Dear Editor,
Most recently, Chan et al.1 reported, in this journal, that convalescent SARS patients' sera may contain cross-reactive antibodies against the emerging novel human coronavirus EMC/2012 (hCoV-EMC) detected by both immunofluorescent and neutralizing antibody tests.1
SARS coronavirus (SARS-CoV), the causative agent of SARS, uses the angiotensin-converting enzyme 2 (ACE2) as its cellular receptor to bind to the target cells,2 and a 193-amino acid fragment (residues 318–510) in the S1 subunit of vial spike (S) protein is the identified receptor-binding domain (RBD).3 The recent emergence of hCoV-EMC has caused 17 people infected including 11 deaths (http://www.who.int/csr/don/2013_03_26/en/), raising serious concern about its potential pandemic. Unlike SARS-CoV, hCoV-EMC uses a different receptor, dipeptidyl peptidase-4 (DPP4),4 for its binding and entry into the target cell. We have predicted that a 286-amino acid fragment (residues 377–662) of hCoV-EMC S1 region contains the viral RBD.5
Previously we reported that the RBD of SARS-CoV S protein contains multiple neutralizing epitopes that induce potent neutralizing antibodies and protection against SARS-CoV infection in animal models.6, 7, 8 Thus, neutralizing antibodies targeting the S protein, particularly the RBD, play the most important roles in the inhibition of viral infection. Since both SARS-CoV and hCoV-EMC genetically belong to the genus betacoronavirus,9, 10 we thus speculate that the antibodies induced by the RBD of SARS-CoV may have cross-reactivity or cross-neutralizing activity against hCoV-EMC.
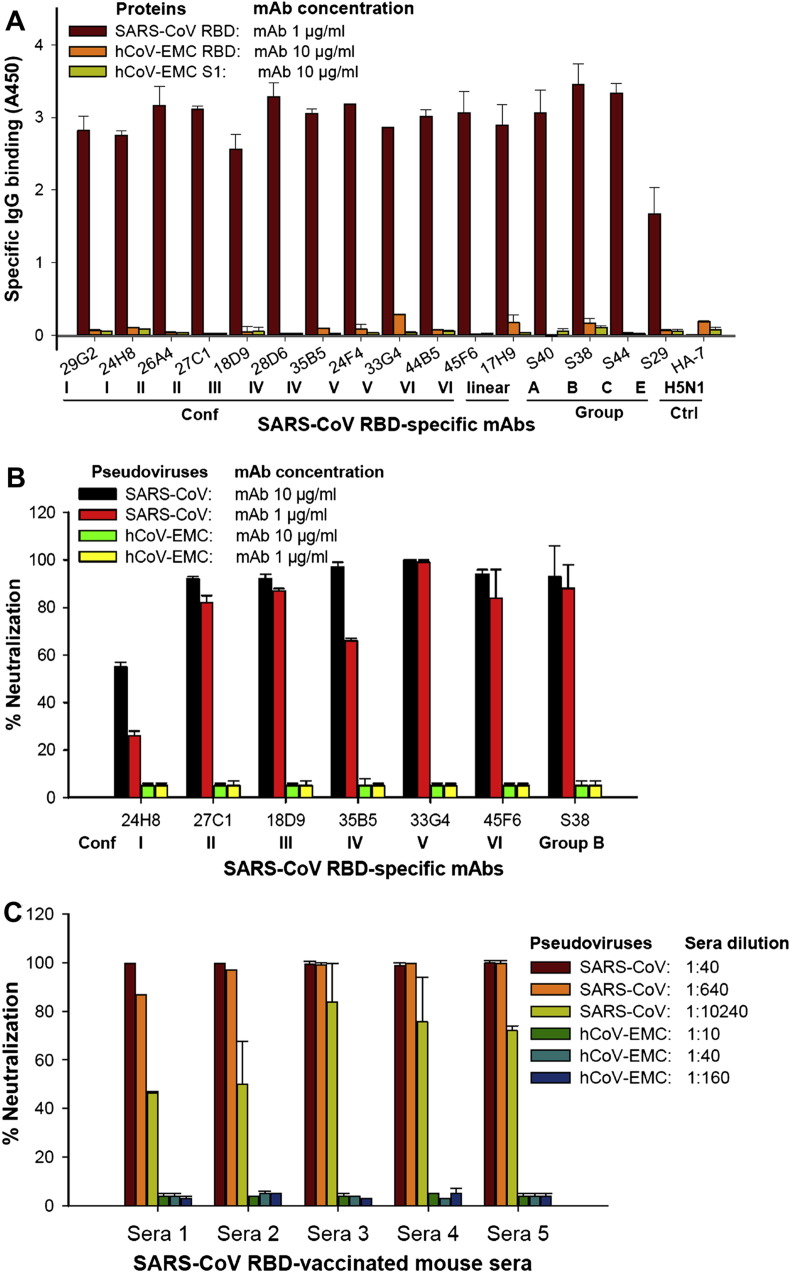
To prove this, we first tested the reactivity of a series of SARS-CoV RBD-specific monoclonal antibodies (mAbs)6, 11 with recombinant proteins containing S1 (residues 18–725) and putative RBD (residues 377–662) in S of hCoV-EMC. We found that all of these mAbs that can recognize the conformational (Conf I–VI, Group A–E) or linear epitopes in RBD of SARS-CoV had low to no binding (A450 < 0.3) to the RBD and S1 proteins of hCoV-EMC