poliovirus | (+)ssRNA | No | Monkey kidney (CMK) cells; macrophage in lumbar cord of cynomologus monkey (Macaca irus) | Poliomyelitis | Paracrystalline array | [93] |
| Poliovirus | (+)ssRNA | No | Endothelial cells in monkey spinal cord | Polio-encephalomyelitis | Crystalline array | [94] |
| Foot-and-Mouth virus | (+)ssRNA | No | Swine kidney (PK15) cells | Food-and-mouth disease | Crystalline array | [95] |
| Togaviridae | ||||||
| Rubella virus | (+)ssRNA | Yes | Chinese hamster ovary (CHO) cells | German measles | Tubular smooth network | [11] |
| Rubella virus | (+)ssRNA | Yes | Rhesus monkey kidney cells (LLC-MK2) cells | German measles | Crystal lattice structure associated with annulate lamellae | [96] |
| Alphavirus (Semliki Forest virus) | (+)ssRNA | Yes | Neuron of weanling mouse brain | Infectious arthritis, encephalitis, rashes and fever | Membrane tubules | [97] |
| Paramyxoviridae | ||||||
| Canine Distemper virus | (–)ssRNA | Yes | Canine cerebrum tissue | Demyelinating encephalitis | Tubular structure | [98] |
| Reoviridae | ||||||
| Rotavirus | dsRNA | No | Human adenocarcinoma (HT-29) cell line | Rotavirus gastroenteritis | Honeycomb-like mesh of TRS | [99] |
| Rotavirus | dsRNA | No | Fetal rhesus monkey kidney (MA104) cells | Rotavirus gastroenteritis | Smooth-membrane vesicles | [100] |
Induction of cubic membranes: the viral factor(s)
A large part of a typical viral genome encodes non-structural (NS) proteins, which are not part of the mature virion but carry out important functions during viral replication. In the case of Flaviviruses, the (+)ssRNA genome encodes a single polyprotein that is subsequently processed into three structural and seven NS proteins. Following translation and protein processing, the viral replicase complex is assembled from NS proteins, viral RNA and, presumably, several host factors, in close association with perinuclear membranes [2]. Early infection induces rearrangements of cytoplasmic membranes which most probably originate from the endoplasmic reticulum and proliferate into smooth convoluted, vesicle-like, also termed paracrystalline or sponge-like, membrane structures, which are believed to host the viral replication machinery. Such ‘induced’ membranes were only found in cells that harbored efficiently replicating replicons [8], suggesting that at least one or more of the viral NS proteins are required to induce host membrane rearrangement and proliferation 8, 9, 10, 11. In particular, the NS1 protein of dengue virus associates with the membrane on the cell surface, where it co-localizes with raft lipid markers, and with the RNA replication complex [12]. Virion particles become visible first in the lumen of the endoplasmic reticulum 13, 14, from where they are released by a budding process and delivered to the cell surface, resembling the typical fate of a secretory protein, that is, transport via the Golgi apparatus and release from the cell by exocytosis, or taking the fast track, bypassing the Golgi along a route that is insensitive to the inhibitor brefeldin A [15]. A central role of the metabolism of the lipid – in particular cholesterol – components of the host cell in the life cycle of a virus is evident; however, most of the morphological evidence about virus particle formation is derived from electron microscopic studies (Box 1, Figure 1 ), and therefore the specific molecular mechanisms triggering intracellular membrane proliferation and their morphological rearrangements and their contribution to virion maturation are unclear.
Figure 1.
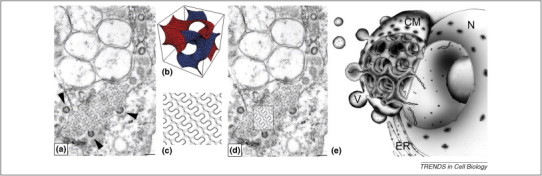
SARS-CoV-induced cubic membranes in Vero cells. (a) The original TEM micrograph is reproduced, with permission, from Figure 3C of Goldsmith et al. [41]. The arrow points at the regular TRS of interest; (b) mathematical 3D simulation to describe a gyroid (G)-based minimal surface; (c) a corresponding computer-simulated 2D projection map (0.4 of a unit cell thickness) derived from the 3D model in (b); (d) the theoretical projection of a slice through a G-based morphology exactly matches the TRS structure in the TEM image (see boxed area), unequivocally identifying it as ‘cubic membrane structure’. Note the spherical virion particles budding off this cubic membrane segment (arrowheads). Scale bar, 100 nm (reproduced, with permission, from Ref. [40]; arrows were removed from the original image); (e) illustration of cubic membrane organization in a virus-infected cell and its possible function as a virus factory. The characteristic interconnected channels of the cubic membrane (CM) provide a transport conduit between the viral replication complex and the cytoplasm and/or nucleus (N). The pores at the outer surface could act as regulators that allow the entry of the essential virus precursors yet inhibit the entry of host defense proteins. The highly curved nature of cubic membranes might support viral assembly and budding. ER, endoplasmic reticulum; V, virus particle.
Induction of cubic membranes: the host factors – proteins or lipids, or both?
