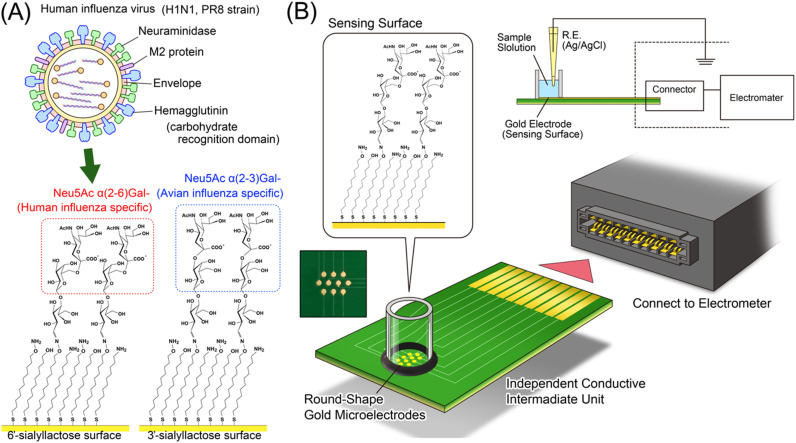
Abstract
A system to discriminate human or avian influenza A remains a highly sought-after tool for prevention of influenza pandemics in humans. Selective binding of the influenza A viral hemagglutinin (HA) to specific sialic acid (SA) receptors (Neu5Acα(2-6)Gal in humans, Neu5Acα(2-3)Gal in birds) is determined by the genotype of the HA and neuraminidase (NA) segments, making it one of the key characteristics that distinguishes human or avian influenza A virus. Here we demonstrate the direct detection of whole H1N1 influenza A virus using 6′-sialyllactose (Neu5Acα(2-6)Galβ(1-4)Glc, 6SL)-immobilized gold electrodes as biosensing surfaces. The sensitivity was higher than that of conventional immunochromatographic technique (ICT) for influenza virus and not restricted by genetic drift. The label-free detection technology via direct attachment of a whole virus using a chemically modified electrode is a promising means to provide a simple and rapid diagnostic system for viral infections.
Keywords: Biosensor, Biointerface, Influrenza virus, Molecular recognition, Label-free
1. Introduction
Protection against infectious diseases such as influenza virus is a sought-after goal, which may be more attainable if also addressed from a viewpoint of diagnostic performance. For example, highly pathogenic avian influenza (HPAI) infections in humans, such as the H5N1 subtype, have been identified as emerging infectious diseases with a high mortality rate compared with human influenza viruses (Peiris et al., 2007, Subbarao et al., 1998, Yuen et al., 1998). There are two main infection mechanisms. One mechanism is the expression of avian influenza receptors in human cells such as type II pneumocytes, which promotes infection with the predominantly avian-restricted H5N1 subtype (Kogure et al., 2006, Yao et al., 2008). The other is mutation of HA, which results in the ability to bind to human SA (Neu5Acα(2-6)Gal). It has been reported that mutations in several key amino acids in the antigenic epitope of HA create an interphyletic type of HA that can bind both avian SA (Neu5Acα(2-3)Gal) and human SA moieties (Sawada et al., 2010, Yamada et al., 2006). This genetic drift can lead to species jump and human-to-human transmission in a previously avian-restricted virus, and may ultimately cause a serious influenza pandemic. For example, despite preferentially binding to avian SA, such rapid mutation of the HA gene has resulted in subtypes H7N7 and H9N2 being capable of binding to the human form, albeit frequently with reduced efficacy, and thus represent emerging pandemic threats. Therefore, the ability to rapidly detect the host restriction of emerging viruses with high sensitivity may enhance surveillance and indicate imminent species jump events.
To identify the subtype of a given influenza virus, biosensing systems bearing specific antibodies (Rowe et al., 1999, Yuen et al., 1998) and nucleic acid aptamers (Gopinath and Kumar, 2013, Jeon et al., 2004, Wang et al., 2013) for HA, which plays a role in the process of virus attachment to the cell membrane during infection (Hamilton et al., 2012, Skehel and Wiley, 2000, Wiley and Skehel, 1987), have been reported. However, antigenic drift poses a problem to the recognition of HA (Carrat and Flahault, 2007), which may cause serious false negative results because of the escape from specific binding. On the other hand, human SA and avian SA are essential for infection with human and avian influenza viruses, respectively, and therefore represent suitable moieties to identify the subtype of virus (Watanabe et al., 2015). However, the detection sensitivity of conventional rapid assays such as ICT is over 5 hemagglutinating units (HAU) (Watanabe et al., 2015, Yeo et al., 2014), which occasionally leads to false negative results in the early stage of infection. If the sensitivity of diagnostic techniques could be improved using SA receptors, it may help prevent the occurrence of a serious influenza pandemic.
