Abstract
Severe acute respiratory syndrome (SARS) is a zoonotic infectious disease caused by a novel coronavirus (CoV). The tissue tropism of SARS-CoV includes not only the lung, but also the gastrointestinal tract, kidney and liver. Angiotensin-converting enzyme 2 (ACE2), the C-type lectin CD209L (also known L-SIGN), and DC-SIGN bind SARS-CoV, but ACE2 appears to be the key functional receptor for the virus. There is a prominent innate immune response to SARS-CoV infection, including acute-phase proteins, chemokines, inflammatory cytokines and C-type lectins such as mannose-binding lectin, which plays a protective role against SARS. By contrast there may be a lack of type 1 interferon response. Moreover, lymphopenia with decreased numbers of CD4+ and CD8+ T cells is common during the acute phase. Convalescent patients have IgG-class neutralizing antibodies that recognize amino acids 441–700 of the spike protein (S protein) as the major epitope.
Introduction
Severe acute respiratory syndrome (SARS), a newly emerged infectious disease, appeared in Guangdong Province, People's Republic of China, in November 2002 and spread globally to infect more than 8,000 people with over 770 deaths in a few months [1]. The etiological agent was identified as a coronavirus (CoV) [2••, 3, 4] and the genome sequence established it as a novel member of the family [5, 6]. This novel CoV has satisfied Koch's postulates for causation, including the consistent detection of the virus and an antibody response to the virus in patients with SARS [2••] and the reproduction of the disease in non-human primates after experimental inoculation [7, 8]. Previous reviews have addressed issues on the clinical presentation [9], aetiology and laboratory diagnosis [10], epidemiology and viral genetics [11], vaccines and therapeutics [1, 12], and public health [13]. Here we will focus on the host–pathogen interaction, including tropism of SARS-CoV, viral entry into host cells, innate and adaptive immune responses, and host genetic susceptibility to SARS.
Tropism of SARS-CoV
Tropism from animals to humans
Markets selling live poultry and fish for human consumption (‘wet markets’) are common place across South-East Asia, and they service the cultural demand for freshly killed meat and poultry products. In parts of southern China, Guangdong province in particular, increasing affluence has resulted in large markets, which house a diversity of animals, including reptile and mammalian species, to serve the restaurant trade for exotic foods.
SARS-CoV-like viruses were isolated from Himalayan palm civets found in a wildlife wet market in Guangdong (Figure 1a; [14••]). Serological evidence of infection in raccoon dogs and humans in the same market was also found, supporting a zoonotic origin of SARS. The animal CoV isolates were different from the human SARS-CoV in that they had an additional 29-nucleotide sequence not found in most human isolates. This additional 29-nucleotide sequence in the animal CoV results in merging opening reading frames (ORF) 10 and 11 into a new ORF encoding a putative protein of 122 amino acids of unknown biological significance. How the precursor animal CoV adapted itself in humans to achieve efficient man-to-man transmission remains unknown, but molecular epidemiological study of SARS-CoV has suggested that several introductions of the animal CoV into humans had occurred, with only the one with the 29-nucleotide deletion associated with the subsequent outbreak in Hong Kong spreading globally [15]. Phylogenetic analysis of the few human cases in Guangdong in December 2003 revealed that this SARS-CoV is much closer to the palm civet CoV than to any human SARS-CoV detected in the epidemic in early 2003 [16]. This again strengthens the contention that SARS is of zoonotic origin, and that the wet markets serve as a continuing source of SARS-CoV crossing species — from animals to humans [17].
Figure 1.