Cubic membranes not only occur upon viral infection but are also frequently associated with deregulated protein synthesis, cellular stress and a variety of other pathological conditions [16]. Indeed, both lipid alterations and overexpression of membrane-resident proteins can induce periodic two-dimensional (i.e. hexagonal) and three-dimensional (cubic) membrane transitions: for instance, over-expression of cytochrome b(5) tagged with green fluorescent protein induces cubic membranes, originally termed as ‘organized smooth endoplasmic reticulum’ (OSER), in COS-7 and CV-1 cells 17, 18. In this system, formation of cubic membranes is a consequence of weak protein–protein interactions as point mutations preventing such protein–protein interactions also prevent the formation of OSER [18]. Similarly, expression of msALDH in COS-1 cells induces ‘crystalloid’ (cubic) endoplasmic reticulum (ER) membranes as a consequence of weak interactions between the membrane-resident proteins [19]. Notably, the observed dimensions of the cubic membrane lattice size in all analyzed TEM images, typically in the range of 50–1000 nm, greatly exceeds the lattice size in artificial cubic phases of pure lipids in vitro (<20 nm), indicating that both lipids and proteins and their interactions are responsible for cubic membrane formation in vivo. Thus, although certain lipid mixtures show a high propensity to form non-lamellar membrane structures in vitro, in vivo these membranes are decorated with integral or peripheral proteins that might stabilize cubic membranes through protein–lipid and protein–protein interactions [20].
Support for an important contribution of proteins to the biogenesis of cubic membranes also comes from a recent study that makes use of controlled dimerization of artificial membrane proteins in mammalian tissue culture cells 21, 22. By contrast, overexpression of HMG-CoA reductase, a key enzyme of cholesterol synthesis, in UT-1 [23] or CHO cells 24, 25 induces formation of crystalloid ER, which accommodates most of the HMG-CoA reductase proteins 26, 27. Whether increased enzyme activity and subsequent over-production of cholesterol is responsible for the observed morphological changes, or whether cubic membrane formation is as a result of induced protein–protein interactions of excess HMG-CoA reductase (or associated proteins) remains to be determined. In budding yeast, overexpression of both enzymatic active and truncated variants of HMG-CoA reductase induces formation of stacked layers of membrane proliferations termed karmellae, which, however, are morphologically unrelated to cubic membranes but could represent precursors thereof; although this observation suggests a sterol-independent process for membrane proliferation in that system it cannot be excluded that even non-functional fragments of the enzyme might interfere with the sterol regulatory network [28].
Cholesterol has profound effects on membrane lipid packing and changes membrane bilayer thickness. Thus, modulation of membrane cholesterol content in combination with phospholipid molecular species that prefer non-lamellar phases could represent the means of regulating sorting processes associated with protein secretion and membrane proliferation. For instance, a mixture of cholesterol with unsaturated phosphatidylcholines readily forms inverted bicontinuous cubic phases in vitro, under physiological conditions [29]. In a recent study, Deng and coworkers [30] unveiled significant changes in membrane lipid composition in the course of cubic membrane formation in amoeba Chaos mitochondria: the inducible transition of inner mitochondrial membranes from random tubular to cubic morphology under starvation conditions correlates with a major increase in docosapentaenoic acid (DPA)-containing phospholipids [30]. Indeed, supplementation of non-starving cells, which typically do not generate cubic mitochondrial membranes, with exogenous ω-6 DPA, but not with ω-3 DPA, induced cubic membranes, indicating a structural role for DPA-modified phospholipids in promoting this morphological transition. Inhibition of fatty acid de novo synthesis in poliovirus-infected cells inhibited membrane proliferation and, subsequently, also viral RNA synthesis, which is consistent with an important function of ongoing lipid and fatty acid synthesis in the formation of cubic membranes [31]. Such a requirement for (unsaturated) fatty acids for viral replication is remarkably conserved: replication of the brome mosaic virus (BMV) RNA in budding yeast is severely inhibited by a mutation in Ole1p, the only and essential Δ9 fatty acid desaturase in this organism [32]. A slight decrease in unsaturated fatty acid content that was without phenotypic consequences for the yeast cell but inhibited BMV RNA replication by more than 95% 32, 33, indicating that this process is highly sensitive to cellular lipid content and composition. Thus, a picture emerges that formation of cubic membranes in the course of cell stress – and potentially in conjunction with viral infection – is a defined and highly regulated cellular process that involves major rearrangements of cellular lipid and perhaps protein content. Whether cubic membrane formation during viral infection occurs as a defense and protection mechanism or as part of the viral re-programming of cellular physiology to promote viral replication, remains to be shown. Notably, cubic membranes were recently identified in cells during autophagy 21, 22, a cellular program that is induced upon starvation to eliminate cellular components and even entire organelles via lysosomal degradation. However, some viruses could subverse the mechanism to induce formation of autophagosomes [34], and it was argued that autophagosomal structures provide indeed membranous support for viral RNA replication complexes and could enable the release of progeny virions, from infected cells 35, 36.
To which extent an altered cellular lipid environment also contributes to the membrane composition of enveloped viruses is unclear: such viruses acquire all their lipids from the host cell membranes, but it remains unclear how their lipid composition is regulated. Although the lipid composition of enveloped viruses has been extensively investigated, only recent advances in high-resolution lipid mass spectrometry promise to yield a more detailed understanding of the origin of virus lipids [37]. Whether the assembly of the virus membrane is a rather passive process, thus reflecting the composition of the host membrane from which the virus buds off, or whether lipids are selectively included in or excluded from the virus envelope, remains to be determined. Accordingly, detailed lipidomic studies comparing the lipid composition of host cubic membranes and the virus envelope might provide further hints as to the specific (subcellular) lipid requirements for virus assembly, and might unveil whether the viral membrane composition specifically reflects that of the cubic membrane environment. Notably, numerous non-enveloped viruses also induce formation of cubic membranes (Table 1).