To realize this detection system, we demonstrate the label-free detection of influenza A virus subtype H1N1 using 6SL immobilized on a self-assembled monolayer (SAM) on gold electrodes ( Fig. 1A). To form the densely immobilized sialyllactose layer on the sensing surface, aminooxyundecyl disulfide (AOUD) was first immobilized as the SAM layer. Aminooxy groups on the SAM surface can react with hemiacetal (aldehyde) moieties of sialyllactose to form stable oxime bonds (Wu et al., 2010).
Fig. 1.
(A) Conceptual rendering of the virus detection using saccharide recognition. (B) Illustration of the influenza virus detection system using electrical detection. The sensing surfaces are isolated from the measurement unit by a conductive intermediate, which can be used as a disposable sensor chip. The FET sensor is mounted in the electrometer with an operational amplifier, differential amplifier, amplifier circuit and capacitor for phase compensation.
Recently, label-free immunosensing systems such as surface plasmon resonance sensors, quartz crystal microbalance (QCM) and shear horizontal surface acoustic wave sensors (Horiguchi et al., 2013) have been widely used as biosensors, which can detect specific interactions via direct attachment to the target without any pretreatment of the sample. In addition, biosensing systems using electrical detection, such as field effect transistors (FETs) have also attracted attention as label-free biosensors (Goda and Miyahara, 2010, Goda and Miyahara, 2013, Hideshima et al., 2013). In this report, the specific binding between human influenza A virus subtype H1N1 and 6SL-immobilized electrodes was first confirmed with QCM and then further confirmed using electrical detection techniques (Fig. 1B). Advantageously, the electrical detection techniques do not require optical devices or any other optional input transducers for biosensing. Therefore, the system can be miniaturized and the procedure simplified. In this report, a systematically fabricated SA receptor layer on a gold electrode for direct and label-free rapid detection of H1N1 influenza A virus was evaluated by QCM and electrical detection.
2. Materials and methods
2.1. Materials, reagents and equipment
11-Bromoundecane-1-thiol was purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). Sodium hydrogen carbonate, magnesium sulfate, gelatin, ethyl acetate (EA), n-hexance, N,N-dimethylformamide (DMF) and dichloromethane (DCM) were purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan). N-Hydroxyphthalimide, 6′-sialyllactose sodium salt and 3′-sialyllactose sodium salt were purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). All chemicals and reagents were used as received. Influenza A H1N1 (A/California/04/2009) hemagglutinin and influenza A virus subtype H5N1 (A/Vietnam/1194/2004) hemagglutinin were purchased from Sino Biological Inc. (Beijing, China). Though influenza A/Vietnam/1194/2004 was isolated from human, the specific interaction between HA and avian SA have been confirmed at previous report (Xiong et al., 2013). Human influenza A virus subtype H1N1 (A/PR/8/34) was cultivated in chicken embryos and then detoxified using 0.05% formalin solution. The size distribution of H1N1 influenza A virus particles in solution was measured using a qNano nanoparticle analyzer from Izon Science Ltd., Christchurch, New Zealand. Zeta potential measurement was performed using a Zetasizer Nano ZS from Malvern Instruments Ltd., (Worcestershire, UK).
2.2. Synthesis of 11-hydroxyundecyl 11-aminooxyundecyl disulfide, 11-aminoundecyl 11-aminooxyundecyl disulfide (aminooxyundecyl disulfide, AOUD)
Functional group transformation from a bromo moiety to oxylamine was reported previously (Park and Yousaf, 2008). Two hundred and fifty milligrams of 11-Bromoundecane-1-thiol (9.33×10−4 M) were dissolved in 5 mL of DMF. For the thiol oxidation reaction, the solution was refluxed for 24 h at 100 °C (solution 1). Separately, 396 mg of N-hydroxyphthalimide (2.43 mM) and 198 mg of sodium hydrogen carbonate (2.36 mM) were dissolved in 5 mL of DMF. The solution was heated to 80 °C (solution 2). After the mixture turned dark brown (15 min), solutions 1 and 2 were mixed and then refluxed for 17 h at 100 °C. After the reaction, the obtained product was extracted by water and EA. The solution was mixed with 20 mL water and then with 20 mL EA in a separating funnel. After collection of the EA product, another 20 mL of EA was added for further extraction. This extraction procedure was repeated four times then the water fraction, including unreacted hydroxyphthalimide, was discarded. The EA fraction was dehydrated with magnesium sulfate, concentrated using a rotary evaporator, and then purified by column chromatography (1:3 ethyl acetate/n-hexane). After evaporation and drying under reduced pressure, a lipid-like white solid product was obtained.