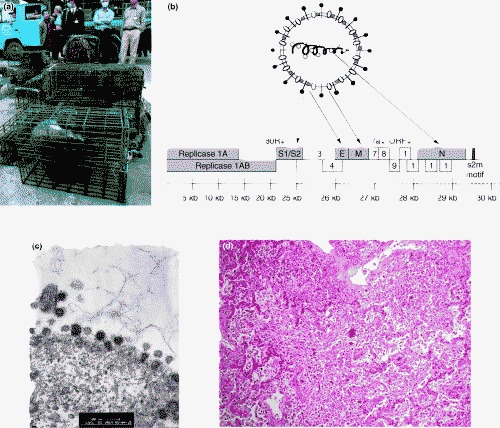
The SARS coronavirus. (a) Wet markets are the interface where animal to human inter-species transmission of SARS-CoV occurred. Guangzhou government officers seize civet cats in Xinyuan wildlife market in Guangzhou to prevent the spread of SARS-CoV. (Reproduced with permission from South China Morning Post). (b) The SARS-CoV expresses structural proteins — spike (S) protein, membrane (M), envelope (E) and nucleocapsid (N) and 14 open reading frames (ORFs). The spike protein determines the virus–host cell receptor interaction and is critical for host species restriction as well as being an important target for neutralizing antibody. (c) Transmission electron micrograph of FRhK cells infected with SARS-CoV. Viral particles are present on the surface of the cell with the particles showing the typical spikes or corona. (Courtesy of John Nicholls, Department of Pathology, The University of Hong Kong). (d) Organizing diffuse alveolar damage with giant cell formation in a patient who died of SARS. (Courtesy of John Nicholls, Department of Pathology, The University of Hong Kong).
Tropism from lung to multiple organs
SARS is largely a viral pneumonia and the lung pathology shows diffuse alveolar damage with multinucleate giant-cells and a prominent increase of macrophages in the alveoli and lung interstitium [18]. Apart from the pneumonia, 38% and 73% of patients, respectively, had diarrhoea in two separate cohorts of patients [19, 20]. Despite the active viral replication in the enterocytes, there is minimal disruption of the intestinal architecture or cellular infiltrate, and the mechanism of pathogenesis of the diarrhoea remains unclear. The absence of intestinal inflammation and destruction has been speculated to be a result of upregulation of potent immunosuppressive cytokine TGF-β [21] and an antiapoptotic host cellular response in the intestinal epithelial cells respectively [1].
Using immunohistochemistry and in situ hybridization to examine organs from four SARS patients who died, SARS-CoV was found not only in the lung and intestine, but also in liver, distal convoluted renal tubules, sweat glands, parathyroid, pituitary, pancreas, adrenal gland and cerebrum [22]. Using reverse-transcriptase polymerase chain reaction to examine 19 fatal SARS cases, SARS-CoV RNA was detected in lung, small and large bowels, lymph nodes, spleen, liver, heart, kidney and skeletal muscle, in descending order of viral load per gram of tissue [23]. These findings suggest that SARS is a systemic disease with widespread extrapulmonary dissemination, resulting in viral shedding in respiratory secretions, stools, urine and possibly even in sweat [22, 23]. The organ damage in patients with SARS could be due to both local viral replication and the immunopathologic consequences of the host response, hence it is important to delineate what human cells the SARS-CoV can infect and replicate in as well as the subsequent host immune response.
Cellular receptors for SARS-CoV to gain entry into target cells
Coronaviruses, including SARS-CoV, associate with cellular receptors to mediate infection of their target cells via the surface spike protein (S protein; Figure 1; [24]). A metallopeptidase, angiotensin-converting enzyme 2 (ACE2), has been shown to be a functional receptor for SARS-CoV [25••, 26], with amino acids 270–510 of the SARS-CoV S protein critical for interaction with ACE2 [27].
The SARS-CoV S protein has striking similarities with class I fusion proteins, which include the influenza virus haemagglutinin and paramyxovirus F protein. The N-terminal half of the S protein (S1) contains the receptor-binding domain whereas the C-terminal half (S2) is the membrane-anchored membrane-fusion subunit, which contains two heptad repeat regions (HR1 and HR2) [28, 29]. After binding to ACE2 on the target cells, the transmembrane S protein changes conformation by association between the HR1 and HR2 regions to form a six-helix oligomeric complex, leading to fusion between the viral and target-cell membranes. The X-ray crystallographic structure of the SARS-CoV S2 fusion-protein core provides a framework for the design of entry inhibitors [30].
Apart from direct membrane fusion at the target cell surface, SARS-CoV might gain cell entry via pH-dependent endocytosis, which is also mediated by the S protein [31]. Furthermore the glycosylated S protein has been shown to bind to the C-type lectin DC-SIGN (DC-specific ICAM-3-grabbing non-integrin) expressed on the DCs; these cells then mediate SARS-CoV infection in trans of cells that express human ACE2, but the DC-SIGN does not initiate SARS-CoV entry into the DCs. Human CD209L (also known as liver/lymph-node-specific [L]-SIGN), which is 77% identical to human DC-SIGN, can also bind to S protein and mediate virus entry [32•] but its role in initiating productive virus replication remains unclear.