at the concentration as high as 10 μg/ml, while they had a strong binding to a recombinant RBD protein of SARS-CoV at the tested concentration of 1 μg/ml (Fig. 1 A).7 These results suggest that the antibodies induced by the RBD of SARS-CoV S protein did not cross-react with the RBD and S1 protein of hCoV-EMC.
Figure 1.
Cross-reactivity and cross-neutralization activity of SARS-CoV S-RBD-specific antibodies against hCoV-EMC. (A) Reactivity of SARS-CoV S-RBD-specific mAbs with RBD and/or S1 protein of hCoV-EMC and SARS-CoV as detected by ELISA. Conf I–VI, Group A–E, and linear mAbs represent the mAbs targeting the conformational and linear epitopes in RBD of SARS-CoV S protein, respectively. HA-7 mAb specific to hemagglutinin (HA) of H5N1 influenza virus was used as the negative control. The data are presented as mean A450 ± standard deviation (SD) of duplicate wells. Neutralization of SARS-CoV S-RBD-specific mAbs (B) and SARS-CoV S-RBD protein-vaccinated mouse antisera (C) against hCoV-EMC and SARS-CoV infection by pseudovirus neutralization assay. The data are presented as mean percentages of neutralization ± SD of duplicate wells.
We next detected the neutralizing activity of the representative SARS-CoV S-RBD-specific neutralizing mAbs against hCoV-EMC infection in Huh-7 cells that express DPP4 receptor for hCoV-EMC4 and against SARS-CoV infection in ACE2/293T cells expressing the receptor for SARS-CoV,7 using our established pseudovirus neutralization assay. As shown in Fig. 1B, in an exception of the mAb 24H8 (Conf I) that had a lower neutralization, all other mAbs including 27C1, 18D9, 35B5, 33G4, 45F6, and S38, which recognize conformational epitopes Conf II–VI and Group B of RBD of SARS-CoV,6, 11 had >90% and ≥70% neutralization of SARS-CoV pseudovirus at the concentration of 10 and 1 μg/ml, respectively. However, all these mAbs could not neutralize hCoV-EMC pseudovirus at the concentration as high as 10 μg/ml, suggesting that the SARS-CoV RBD-specific neutralizing mAbs had low to no cross-neutralization against hCoV-EMC.