One hundred and forty two point five milligrams of the solid product was dissolved in 5 mL of DCM. Two milliliters of 1 M hydrazine in tetrahydrofuran was added then stirred for 5 h at room temperature. After the reaction, the precipitated phthalic hydrazide was filtered and then concentrated for purification by column chromatography (1:3 ethyl acetate/n-hexane). After purification, evaporation and drying under reduced pressure, a lipid-like white solid product was obtained. The product (AOUD) was analyzed by 1H NMR in CDCl3 to confirm the structure of product. The detailed analytical result was shown in Supporting Information (Fig. S1).
2.3. Formation procedure of the self-assembled monolayer (SAM) and evaluation of packing density
Gold electrodes were washed with acetone, 1 M sodium hydroxide and 1 M hydrochloric acid before immobilization. The electrodes were immersed in 1 mM of AOUD in ethanol for 24 h. After the formation of the SAM, the electrodes were washed with ethanol. The evaluation procedure for packing density of the SAM was reported previously (Poirier et al., 1994). Cyclic voltammetry (CV) was carried out using an Autolab PGSTAT302 Potentiostat (Metrohm, Herisau, Switzerland). To measure the oxidation reaction on the SAM, an AOUD-modified gold electrode was used as a working electrode for CV. A platinum wire (φ=1 mm) and a silver/silver chloride wire in 3.3 M KCl solution with a salt bridge were used as the counter and reference electrode, respectively. A 0.5 M potassium hydroxide solution was degassed for CV previously. Cyclic voltammetry scans were performed from −0.1 to −1.3 V with a scan rate of 0.5 V/s.
2.4. Immobilization of 6SL or 3SL to the SAM
The condensation reaction between aminooxyl groups and glucose was performed as previously described (Hideshima et al., 2013). Gold electrodes with SAM were immersed in 1 mM of 6SL or 3SL solution (pH 5.3 acetic acid) and heated at 60 °C for 240 min. After the reaction, the gold electrodes were washed with water.
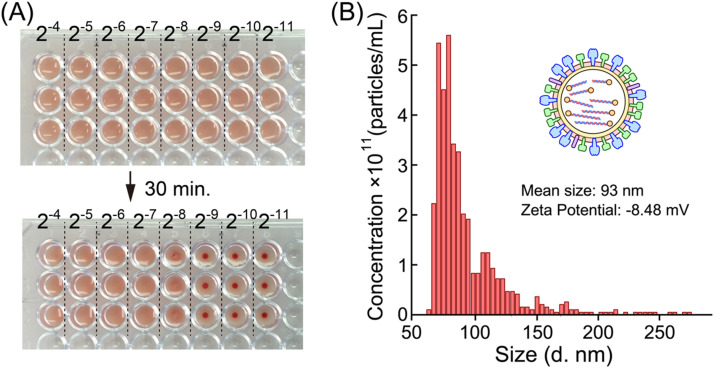
2.5. Determination of virus titer of detoxified influenza A virus subtype H1N1
To quantify the virus infectious ability, HA assay was performed to measure the HA titer of the detoxified H1N1 influenza virus as hemagglutinating units (HAU). A 16- to 2048-fold dilution series of virus solution was prepared using 0.01% gelatin DPBS solution. Then 50 μL of each solution was added to a 96-well round-bottom plate. Fifty microliters of 0.5% chicken red blood cell suspension was mixed with each virus dilution. After 30 min, blood sedimentation was confirmed. According to this result, the virus titer of original virus solution was determined as 256 HAU ( Fig. 2A). The sized distribution and zeta potential of virus were measured by qNano and Zetasizer Nano ZS (Fig. 2B).
Fig. 2.