ACE2 protein is reportedly present in type 1 and type 2 pneumocytes, enterocytes of all parts of the small intestine, the brush border of the proximal tubular cells of the kidney, as well as the endothelial cells of small and large arteries and veins of all tissues studied and arterial smooth muscle cells [33]. This localization of ACE2 explains the tissue tropism of SARS-CoV for the lung, small intestine and kidney; however, notable discrepancies include virus replication in colonic epithelium, which has no ACE2, and no virus infection in endothelial cells, which have ACE2. Other receptors or co-receptors such as L-SIGN may explain such discrepancies. Since only the basal layer of the non-keratinized squamous epithelium of the upper respiratory tract expresses ACE2, undamaged epithelium of the nasopharynx is not likely to support SARS-CoV replication. This raises the interesting possibility that a preceding infection by another pathogen may expose the basal layer and facilitate SARS-CoV infecting the damaged upper respiratory tract before spreading to involve the lung.
Furthermore, this may partly explain the longer incubation period (mean 4.6 days, variance 15.9 days) of SARS [34] and the gradual increase of viral load in nasopharyngeal aspirates of SARS patients in the first 10 days of illness [19], as compared with influenza, since SARS-CoV may take a longer time than influenza virus does to establish infection in the respiratory tract.
The innate immune response to SARS-CoV
The gradual increase in viral load in the first 10 days of SARS also suggests that the virus may be evading some of the innate immune defences of the host. It may be relevant that SARS-CoV infection of human macrophages in vitro leads to initiation of viral replication and viral protein synthesis without a detectable type-1 interferon response [35]. Furthermore, in marked contrast to patients with influenza, there was a lack of expression of mRNA for the type-1 interferons, interferon α and β in peripheral blood mononuclear cells of patients with SARS [36]. On the other hand, other innate immune defences such as the collectins that can bind the glycosylated SARS-CoV S protein may play an important role in host defence. Such collectins include the lung surfactant proteins A and D as well as the mannose-binding lectin (MBL), which is present in serum. In a case-control gene-association study of 569 Hong Kong Chinese SARS patients and 1188 healthy controls, low-MBL-producing genotypes have been shown to be associated with increased risk of SARS [37•]. Moreover MBL can bind SARS-CoV, resulting in protective biological effects in a calcium-dependent and mannan-inhibitable fashion, suggesting the binding is through the carbohydrate-recognition domains of MBL with the S protein. Plasma proteomic analysis of four patients revealed active innate immune responses to SARS-CoV, including increased acute-phase proteins such as serum amyloid A, and C-type lectins such as MBL and tetranectin [38•]. These observations suggest that MBL plays a protective role against development of SARS.
High-density oligonucleotide array analysis of gene-expression changes in peripheral blood mononuclear cells from normal healthy donors inoculated in vitro with SARS-CoV showed an early activation of the innate immunity pathway in the first 12 h, including enhanced expression of CD14, Toll-like receptor 9 (TLR9), CC chemokines (CCL4, CCL20, CCL22, CCL25, CCL27) and their receptors (CCR4, CCR7), IL-8 and IL-17 [39]. This pattern indicates a rapid mobilization and increased trafficking of the monocyte-macrophage lineage into the lung very early on in the infection. In another study, the SARS-CoV was shown to induce chemokine release in lung epithelial cells and fibroblasts via amino acids 324–688 of the S protein [40]. In addition to S protein, the structural N protein was found to activate the IL-8 promoter in lung cells as well.
A model of SARS-CoV infection in C57BL/6 mice showed increased production of proinflammatory chemokines and their receptors in the lung but, surprisingly, Th1 cytokines (IL12 p70 and interferon-γ) were not detectable and there was little pulmonary infiltration [41]. Moreover beige, CD1−/−, or RAG1−/− mice cleared SARS-CoV normally. These observations suggest proinflammatory chemokines may co-ordinate a highly effective innate antiviral response in the lung, and NK cells and adaptive cellular immunity are not required for viral clearance in this species [41]. On the other hand, Stat-1 appears to contribute to host defence in SARS-infected mice [42]. Deficiency of Stat-1 leads to resistance to the antiviral effects of interferons and this may be relevant to the above observation.