Functional roles of host cubic membranes for viral replication and assembly
Viral entry, replication, and excretion from the infected cell are strongly dependent on cellular lipid – in particular cholesterol – homeostasis (Box 2 ). Also, the importance of virus-induced intracellular membrane rearrangements for viral replication and survival is evident [1], but the mechanisms of host cubic membrane formation and the specific roles of such membranes in the life cycle of a virus are not understood. Because both enveloped and non-enveloped viruses might induce cubic membranes these could serve a broader purpose than providing membrane lipids to the virus. Cubic membranes are frequently observed in cells exposed to oxidative stress conditions [16], and there is evidence suggesting that the formation of cubic mitochondrial cristae in the amoeba Chaos indeed plays a protective role against oxidative damage through enhancing the efflux of H2O2 and reactive oxygen species (ROS) and by reducing the susceptibility of membrane lipids to oxidants [38]. Cubic membranes might also generate the environment to accommodate the newly translated viral NS proteins and provide an organizing platform for viral RNA replication and virion assembly. In addition, the interconnected convoluted channel structure of cubic membranes could provide an ideal architecture for local concentration and storage of viral precursor molecules, with adequate interaction with the host cytoplasm through numerous surface pores, as illustrated in Figure 1e. The three-dimensional continuous membrane network with surface pores that are 50–80 nm in diameter [39] might offer a preferable environment as a transport system or a passageway for selective entry of viral proteins, nucleic acids, and other precursors for de novo assembly of virion particles. Recent data obtained from studies on cubic membranes in the amoeba Chaos system in vivo (Deng et al., unpublished) and in vitro [39] indeed suggest a role in (small) nucleic acid trafficking: isolated cubic membranes efficiently take up short oligonucleotides into their lumen in vitro, which might even make them practically useful as a novel nucleic-acid-delivery system. The model for cubic membranes as a virus assembly site (‘virus factory’) [2] is strongly supported by TEM images of virus particles captured in the process of budding off from cubic membranes 40, 41 (Figure 1). The time-course of host cubic membrane formation that occurs immediately before RNA virus maturation further supports a functional role for cubic membranes as a specialized site for viral replication and assembly [42]. The extended surface-to-volume ratio of cubic membranes provides a large surface area for controlled metabolite exchange, and could also protect the virus assembly from host defense mechanisms such as proteolytic degradation. Notably, viruses could subvert the cellular ‘clean-up program’, autophagy, which ultimately directs cellular components including cubic membranes to lysosomal degradation, for their replication cycle [34].
Box 2. Role of cholesterol in virus entry, replication, and assembly.
Lipids, in particular cholesterol, are crucial for the virus life cycle, and inhibitors of cholesterol, fatty acid, and sphingolipid biosynthesis inhibit virus replication, maturation, and secretion 60, 61. Although a role for cholesterol in these processes is undisputed, controversial observations indicate multiple mechanisms of its action, dependent on the type of virus and cell system. Notably, most documented reports on the effects of cholesterol on viral infectivity imply a function in their cellular entry or exit 4, 5, 6, 62, 63, 64, 65, 66, 67, 68, rather than formation of intracellular membrane-associated assembly sites related to cubic membrane structures. These cholesterol effects are primarily discussed in the context of its propensity to concentrate in certain membrane microdomains, termed ‘rafts’ that are characterized by specific lipid and protein compositions in post-Golgi membranes, including the plasma membrane [69].
As illustrated in Figure 2, we propose that viruses reprogram and hijack cellular metabolism to stimulate availability of crucial compounds, including lipids, and to provide the membrane environment for efficient virus assembly. First, the morphological similarities in cubic membrane proliferations during viral infection and virus-independent HMG-CoA reductase overexpression indicate an interference with the regulatory machinery controlling cholesterol content in the ER through SREBP. Based on current evidence, however, it is unclear whether cubic membranes are the result of over-expressed HMG-CoA reductase enzyme that could trigger membrane folding by protein–protein interactions, or whether it is indeed a ‘cholesterol’ effect owing to its over-production and lateral rearrangement to form membrane domains. Cholesterol has profound effects on lateral lipid packing and prefers interaction with saturated acyl-chain containing phospholipids and sphingolipids, which might also sequester transmembrane proteins that would normally not appear in such clusters in the ER. Because it is also evident that cubic membranes could be induced by weak protein–protein interactions in the absence of altered lipid compositions, rearrangement of membrane proteins in the ER could give rise to altered membrane morphology. Such protein rearrangements in turn are likely to trigger the unfolded protein response (UPR), which is a signaling pathway in response to accumulation of (mis-folded) proteins and perhaps lipids, in the ER. UPR and the induction of autophagy under starvation conditions are also functionally linked. Thus, it appears that viral components stimulate multiple molecular mechanisms involved in maintaining cellular physiology and membrane integrity, and trigger cellular programs to regulate enzymatic activity, membrane quality control, and repair processes, such as autophagy. As a common feature of these processes, viruses could thus subvert these cellular programs culminating in the formation of cubic membranes that provide a suitable environment for their efficient replication.
It thus appears that viruses have adopted several cellular programs that could lead to the formation of cubic membranes to facilitate their replication, assembly, and perhaps delivery to the cell surface. Common to these processes are regulatory circuits involved in maintaining membrane lipid homeostasis, including the unfolded protein response, starvation signals triggering autophagy or regulation of cholesterol synthesis through sterol regulatory element binding protein (SREBP; Figure 2 ). Thus, it will be of great interest to identify the viral components that could specifically trigger these cellular programs, which might lead to a better understanding of the underlying molecular mechanisms and offer strategies for pharmacological intervention.
Figure 2.