Characterization of influenza A virus subtype H1N1. (A) Determination of HA titer by hemagglutination assay. A 16- to 2048-fold dilution series (2−4 to 2−11) of virus solution was prepared using 0.01% gelatin DPBS solution. Then, 50 μL of each dilution was added to a 96-well round-bottom plate. Fifty microliters of 0.5% chicken red blood cell suspension was mixed with each virus solution (n=3). Top panel: 96-well plate just after mixing. Lower panel: 96-well plate after 30 min. The blood cells were precipitated from 1/512 dilution of virus solution, giving an HA titer of the original virus of 256. (B) The size distribution, mean size of particle diameter and zeta potential of virus (32 HAU).
2.6. The virus adsorption measurement using Quartz crystal microbalance
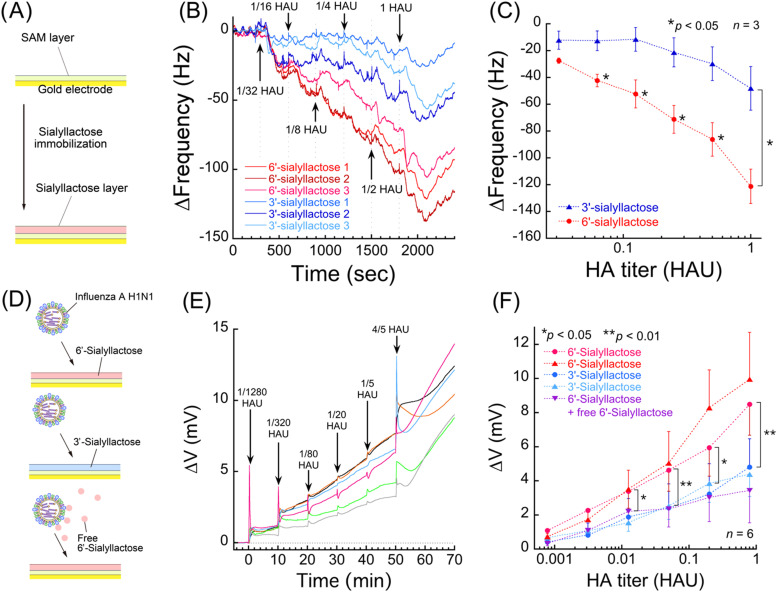
Quartz crystal microbalance (QCM) measurement was carried out using NAPiCOS® (Nihon Dempa Kogyo Co., Ltd., Tokyo, Japan). 6SL or 3SL immobilized gold electrodes on quartz crystal sensor chips had been prepared previously. Dulbecco's phosphate-buffered saline (×0.01) solution was used as a running buffer with flow rate of 0.3 mL min–1. After the frequency response had been stabilized, 200 μL of 1/32 HAU virus solution was injected. Then, 1/16, 1/8, 1/4, 1/2 and 1 HAU virus solutions were injected at 5-min intervals.
2.7. The device for electrical detection
The measurement procedure was reported previously (Goda and Miyahara, 2010). Schematic illustration of sensing system is shown in Fig. 1B. To perform the potentiometry, a sensor chip possessing 10-channel round-shaped gold microelectrodes (500 µm in diameter; from Towa Tech, Shizuoka, Japan) was used as a working electrode. A glass tube (5 mm in inner diameter) was fixed around the electrodes using a thermosetting resin (ZC-203; Pelnox Limited, Kanagawa, Japan). An Ag/AgCl electrode was used as a reference.
2.8. Confirmation by electrical detection
The procedure of SA receptor immobilization has been described in the Section 2.4. The 6SL or 3SL immobilized gold electrodes were immersed in 200 μL of ×0.01 DPBS for 6 h to stabilize the potentiometric response. The running buffer was exchanged for fresh buffer several times until steady-state potential with constant drift was obtained. To minimize the potentiometric noise by pipetting, the buffer solution was very carefully exchanged for virus solution. The final concentrations of 1/1280, 1/320, 1/80, 1/20, 1/5 and 4/5 HAU virus solution were applied to 6SL or 3SL immobilized gold electrodes.
3. Results and discussion
3.1. Label-free influenza A virus subtype H1N1 detection using QCM measurement
The virus detection was performed on densely immobilized sialiyllactose surface based on SAM layer. The characterization of SAM formation and sialyllactose immobilization was confirmed before the virus detection experiments. According to the CV measurement result, the SAM density was confirmed as 4.7 molecules nm–2. The detailed analysis was shown in Supporting Information (Fig. S2). The density of immobilized sialyllactose was 1.46±0.33 molecules/nm2, as determined by QCM when the reaction time was 240 min (Supporting Information, Fig. S3). All of the sensor chips were prepared at same condition.