Cytokines and chemokines measurements in the plasma from SARS patients are difficult to interpret partly because of the many confounding factors during the disease course [43]. Nevertheless both chemokines (IL-8, MCP-1 and IP-10) and inflammatory cytokines (IL-1, IL-6 and IL-12) were found to be elevated in patients. Interestingly, prolonged disturbances of in vitro cytokine production in SARS patients have also been demonstrated [44].
The adaptive immune response to SARS-CoV
Lymphopenia with a rapid decrease in both CD4 and CD8 T cells is common during the acute phase of SARS and may be associated with an adverse outcome [45, 46]. In recovering patients, rapid restoration of T cell subsets was seen [46]. The mechanism underlying the acute lymphopenia remains unclear, and may be related to induced apoptosis of uninfected lymphocytes as in measles, since the consistent absence of ACE2 in T and B lymphocytes and macrophages in all haemato-lymphoid organs suggests that direct viral infection may not be the cause [33]. Overexpression of 7a, a protein specifically encoded by SARS-CoV, can induce apoptosis via a caspase-dependent pathway and in cell lines derived from different organs, including lung, liver and kidney. Expression of 7a may be one of the underlying mechanisms for the pathogenesis of SARS-CoV infection [47].
Detection of serum IgG, IgM and IgA against SARS-CoV using immunofluorescent assays and by ELISA against nucleocapsid antigen occurs around the same time with most patients seroconverted by day 14 after onset of illness [48]. IgG can be detected as early as 4 days after the onset of illness. The kinetics of neutralization antibodies nearly parallel those for IgG [48] and most of the neutralizing-antibody activity is attributed to IgG [49]. In a study of 623 SARS patients, the neutralizing-antibody levels peaked at 20–30 days and were sustained for over 150 days. These antibodies can neutralize the pseudotype particles bearing the S protein from different SARS-CoV strains, suggesting that these antibodies are broadly active and that the S protein is highly immunogenic [49]. Indeed the S protein, among the other structural proteins, such as M, E or N, is the only significant SARS-CoV neutralization antigen and protective antigen [50], with amino acids 441–700 as the major immunodominant epitope [51].
An anti-S1 human monoclonal antibody 80R has been shown to have nanomolar affinity, to potently neutralize SARS-CoV infection and to inhibit syncytia formation through blocking of receptor binding [52]. The 80R epitope is located within the N-terminal 261–672 amino acids of S protein and is not glycosylation-dependent. Immortalization of memory B cells from a SARS patient who recovered also enabled the isolation of human monoclonal antibodies with potent neutralization of SARS-CoV [53]. These antibodies have two patterns of staining the S protein and neutralizing activity: some antibodies have high-avidity binding to spike transfectants and a neutralizing titer proportional to the degree of binding, whereas others have low-avidity binding in spite of efficient viral neutralization. The B cell memory repertoire of patients can be analyzed by this approach.
In a study of the functional effects of S-protein mutations in terms of their affinity for cellular receptor ACE2 and their sensitivity to antibody neutralization with viral pseudotypes, substantial functional changes were found in S-proteins derived from a case in late 2003 from Guangdong [S(GD03T0013)] [54]. S(GD03T0013) depends much less on ACE2 as receptor for cell entry and is markedly resistant to antibody neutralization in comparison to eight strains transmitted during human outbreaks in early 2003. Moreover, antibodies that neutralize most human S proteins can enhance entry mediated by the palm civet virus S proteins [54]. This underscores the need to address the evolving diversity of SARS-CoV for vaccine development; progress in experimental vaccine candidates for SARS has been previously reviewed [1]. Insight into the mechanisms of antibody-dependent enhancement will help to avoid complications during vaccine development. The inability to maintain, using in vitro culture, the animal SARS-CoV precursor virus from palm civets makes it difficult to confirm whether these findings with pseudotyped virus are relevant to infectious SARS-CoV-like virus isolates. An alternative approach will be to address this question using techniques of SARS-CoV reverse genetics, which are now becoming available.
The cytotoxic T-cell response is the other major specific defence against viral infection. Two HLA-A2-restricted T-cell epitopes capable of eliciting a CD8 T-cell response in patients who had recovered from SARS-CoV infection have been identified [55].