To further confirm our conclusion, we performed another experiment to test the neutralizing activity of antibodies in the sera of SARS-CoV S-RBD protein-vaccinated mice. As shown in Fig. 1C, none of the tested sera neutralized hCoV-EMC pseudovirus at the dilution of 1:10, while they could potently neutralize SARS-CoV pseudovirus infection in ACE2/293T cells at the dilution of 1:10,240. These results confirm that the antibodies induced by the RBD of SARS-CoV S1 protein cannot cross-neutralize hCoV-EMC infection. Therefore, the epitopes in SARS-CoV S protein that elicit the antibodies with cross-reactivity and cross-neutralizing activity against hCoV-EMC may not be located in the RBD in S1 subunit of SARS-CoV.
By bioinformatic analysis of S proteins of SARS-CoV and hCoV-EMC, Chan et al.1 found that an immunogenic region hCoV-EMC S (emc-II) and that in SARS-CoV S (sars-I) overlapped the heptad repeat 2 (HR2) region of the S2 domain of both hCoV-EMC and SARS-CoV, while SARS-CoV S-HR2 harbors an epitope for broadly neutralizing antibody in the case of SARS-CoV.12 They thus believed that the epitope located in this region may be responsible for inducing cross-neutralizing antibodies against both hCoV-EMC and SARS-CoV. However, an experiment to prove this hypothesis is warranted.
Potentials conflicts of interest
No reported conflicts.
Acknowledgments
The authors were partially supported by the grant from the National Institutes of Health (R01AI098775).
References
- 1.Chan K.H., Fuk-Woo C.J., Tse H., Chen H., Choi-Yi L.C., Cai J.P. Cross-reactive antibodies in convalescent SARS patients' sera against the emerging novel human coronavirus EMC (2012) by both immunofluorescent and neutralizing antibody tests. J Infect. 2013 Apr 10 doi: 10.1016/j.jinf.2013.03.015. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong S.K., Li W., Moore M.J., Choe H., Farzan M.A. 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raj V.S., Mou H., Smits S.L., Dekkers D.H., Muller M.A., Dijkman R. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang S., Lu L., Du L., Debnath A.K. A predicted receptor-binding and critical neutralizing domain in S protein of the novel human coronavirus HCoV-EMC. J Infect. 2013;66:464–466. doi: 10.1016/j.jinf.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He Y., Lu H., Siddiqui P., Zhou Y., Jiang S. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J Immunol. 2005;174:4908–4915. doi: 10.4049/jimmunol.174.8.4908. [DOI] [PubMed] [Google Scholar]
- 7.Du L., Zhao G., Chan C.C., Sun S., Chen M., Liu Z. Recombinant receptor-binding domain of SARS-CoV spike protein expressed in mammalian, insect and E. coli cells elicits potent neutralizing antibody and protective immunity. Virology. 2009;393:144–150. doi: 10.1016/j.virol.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV – a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaki A.M., van B.S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 10.Chan J.F., Li K.S., To K.K., Cheng V.C., Chen H., Yuen K.Y. Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) the beginning of another SARS-like pandemic? J Infect. 2012;65:477–489. doi: 10.1016/j.jinf.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Y., Li J., Du L., Yan X., Hu G., Zhou Y. Identification and characterization of novel neutralizing epitopes in the receptor-binding domain of SARS-CoV spike protein: revealing the critical antigenic determinants in inactivated SARS-CoV vaccine. Vaccine. 2006;24:5498–5508. doi: 10.1016/j.vaccine.2006.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elshabrawy H.A., Coughlin M.M., Baker S.C., Prabhakar B.S. Human monoclonal antibodies against highly conserved HR1 and HR2 domains of the SARS-CoV spike protein are more broadly neutralizing. PLoS One. 2012;7:e50366. doi: 10.1371/journal.pone.0050366. [DOI] [PMC free article] [PubMed] [Google Scholar]