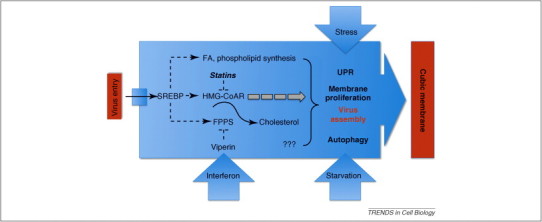
Model illustrating the interference of viruses with components of the regulatory machinery controlling cholesterol homeostasis. Dashed lines indicate regulatory processes. FPPS, farnesyl pyrophosphate synthase; SREBP, sterol regulatory element binding protein; UPR, unfolded protein response. See text for details.
Cubic membrane formation during viral infection: potential mechanisms
The multiple roles of cholesterol in the life cycle of a virus is undisputed (Box 2, Figure 2), and many viruses require cholesterol for their replication [43]. Viral infection also often leads to an alteration of cellular cholesterol content. The striking similarities between cubic membranes induced by viral infection and as a consequence of deregulated HMG-CoA reductase, and the strong dependence of viral replication on cellular cholesterol homeostasis indicate that these processes might be mechanistically linked. The overexpression of HMG-CoA reductase in UT-1 cells as well as administration of HMG-CoA reductase inhibitors, such as compactin or statin, induce the formation of a crystalloid ER 23, 26, 44. Because these membrane proliferations disappear upon adding cholesterol to UT-1 cells [24], crystalloid membrane formation might be a consequence of an altered feedback control of cholesterol biosynthesis. Indeed, West Nile virus infection appears to be mechanistically linked to dysregulation of HMG-CoA reductase. Depletion of cholesterol in the plasma membrane triggers the upregulation of the enzyme and subsequently leads to membrane proliferations similar to crystalloid ER in UT-1 cells in response to experimental overexpression of HMG-CoA reductase[7]. Accordingly, administration of cholesterol disrupts the convoluted membrane formation and significantly suppresses viral replication, strongly suggesting that the virus-induced host membranes are essential for viral replication and survival [7].
In addition to viral infection, cubic membranes (also termed ‘tubuloreticular structures’; TRS) frequently also occur in association with autoimmune and neoplastic diseases [45], and interferon is considered to be a common denominator to induce TRS (even in the absence of viruses) in many human pathological conditions. Thus, membrane morphological transitions could be a universal cellular response to foreign invaders such as viruses and parasites or in tumor cells via an interferon-induced host defense mechanism. One such pathway could operate through the interferon-induced protein viperin (‘virus inhibitory protein, endoplasmic reticulum-associated, interferon-inducible’) [46]. Viperin interacts with and inhibits farnesyl pyrophosphate synthase (FPPS), an enzyme involved in sterol biosynthesis, and FPPS downregulation alters plasma membrane fluidity by affecting the formation of lipid rafts that might play a role in virus entry or budding [47]. Notably, overexpression of viperin induces crystalloid ER (cubic membrane) formation [48]. The gene encoding FPPS is a target of the SREBP transcription factor [49] and subject to negative-feedback regulation by sterols, in coordination with HMG-CoA reductase (Figure 2).
The SREBP pathway is a regulatory circuit controlling more than 30 genes involved in the uptake and synthesis of cholesterol, fatty acids, triacylglycerols, and phospholipids [50]. SREBPs, which exist in three isoforms, are membrane-bound transcription factors that are proteolytically processed as a consequence of altered cholesterol levels in the ER. In the presence of cholesterol, SREBP tightly binds to SREBP cleavage activation protein (SCAP), and is retained in the ER by interacting with the ER-resident protein Insig. Stepwise proteolytic cleavage that takes place in the Golgi by the site-specific proteases S1P and S2P leads to the release of the N-terminal fragment that might enter the nucleus to regulate transcription of SRE-dependent genes. SCAP functions as the cholesterol sensor, and, only upon cholesterol depletion in the ER, SREBP/SCAP exits via COPII vesicles to the Golgi where it is proteolytically processed 49, 50, 51. Sterol synthesis, as a major oxygen consuming pathway, is sensitive to lipid composition, but also to temperature and other factors affecting membrane fluidity, and thus integrates multiple environmental conditions. Several viral NS-proteins are hydrophobic and are found to be associated with endomembranes, giving the potential for multiple mechanisms of interfering with the intricate network that controls sterol homeostasis. Such a dysregulation of cholesterol synthesis during viral infection thus could culminate in a reprogramming of HMG-CoA reductase expression, leading to cubic membrane formation to promote viral replication and assembly. Thus understanding the molecular interactions between viral NS proteins and the sterol regulatory machinery that lead to the induction of cubic membranes might open novel therapeutic antiviral strategies through inhibiting their assembly – without interfering with cellular cholesterol homeostasis (Box 3 ).
Box 3. Future questions.
Induction of cubic membranes (CM) in the course of viral infection provides an excellent experimental system to analyze the underlying molecular mechanisms. Because cholesterol homeostasis and HMG-CoA reductase activity are tightly linked through the SREBP regulatory circuit, it is currently unclear whether changes in cellular cholesterol content or HMG-CoAR enzyme levels are responsible for CM formation. To discriminate between these possibilities, and to identify the role of CM for viral proliferation the following questions need to be addressed:
-
•
What are the viral components involved in inducing CM formation? This could potentially be resolved by analyzing individual viral subclones for their propensity to induce CM.
-
•
Are the SREBP and UPR pathways triggered by specific viral proteins? Some evidence suggests that SREBP target genes are de-regulated in virus-infected cells. This analysis needs to be extended to the analysis of individual viral structural and NS proteins.
-
•
What is the relevance of CM formation during autophagy? This could be tested in LC-3 knock-down models defective in autophagosome formation, to show whether autophagy is important to promote – or inhibit – viral proliferation.