Host genetic susceptibility to SARS
Since SARS is an entirely new disease, it offers a unique opportunity to study the genetics of innate immunity in SARS at population level. As mentioned above, individuals carrying the low-MBL-producing haplotype YB have an odds ratio (OR) of 1.5 in developing SARS [37•]. Association of HLA-B*4601 with SARS-CoV infection has been reported in a study of 37 SARS patients in Taiwan [56], but not confirmed in another study of 90 patients in Hong Kong [57], which instead found significant association of HLA-B*0703 (OR 4.08) and HLA-DR B1*0301 (OR 0.06) with the development of SARS. ACE2 gene polymorphisms were not associated with SARS susceptibility or outcome in a study of 168 patients and 328 healthy controls [58].
Conclusions
Although much has been learnt of the SARS in the two years since its discovery, aspects of the pathogenesis of the disease are still not fully understood. This is because there are no further human cases of SARS and because there is no animal model that accurately reflects the human disease (reviewed in [1]). Although the kinetics and protective role of the host antibody responses are better defined, the roles of the adaptive cell-mediated and the innate immune responses to SARS are still being unravelled. Empirical approaches to vaccine development are progressing rapidly but it remains important to better elucidate the mechanisms of disease pathogenesis so as to minimize the risk of being unpleasantly surprised by unintended consequences of vaccination.
Update
The lack of a β-interferon response in cells infected with SARS CoV has now been documented in two other cell culture models in vitro; human primary myeloid-derived DCs [59•] and the epithelial cell line 293 [60•]. SARS CoV induced the nuclear translocation of interferon regulatory factor (IRF)-3 in 293 cells but it was proposed that the lack of an interferon response is due to a block in subsequent dimerization and hyper-phosphorylation of IRF-3.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We are grateful for the support from the Edward Sai-Kim Hotung Paediatric Education and Research Fund, the SARS Research Fund of The University of Hong Kong and to research funding from the National Institute of Allergy and Infectious Diseases, USA (public health research grant AI95357).
References
- 1.Peiris J.S.M., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat Med (Suppl) 2004;10:S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2••.Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W., Nicholls J., Yee W.K.S., Yan W.W., Cheung M.T. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first report of the isolation and characterization of the etiological agent of SARS. Demonstrated the consistent detection of the virus and immune response to it in a series of patients with the disease and the absence of both virus and antibody in controls.
- 3.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Corner J.A., Lim W. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 4.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 5.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 6.Marra M.A., Jones S.J.M., Astell C.R., Holt R.A., Wilson A.B., Butterfield Y.S.N., Khattra J., Asano J.K., Barber S.A., Chan S.Y. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 7.Foucher R.A.M., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J.J., van den Hoogen B.G., Peiris M., Lim W., Stohr K., Osterhaus A.D.M.E. Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuiken T., Fouchier R.A.M., Schutten M., Rimmeizwaan G.F., van Amerongen G., van Riel D., Laman J.D., de Jong T., van Doornum G., Lim W. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peiris J.S.M., Yuen K.Y., Osterhaus A.D.M.E., Stohr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 10.Poon L.L.M., Guan Y., Nicholls J.M., Yuen K.Y., Peiris J.S.M. The aetiology, origins, and diagnosis of severe acute respiratory syndrome. Lancet Infect Dis. 2004;4:663–671. doi: 10.1016/S1473-3099(04)01172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnelly C.A., Fisher M.C., Fraser C., Ghani A.C., Riley S., Ferguson N.M., Anderson R.M. Epidemiological and genetic analysis of severe acute respiratory syndrome. Lancet Infect Dis. 2004;4:672–683. doi: 10.1016/S1473-3099(04)01173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau Y.L. SARS: future research and vaccine. Paediatr Respir Rev. 2004;5:300–303. doi: 10.1016/j.prrv.2004.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinstein R.A. Planning for epidemics — the lessons of SARS. N Engl J Med. 2004;350:2332–2334. doi: 10.1056/NEJMp048082. [DOI] [PubMed] [Google Scholar]
- 14••.Guan Y., Zhen B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J. Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]; First demonstration of the animal precursor virus of SARS-CoV (severe acute respiratory syndrome coronavirus) in civet cats and other small mammals in a live game animal market. Many workers associated with these markets had antibody to the virus.
- 15.Guan Y., Peiris J.S.M., Zheng B., Poon L.L.M., Chan K.H., Zeng F.Y., Chan C.W.M., Chan M.N., Chen J.D., Chow K.Y.C. Molecular epidemiology of the novel coronavirus that causes severe acute respiratory syndrome. Lancet. 2004;363:99–104. doi: 10.1016/S0140-6736(03)15259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Chinese SARS Molecular Epidemiology Consortium: Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 2004, 303:1666-1669. [DOI] [PubMed]
- 17.Webster R.G. Wet markets — a continuing source of severe acute respiratory syndrome and influenza? Lancet. 2004;363:234–236. doi: 10.1016/S0140-6736(03)15329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicholls J.M., Poon L.L.M., Lee K.C., Ng W.F., Lai S.T., Leung C.Y., Chu C.M., Hui P.K., Mak K.L., Lim W. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F.N., Poon L.L.M., Law K.I., Tang B.S.F., Hon T.Y.W., Chan C.S. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:767–772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leung W.K., To K.F., Chan P.K.S., Chan H.L.Y., Wu A.K.L., Lee N., Yuen K.Y., Sung J.J.Y. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125:1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng V.C.C., Hung I.F.N., Tang B.S.F., Chu C.M., Wong M.M.L., Chan K.H., Wu A.K.L., Tse D.M.W., Chan K.S., Zheng B.J. Viral replication in the naspharynx is associated with diarrhea in patients with severe acute respiratory syndrome. Clin Infect Dis. 2004;38:467–475. doi: 10.1086/382681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding Y.Q., He L., Zhang Q.L., Huang Z.X., Che X.Y., Hou J.L., Wang H.J., Shen H., Qiu L.W., Li Z.G. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J Pathol. 2004;203:622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farcas G.A., Poutanen S.M., Mazzulli T., Willey B.M., Butany J., Asa S.L., Faure P., Akhavan P., Low D.E., Kain K.C. Fatal severe acute respiratory syndrome is associated with multiorgan involvement by coronavirus. J Infect Dis. 2005;191:193–197. doi: 10.1086/426870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc Natl Acad Sci USA. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Li W.H., Moore M.J., Vasilieva N., Sui J.H., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first report of a functional receptor for the SARS-CoV, angiotensin-converting enzyme 2 (ACE2), which efficiently binds the S1 domain of the SARS-CoV spike protein. SARS-CoV can replicate efficiently on ACE2-transfected but not mock-transfected 293T cells.
- 26.Wang P.G., Chen J., Zheng A.H., Nie Y.C., Shi X.L., Wang W., Wang G.W., Luo M., Liu H.J., Tan L. Expression cloning of functional receptor used by SARS coronavirus. Biochem Biophys Res Commun. 2004;315:439–444. doi: 10.1016/j.bbrc.2004.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Babcock G.J., Esshaki D.J., Thomas W.D., Ambrosino D.M. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J Virol. 2004;78:4552–4560. doi: 10.1128/JVI.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu S., Xiao G.F., Chen Y.B., He Y.X., Niu J.K., Escalante C.R., Xiong H.B., Farmar J., Debnath A.K., Tien P. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosch B.J., Martina B.E.E., van der Zee R., Lepault J., Haijema B.J., Versiuis C., Heck AJR, de Groot R., Osterhaus A.D.M.E., Rottier J.M. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc Natl Acad Sci USA. 2004;101:8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Supekar V.M., Bruckmann C., Ingallinella P., Bianchi E., Pessi A., Carfi A. Structure of a proteolytically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein. Proc Natl Acad Sci USA. 2004;101:17958–17963. doi: 10.1073/pnas.0406128102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z.Y., Huang Y., Ganesh L., Leung K.Y., Kong W.P., Schwartz O., Subbarao K., Nabel G.J. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol. 2004;78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J., Thomas W.D., Thackray L.B., Young M.D., Mason R.J. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first demonstration that a C-type lectin, CD290L (also called L-SIGN), is a binding receptor for SARS-CoV). CD290L is expressed in human lung in type II alveolar cells and endothelial cells. Its role as a functional receptor for productive virus replication is unclear.