-
•
Is CM formation essential for virus replication and assembly? The answer to this question will allow to discriminate whether CMs are a cellular defense program to deal with virus infections, or rather that viruses are subversing such mechanisms for their efficient assembly and proliferation. The major challenge to this question is the inhibition of CM formation without affecting cellular lipid metabolism, which, however, might be intricately linked. Identifying such strategies could provide novel and unique approaches to combating viral infections.
Concluding remarks
The frequent appearance of cubic membranes in a wide variety of cells from all kingdoms of life under normal, stressed, and pathological conditions points to a fundamental cellular mechanism for membrane morphology conversion in response to internal or external stimuli. Although virus-induced intracellular membrane rearrangements and their importance in the context of viral replication are now well recognized, their function and the molecular mechanisms involved in their biogenesis are poorly understood. We propose that cubic membranes serve as a platform for virus assembly, and which are induced by interfering with the cellular machinery controlling lipid and membrane homeostasis. The potential reprogramming of cellular cholesterol metabolism by the interference of viral nonstructural proteins is an intriguing hypothesis that might shed light onto the molecular basis of virus–host interactions, with a great potential for identifying novel regulatory mechanisms controlling membrane biogenesis. Inhibiting cubic membrane formation without altering membrane lipid composition is a major experimental challenge, however, it might represent a novel and unique strategy for combating viral infections.
Acknowledgments
We thank Akik Kia Khaw for his illustrative work presented in Figure 1e. We apologize to our colleagues whose relevant research could not be cited here owing to space limitations. Research in the authors’ laboratories is supported by research grants NMRC (R-185-000-058-213) and BMRC (R-185-000-197-305) from Singapore to Y.D., and the Austrian Federal Ministry for Science and Research (Project GOLD) in the framework of the genome project GEN-AU, and the Austrian Science Fund, FWF (project SFB Lipotox F3005) to S.D.K.
References
- 1.Miller S., Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Novoa R.R. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell. 2005;97:147–172. doi: 10.1042/BC20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marquart K.H. Occurrence of tubuloreticular structures and intracisternal paracrystalline inclusions in endothelial cells of tissue from different epidemiological types of Kaposi's sarcoma. Ultrastruct. Pathol. 2005;29:85–93. doi: 10.1080/01913120590912205. [DOI] [PubMed] [Google Scholar]
- 4.Lee C.J. Cholesterol effectively blocks entry of flavivirus. J. Virol. 2008;82:6470–6480. doi: 10.1128/JVI.00117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medigeshi G.R. West Nile virus entry requires cholesterol-rich membrane microdomains and is independent of alphavbeta3 integrin. J. Virol. 2008;82:5212–5219. doi: 10.1128/JVI.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothwell C. Cholesterol biosynthesis modulation regulates dengue viral replication. Virology. 2009;389:8–19. doi: 10.1016/j.virol.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 7.Mackenzie J.M. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe. 2007;2:229–239. doi: 10.1016/j.chom.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Mackenzie J.M. Stable expression of noncytopathic Kunjin replicons simulates both ultrastructural and biochemical characteristics observed during replication of Kunjin virus. Virology. 2001;279:161–172. doi: 10.1006/viro.2000.0691. [DOI] [PubMed] [Google Scholar]
- 9.Teterina N.L. Induction of intracellular membrane rearrangements by HAV proteins 2C and 2BC. Virology. 1997;237:66–77. doi: 10.1006/viro.1997.8775. [DOI] [PubMed] [Google Scholar]
- 10.Moradpour D. Membrane association of hepatitis C virus nonstructural proteins and identification of the membrane alteration that harbors the viral replication complex. Antiviral. Res. 2003;60:103–109. doi: 10.1016/j.antiviral.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 11.Hobman T.C. The rubella virus E1 glycoprotein is arrested in a novel post-ER, pre-Golgi compartment. J. Cell Biol. 1992;118:795–811. doi: 10.1083/jcb.118.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noisakran S. Association of dengue virus NS1 protein with lipid rafts. J. Gen. Virol. 2008;89:2492–2500. doi: 10.1099/vir.0.83620-0. [DOI] [PubMed] [Google Scholar]
- 13.Brinton M.A. The molecular biology of West Nile Virus: a new invader of the western hemisphere. Annu. Rev. Microbiol. 2002;56:371–402. doi: 10.1146/annurev.micro.56.012302.160654. [DOI] [PubMed] [Google Scholar]
- 14.Ng M.L. Cryosubstitution technique reveals new morphology of flavivirus-induced structures. J. Virol. Methods. 1994;49:305–314. doi: 10.1016/0166-0934(94)90145-7. [DOI] [PubMed] [Google Scholar]