- 33.Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leung G.M., Hedley A.J., Ho L.M., Chau P., Wong O.L., Thach T.Q., Ghani A.C., Donnelly C.A., Fraser C., Riley S. The epidemiology of severe acute respiratory syndrome in the 2003 Hong Kong epidemic: an analysis of all 1755 patients. Ann Intern Med. 2004;141:662–673. doi: 10.7326/0003-4819-141-9-200411020-00006. [DOI] [PubMed] [Google Scholar]
- 35.Cheung CY, Poon LLM, Ng IHY, Luk WM, Sia SF, Wu MHS, Chan KH, Yuen KY, Gordon S, Guan Y et al.: Cytokine responses to SARS coronavirus infected macrophages in vitro: Possible relevance to pathogenesis. J Virol 2005, in press. [DOI] [PMC free article] [PubMed]
- 36.Reghunathan R., Jayapal M., Hsu L.Y., Chng H.H., Tai D., Leung B.P., Melendez A.J. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 2005;6:2–12. doi: 10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Ip W.K., Chan K.H., Law H.K.W., Tso G.H.W., Kong E.K.P., Wong W.H.S., To Y.F., Yung R.W.H., Chow E.Y., Au K.L. Mannose-binding lectin in SARS coronavirus infection. J Infect Dis. 2005;191:1697–1704. doi: 10.1086/429631. [DOI] [PMC free article] [PubMed] [Google Scholar]; An elegant demonstration of the protective role of an innate immune molecule (MBL) in a novel infection (SARS), for which the human population had no previous memory. MBL can bind SARS-CoV in a calcium-dependent and mannan-inhibitable fashion, resulting in complement activation and inhibition of SARS-CoV infection of FRhK-4 cells. The low-MBL-producing haplotype is associated with SARS.
- 38•.Chen J.H., Chang Y.W., Yao C.W., Chiueh T.S., Huang S.C., Chien K.Y., Chen A., Chang F.Y., Wong C.H., Chen Y.J. Plasma proteome of severe acute respiratory syndrome analyzed by two-dimensional gel electrophoresis and mass spectrometry. Proc Natl Acad Sci USA. 2004;101:17039–17044. doi: 10.1073/pnas.0407992101. [DOI] [PMC free article] [PubMed] [Google Scholar]; A complete proteomic analysis was performed on four SARS patients. The authors demonstrated that the active innate immune responses, including acute-phase proteins and C-type lectins, such as MBL, along with oxidation-associated injuries, may play a role in the pathogenesis of SARS.
- 39.Ng L.F.P., Hibberd M.L., Ooi E.E., Tang K.F., Neo S.Y., Tan J., Murthy K.R.K., Vega V.B., Chia J.M., Liu E.T. A human in vitro model system for investigating genome-wide host responses to SARS coronavirus infection. BMC Infect Dis. 2004;4:34. doi: 10.1186/1471-2334-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang Y.J., Liu C.Y.Y., Chiang B.L., Chao Y.C., Chen C.C. Induction of IL-8 release in lung cells via actiator protein-1 by recombinant baculovirus displaying severe acute respiratory syndrome-coronavirus spike proteins: identification of two functional regions. J Immunol. 2004;173:7602–7614. doi: 10.4049/jimmunol.173.12.7602. [DOI] [PubMed] [Google Scholar]
- 41.Glass W.G., Subbarao K., Murphy B., Murphy P.M. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J Immunol. 2004;173:4030–4039. doi: 10.4049/jimmunol.173.6.4030. [DOI] [PubMed] [Google Scholar]
- 42.Hogan R.J., Gao G., Rowe T., Bell P., Flieder D., Paragas J., Kobinger G.P., Wivel N.A., Crystal R.G., Boyer J. Resolution of primary severe acute respiratory syndrome-associated coronavirus infections requires Stat-1. J Virol. 2004;78:11416–11421. doi: 10.1128/JVI.78.20.11416-11421.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong C.K., Lam C.W.K., Wu A.K.L., Ip W.K., Lee N.L.S., Chan H.I.S., Lit L.C.W., HuI D.S.C., Chan M.H.M., Chung S.S.C. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones B.M., Ma E.S.K., Peiris J.S.M., Wong P.C., Ho J.C.M., Lam B., Lai K.N., Tsang K.W.T. Prolonged disturbances of in vitro cytokine production in patients with severe acute respiratory syndrome (SARS) treated with ribavirin and steroids. Clin Exp Immunol. 2004;135:467–473. doi: 10.1111/j.1365-2249.2003.02391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong R.S.M., Wu A., To K.F., Lee N., Lam C.W.K., Wong C.K., Chan P.K.S., Ng M.H.L., Yu L.M., Hui D.S. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ. 2003;326:1358–1362. doi: 10.1136/bmj.326.7403.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li T.S., Qiu Z.F., Zhang L.Q., Han Y., He W., Liu Z.Y., Ma X.J., Fan H.W., Lu W., Xie J. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J Infect Dis. 2004;189:648–651. doi: 10.1086/381535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan Y.J., Fielding B.C., Goh P.Y., Shen S., Tan T.H.P., Lim S.G., Hong W. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J Virol. 2004;78:14043–14047. doi: 10.1128/JVI.78.24.14043-14047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsueh P.R., Huang L.M., Chen P.J., Kao C.L., Yang P.C. Chronological evolution of IgM, IgA, IgG and neutralization antibodies after infection with SARS-associated coronavirus. Clin Microbiol Infect. 2004;10:1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nie Y.C., Wang G.W., Shi X.L., Zhang H., Qiu Y., He Z.P., Wang W., Lian G.W., Yin X.L., Du L.Y. Neutralizing antibodies in patients with severe acute respiratory syndrome-associated coronavirus infection. J Infect Dis. 2004;190:1119–1126. doi: 10.1086/423286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buchholz U.J., Bukreyev A., Yang L., Lamirande E.W., Murphy B.R., Subbarao K., Collins P.L. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc Natl Acad Sci USA. 2004;101:9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu L., Manopo I., Leung B.P., Chng H.H., Ling A.E., Chee L.L., Ooi E.E., Chan S., Kwang J. Immunological characterization of the spike protein of the severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004;42:1570–1576. doi: 10.1128/JCM.42.4.1570-1576.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sui J.H., Li W.H., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., St Clair Tallarico A., Olurinde M., Choe H. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci USA. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Traggiai E., Becker S., Subbarao K., Kolesnikova L., Uematsu Y., Gismondo M.R., Murphy B.R., Rappuoli R., Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Z., Werner H.C., Kong W., Leung K., Traggiai E., Lanzavecchia A., Nabel G.J. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc Natl Acad Sci USA. 2005;102:797–801. doi: 10.1073/pnas.0409065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y., Sin W., Xu G., Yang H., Wong T., Pang X., He X., Zhang H., Ng J.N.L., Cheng C. T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus spike protein elicit a specific T-cell immune response in patients who recover from SARS. J Virol. 2004;78:5612–5618. doi: 10.1128/JVI.78.11.5612-5618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin M., Tseng H., Trejaut J.A., Lee H., Loo J., Chu C., Chen P., Su Y., Lim K.H., Tsai Z. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med Genetics. 2003;4:9. doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ng M.H.L., Lau K., Li L., Cheng S., Chan W.Y., Hui P.K., Zee B., Leung C., Sung J.J.Y. Association of human-leukocyte-antigen class I (B*0703) and class II (DRB1*0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J Infect Dis. 2004;190:515–518. doi: 10.1086/421523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chiu R.W.K., Tang N.L.S., Hui D.S.C., Chung G.T.Y., Chim S.S.C., Chan K.C.A., Sung Y., Chan L.Y.S., Tong Y., Lee W. ACE2 gene polymorphisms do not affect outcome of severe acute respiratory syndrome. Clin Chem. 2004;50:1683–1686. doi: 10.1373/clinchem.2004.035436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59•.Law HK, Cheung CY, Ng HY, Sia SF, Chan YO, Luk W, Nicholls JM, Peiris JS, Lau YL: Chemokine upregulation in SARS coronavirus infected human monocyte derived dendritic cells. Blood 2005, in press. (DOI: 10.1182/blood-2004-10-4166). [DOI] [PMC free article] [PubMed]; This is the first demonstration of the effect of SARS-CoV on human monocyte-derived DCs, and the authors propose a mechanism of immune evasion by SARS-CoV. In SARS-CoV-infected DCs, there is low expression of antiviral cytokines (IFN-α, IFN-β, IFN-γ and IL-12p40), moderate upregulation of proinflammatory cytokines (TNF-α and IL-6), and significant upregulation of inflammatory chemokines (MIP-1α, RANTES, IP-10 and MCP-1).
- 60•.Spiegel M., Pichlmair A., Martinez-Sobrido L., Cors J., Garcia-Sastre A., Haller O., Weber F. Inhibition of beta interferon induction by Severe Acute Respiratory Syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. Journal of Virology. 2005;79:2079–2086. doi: 10.1128/JVI.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first demonstration that SARS-CoV escapes interferon-mediated growth inhibition by preventing the induction of interferon-β through interfering with the activation of interferon regulatory factor 3 (IRF-3).