- 15.Marie M. Take the ‘A’ train: on fast tracks to the cell surface. Cell Mol. Life Sci. 2008;65:2859–2874. doi: 10.1007/s00018-008-8355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Almsherqi Z.A. Cubic membranes the missing dimension of cell membrane organization. Int. Rev. Cell Mol. Biol. 2009;274:275–342. doi: 10.1016/S1937-6448(08)02006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Almsherqi Z.A. Cubic membranes: a legend beyond the Flatland* of cell membrane organization. J. Cell Biol. 2006;173:839–844. doi: 10.1083/jcb.200603055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snapp E.L. Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 2003;163:257–269. doi: 10.1083/jcb.200306020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto A. Formation of crystalloid endoplasmic reticulum in COS cells upon overexpression of microsomal aldehyde dehydrogenase by cDNA transfection. J. Cell Sci. 1996;109:1727–1738. doi: 10.1242/jcs.109.7.1727. [DOI] [PubMed] [Google Scholar]
- 20.Luzzati V. Biological significance of lipid polymorphism: the cubic phases. Curr. Opin. Struct. Biol. 1997;7:661–668. doi: 10.1016/s0959-440x(97)80075-9. [DOI] [PubMed] [Google Scholar]
- 21.Lingwood D. Morphological homeostasis by autophagy. Autophagy. 2009;5:1039–1040. doi: 10.4161/auto.5.7.9314. [DOI] [PubMed] [Google Scholar]
- 22.Lingwood D. Generation of cubic membranes by controlled homotypic interaction of membrane proteins in the endoplasmic reticulum. J. Biol. Chem. 2009;284:12041–12048. doi: 10.1074/jbc.M900220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chin D.J. Appearance of crystalloid endoplasmic reticulum in compactin-resistant Chinese hamster cells with a 500-fold increase in 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc. Natl. Acad. Sci. U. S. A. 1982;79:1185–1189. doi: 10.1073/pnas.79.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jingami H. Partial deletion of membrane-bound domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase eliminates sterol-enhanced degradation and prevents formation of crystalloid endoplasmic reticulum. J. Cell Biol. 1987;104:1693–1704. doi: 10.1083/jcb.104.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roitelman J. Immunological evidence for eight spans in the membrane domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase: implications for enzyme degradation in the endoplasmic reticulum. J. Cell Biol. 1992;117:959–973. doi: 10.1083/jcb.117.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson R.G. Ultrastructural analysis of crystalloid endoplasmic reticulum in UT-1 cells and its disappearance in response to cholesterol. J. Cell Sci. 1983;63:1–20. doi: 10.1242/jcs.63.1.1. [DOI] [PubMed] [Google Scholar]
- 27.Orci L. Increase in membrane cholesterol: a possible trigger for degradation of HMG CoA reductase and crystalloid endoplasmic reticulum in UT-1 cells. Cell. 1984;36:835–845. doi: 10.1016/0092-8674(84)90033-3. [DOI] [PubMed] [Google Scholar]
- 28.Profant D.A. The role of the 3-hydroxy 3-methylglutaryl coenzyme A reductase cytosolic domain in karmellae biogenesis. Mol. Biol. Cell. 1999;10:3409–3423. doi: 10.1091/mbc.10.10.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tenchov B.G. Cubic phases in phosphatidylcholine-cholesterol mixtures: cholesterol as membrane ‘fusogen’. Biophys. J. 2006;91:2508–2516. doi: 10.1529/biophysj.106.083766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng Y. Docosapentaenoic acid (DPA) is a critical determinant of cubic membrane formation in amoeba Chaos mitochondria. FASEB J. 2009;23:2866–2871. doi: 10.1096/fj.09-130435. [DOI] [PubMed] [Google Scholar]
- 31.Guinea R., Carrasco L. Phospholipid biosynthesis and poliovirus genome replication, two coupled phenomena. EMBO J. 1990;9:2011–2016. doi: 10.1002/j.1460-2075.1990.tb08329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee W.M. Mutation of host delta9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J. Virol. 2001;75:2097–2106. doi: 10.1128/JVI.75.5.2097-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee W.M., Ahlquist P. Membrane synthesis, specific lipid requirements, and localized lipid composition changes associated with a positive-strand RNA virus RNA replication protein. J. Virol. 2003;77:12819–12828. doi: 10.1128/JVI.77.23.12819-12828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee Y.R. Autophagic machinery activated by dengue virus enhances virus replication. Virology. 2008;374:240–248. doi: 10.1016/j.virol.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson W.T. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirkegaard K. Subversion of the cellular autophagy pathway by viruses. Curr. Top. Microbiol. Immunol. 2009;335:323–333. doi: 10.1007/978-3-642-00302-8_16. [DOI] [PubMed] [Google Scholar]
- 37.Kalvodova L. The lipidomes of vesicular stomatitis virus, semliki forest virus, and the host plasma membrane analyzed by quantitative shotgun mass spectrometry. J. Virol. 2009;83:7996–8003. doi: 10.1128/JVI.00635-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng Y. Fasting induces cyanide-resistant respiration and oxidative stress in the amoeba Chaos carolinensis: implications for the cubic structural transition in mitochondrial membranes. Protoplasma. 2002;219:160–167. doi: 10.1007/s007090200017. [DOI] [PubMed] [Google Scholar]
- 39.Almsherqi Z. Cubic membranes: a structure-based design for DNA uptake. J. R. Soc. Interface. 2008;5:1023–1029. doi: 10.1098/rsif.2007.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Almsherqi Z.A. Direct template matching reveals a host subcellular membrane gyroid cubic structure that is associated with SARS virus. Redox Rep. 2005;10:167–171. doi: 10.1179/135100005X57373. [DOI] [PubMed] [Google Scholar]
- 41.Goldsmith C.S. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 2004;10:320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ulasli M. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell Microbiol. 2010 doi: 10.1111/j.1462-5822.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bukrinsky M., Sviridov D. Human immunodeficiency virus infection and macrophage cholesterol metabolism. J. Leukoc. Biol. 2006;80:1044–1051. doi: 10.1189/jlb.0206113. [DOI] [PubMed] [Google Scholar]
- 44.Pathak R.K. Biogenesis of the crystalloid endoplasmic reticulum in UT-1 cells: evidence that newly formed endoplasmic reticulum emerges from the nuclear envelope. J. Cell Biol. 1986;102:2158–2168. doi: 10.1083/jcb.102.6.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luu J.Y. Tubuloreticular structures and cylindrical confronting cisternae: a review. Hum. Pathol. 1989;20:617–627. doi: 10.1016/0046-8177(89)90148-2. [DOI] [PubMed] [Google Scholar]
- 46.Chin K.C., Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 2001;98:15125–15130. doi: 10.1073/pnas.011593298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe. 2007;2:96–105. doi: 10.1016/j.chom.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 48.Hinson E.R., Cresswell P. The N-terminal amphipathic alpha-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J. Biol. Chem. 2009;284:4705–4712. doi: 10.1074/jbc.M807261200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown M.S., Goldstein J.L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 50.Espenshade P.J., Hughes A.L. Regulation of sterol synthesis in eukaryotes. Annu. Rev. Genet. 2007;41:401–427. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- 51.Espenshade P.J. SREBPs: sterol-regulated transcription factors. J. Cell Sci. 2006;119:973–976. doi: 10.1242/jcs.02866. [DOI] [PubMed] [Google Scholar]
- 52.Deng Y., Mieczkowski M. Three-dimensional periodic cubic membrane structure in the mitochondria of amoebae Chaos carolinensis. Protoplasma. 1998;203:16–25. [Google Scholar]
- 53.Landh T. From entangled membranes to eclectic morphologies: cubic membranes as subcellular space organizers. FEBS Lett. 1995;369:13–17. doi: 10.1016/0014-5793(95)00660-2. [DOI] [PubMed] [Google Scholar]
- 54.Deng Y. Cubic membrane structure in amoeba (Chaos carolinensis) mitochondria determined by electron microscopic tomography. J. Struct. Biol. 1999;127:231–239. doi: 10.1006/jsbi.1999.4147. [DOI] [PubMed] [Google Scholar]
- 55.Knoops K. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Welsch S. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schaff Z. Ultrastructure of normal and hepatitis virus infected human and chimpanzee liver: similarities and differences. Acta Morphol. Hung. 1992;40:203–214. [PubMed] [Google Scholar]
- 58.Murphy F.A. St. Louis encephalitis virus infection in mice. Electron microscopic studies of central nervous system. Lab. Invest. 1968;19:652–662. [PubMed] [Google Scholar]
- 59.Marsh D. Protein modulation of lipids, and vice-versa, in membranes. Biochim. Biophys. Acta. 2008;1778:1545–1575. doi: 10.1016/j.bbamem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 60.Syed G.H. Hepatitis C virus hijacks host lipid metabolism. Trends Endocrinol. Metab. 2009;21:33–40. doi: 10.1016/j.tem.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waheed A.A., Freed E.O. Lipids and membrane microdomains in HIV-1 replication. Virus Res. 2009;143:162–176. doi: 10.1016/j.virusres.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang W. Lipid rafts play an important role in the vesicular stomatitis virus life cycle. Arch. Virol. 2009;154:595–600. doi: 10.1007/s00705-009-0348-2. [DOI] [PubMed] [Google Scholar]
- 63.Carter G.C. HIV entry in macrophages is dependent on intact lipid rafts. Virology. 2009;386:192–202. doi: 10.1016/j.virol.2008.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tang Y. Deficiency of niemann-pick type C-1 protein impairs release of human immunodeficiency virus type 1 and results in Gag accumulation in late endosomal/lysosomal compartments. J. Virol. 2009;83:7982–7995. doi: 10.1128/JVI.00259-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu Y. Lipid rafts are involved in SARS-CoV entry into Vero E6 cells. Biochem. Biophys. Res. Commun. 2008;369:344–349. doi: 10.1016/j.bbrc.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hotta K. Effect of cellular cholesterol depletion on rabies virus infection. Virus Res. 2009;139:85–90. doi: 10.1016/j.virusres.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 67.Barman S., Nayak D.P. Lipid raft disruption by cholesterol depletion enhances influenza A virus budding from MDCK cells. J. Virol. 2007;81:12169–12178. doi: 10.1128/JVI.00835-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mohan K.V. Defective rotavirus particle assembly in lovastatin-treated MA104 cells. Arch. Virol. 2008;153:2283–2290. doi: 10.1007/s00705-008-0261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 70.Cheville N.F. The cytopathology of swine pox in the skin of swine. Am. J. Pathol. 1966;49:339–352. [PMC free article] [PubMed] [Google Scholar]
- 71.Casey H.W. Electron microscopy of a benign epidermal pox disease of rhesus monkeys. Am. J. Pathol. 1967;51:431–446. [PMC free article] [PubMed] [Google Scholar]
- 72.Espana C. Electron microscopy of the Tana poxvirus. Exp. Mol. Pathol. 1971;15:34–42. doi: 10.1016/0014-4800(71)90017-7. [DOI] [PubMed] [Google Scholar]
- 73.Pospischil A., Bachmann P.A. Nuclear changes in cells infected with parapoxviruses stomatitis papulosa and orf: an in vivo and in vitro ultrastructural study. J. Gen. Virol. 1980;47:113–121. doi: 10.1099/0022-1317-47-1-113. [DOI] [PubMed] [Google Scholar]
- 74.Baringer J.R., Griffith J.F. Experimental herpes encephalitis: crystalline arrays in endoplasmic reticulum. Science. 1969;165:1381. doi: 10.1126/science.165.3900.1381. [DOI] [PubMed] [Google Scholar]
- 75.Baringer J.R., Griffith J.F. Experimental herpes simplex encephalitis: early neuropathologic changes. J. Neuropathol. Exp. Neurol. 1970;29:89–104. doi: 10.1097/00005072-197001000-00007. [DOI] [PubMed] [Google Scholar]
- 76.Baringer J.R., Swoveland P. Tubular aggregates in endoplasmic reticulum: evidence against their viral nature. J. Ultrastruct. Res. 1972;41:270–276. doi: 10.1016/s0022-5320(72)90069-x. [DOI] [PubMed] [Google Scholar]
- 77.Couch J.A. An enzootic nuclear polyhedrosis virus of pink shrimp: ultrastructure, prevalence, and enhancement. J. Invertebr. Pathol. 1974;24:311–331. doi: 10.1016/0022-2011(74)90139-6. [DOI] [PubMed] [Google Scholar]
- 78.Kartenbeck J. Endocytosis of simian virus 40 into the endoplasmic reticulum. J. Cell Biol. 1989;109:2721–2729. doi: 10.1083/jcb.109.6.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bucciarelli E. Ultrastructure of meningeal tumors induced in dogs with Rous sarcoma virus. J. Natl. Cancer Inst. 1967;38:359–381. [PubMed] [Google Scholar]
- 80.Smith R.D., Deinhardt F. Unique cytoplasmic membranes in Rous sarcoma virus-induced tumors of a subhuman primate. J. Cell Biol. 1968;37:819–823. doi: 10.1083/jcb.37.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kaup F.J. Tubuloreticular structures in rectal biopsies of SIV-infected rhesus monkeys (Macaca mulatta) Ultrastruct. Pathol. 2005;29:357–366. doi: 10.1080/019131290968740. [DOI] [PubMed] [Google Scholar]
- 82.Deneka M. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. J. Cell Biol. 2007;177:329–341. doi: 10.1083/jcb.200609050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Orenstein J.M. Ultrastructural markers in circulating lymphocytes of subjects at risk for AIDS. Am. J. Clin. Pathol. 1985;84:603–609. doi: 10.1093/ajcp/84.5.603. [DOI] [PubMed] [Google Scholar]
- 84.Ruebner B.H. Electron microscopy of the hepatocellular and Kupffer-cell lesions of mouse hepatitis, with particular reference to the effect of cortisone. Am. J. Pathol. 1967;51:163–189. [PMC free article] [PubMed] [Google Scholar]
- 85.Nii S. Crystalline and lamellar structures in LLCMK2 cell cultures persistently infected with dengue viruses. Biken J. 1970;13:43–51. [PubMed] [Google Scholar]
- 86.Matsumura T. Morphogenesis of dengue-1 virus in cultures of a human leukemic leukocyte line (J-111) Microbiol. Immunol. 1977;21:329–334. doi: 10.1111/j.1348-0421.1977.tb00295.x. [DOI] [PubMed] [Google Scholar]
- 87.Parker J.R., Stannard L.M. Intracytoplasmic inclusions in foetal lamb kidney cells infected with Wesselsbron virus. Arch. Gesamte Virusforsch. 1967;20:469–472. doi: 10.1007/BF01275229. [DOI] [PubMed] [Google Scholar]
- 88.Hong S.S., Ng M.L. Involvement of microtubules in Kunjin virus replication. Brief report. Arch. Virol. 1987;97:115–121. doi: 10.1007/BF01310739. [DOI] [PubMed] [Google Scholar]
- 89.Ng M.L., Hong S.S. Flavivirus infection: essential ultrastructural changes and association of Kunjin virus NS3 protein with microtubules. Arch. Virol. 1989;106:103–120. doi: 10.1007/BF01311042. [DOI] [PubMed] [Google Scholar]
- 90.Westaway E.G. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tandler B. Unusual membrane formations in HEp-2 cells infected with Ilheus virus. Lab. Invest. 1973;28:218–223. [PubMed] [Google Scholar]
- 92.Koestner A. Electron microscopic evaluation of the pathogenesis of porcine polioencephalomyelitis. Am. J. Pathol. 1966;49:325–337. [PMC free article] [PubMed] [Google Scholar]
- 93.Hashimoto I. Ultrastructural studies on the pathogenesis of poliomyelitis in monkeys infected with poliovirus. Acta Neuropathol. 1984;64:53–60. doi: 10.1007/BF00695606. [DOI] [PubMed] [Google Scholar]
- 94.Blinzinger K. Poliovirus crystals within the endoplasmic reticulum of endothelial and mononuclear cells in the monkey spinal cord. Science. 1969;163:1336–1337. doi: 10.1126/science.163.3873.1336. [DOI] [PubMed] [Google Scholar]
- 95.Breese S.S., Jr., Graves J.H. Electron microscopic observation of crystalline arrays of foot-and-mouth disease virus. J. Bacteriol. 1966;92:1835–1837. doi: 10.1128/jb.92.6.1835-1837.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim K.S., Boatman E.S. Electron microscopy of monkey kidney cell cultures infected with rubella virus. J. Virol. 1967;1:205–214. doi: 10.1128/jvi.1.1.205-214.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Grimley P.M., Friedman R.M. Development of Semliki forest virus in mouse brain: an electron microscopic study. Exp. Mol. Pathol. 1970;12:1–13. doi: 10.1016/0014-4800(70)90070-5. [DOI] [PubMed] [Google Scholar]
- 98.Koestner A., Long J.F. Ultrastructure of canine distemper virus in explant tissue cultures of canine cerebellum. Lab. Invest. 1970;23:196–201. [PubMed] [Google Scholar]
- 99.Tinari A. Tubuloreticular structures induced by rotavirus infection in HT-29 cells. Ultrastruct. Pathol. 1996;20:571–576. doi: 10.3109/01913129609016361. [DOI] [PubMed] [Google Scholar]
- 100.Quan C.M., Doane F.W. Ultrastructural evidence for the cellular uptake of rotavirus by endocytosis. Intervirology. 1983;20:223–231. doi: 10.1159/000149395. [DOI] [PubMed] [Google Scholar]