Quartz crystal microbalance measurements were carried out to confirm the specific binding of influenza virus to the chemically modified sialyllactose surface ( Fig. 3A). Influenza A virus subtype H1N1 binding assays were compared between the 6SL and 3′-sialyllactose (Neu5Acα(2-3)Galβ(1-4)Glc, 3SL) surfaces. It was revealed that the frequency change was higher on the 6SL surfaces than the 3SL surfaces (Fig. 3B). To determine the specificity of the 6SL surfaces, the average frequency changes 5 min after the injection of virus solution were plotted against each virus titer (Fig. 3C). This result showed that relative difference (specific response) was observed at 1/16 HAU, which was 80 times higher than that of ICT (5 HAU) (Watanabe et al., 2015, Yeo et al., 2014). In this study, the virus was cultivated in chorioallantoic membrane of chicken embryos, then chorioallantoic fluid including virus was collected. The solution was filtered using 0.45 µm membrane filter but no more purification process was performed, which means that many kinds of impurity such as proteins were entirely remained in the original virus solution. The signals of nonspecific binding shown in Fig. 3B and C may be derived from these impurities. Even though this is not real sample from patients, the virus derived from biological fluid can be detected with this technique. The techniques for suppression of non-specific adsorption such as co-immobilization of zwitterion (Goda and Miyahara, 2013, Horiguchi et al., 2014, Ostuni et al., 2001). have been reported, which would be effective for improving the signal to noise ratio.
Fig. 3.
(A) Schematic illustration siallylactose immobilization. (B) Result of label-free influenza A virus subtype H1N1 detection using QCM measurement. Frequency shifts were detected following the application of influenza A H1N1 solution on 6SL or 3SL immobilized QCM sensor surfaces (experiments were performed in triplicate). Virus solutions of 1/32, 1/16, 1/8, 1/4, 1/2 and 1 HAU were injected at 5-min intervals (position of arrows). (C) The plotted data were converted from Fig. 3B. All plots were sampled after 5 min from injection. (D) Schematic illustration of the influenza A virus subtype H1N1 detection experiment. (E) An example of real-time measurements of transition in potential vs. Ag/AgCl reference electrode as potential changes. An average of six cooperative measurement lines were shown as measurement data. Solutions of 1/1280, 1/320, 1/80, 1/20, 1/5 and 4/5 HAU were applied on 6SL modified gold electrodes at 10-min intervals (position of arrows). The original measurement lines are shown in Supporting Information (Fig. S4). (F) Data plots of relative potential changes 10 min after virus introduction. All plots were converted from the original real-time potential measurements (Fig. 2E). The potential measurement of virus binding on 6SL or 3SL surface were performed in duplicate. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Label-free influenza A virus subtype H1N1 detection using electrical measurements
Based on the results obtained with QCM measurements, electrical measurements using chemically modified electrodes were carried out to investigate the specific electrical interactions between influenza A virus subtype H1N1 and 6SL-immobilized surfaces (Fig. 3D). The real-time electrical measurements of virus detection on 6SL-modified gold electrodes showed that the potential changes (∆V) increased with increasing HA titer of H1N1 influenza A virus (Fig. 3E). These measurements revealed significant relative potential differences between 6SL (red) and 3SL (blue) immobilized surfaces at a concentration of 1/80 HAU (Fig. 3F). As we described in the Section 3.1, the virus solution is almost intact biological fluid. The non-specific signal derived from impurities might disturb the specific signal. In this study, the exchange of a solution was performed carefully to minimize the non-specific signal. However, manual introduction of a solution sometimes induced unusual response of the electrode as shown in Fig. 3E. As is often the case with a gold electrode, we observed a potential drift when we exchanged the solutions. To solve this problem, we approximated the drift behavior with a straight line to take away the effect of drifts (Supporting Information, Fig. S4). As shown in Fig. 3F, virus detection on 6SL and 3SL surfaces were performed twice each, and the two red lines and two blue lines showed the same tendency even though these noise signals disturb the specific signals. It would be possible to improve accuracy of detection by optimizing the baseline correction, since the observed drifts are not exactly but slightly curved. For virus detection under competitive binding inhibition conditions with 1 mM free 6SL solution (purple), the potential changes on 6SL-modified electrodes were decreased to around the same level as 3SL immobilized surfaces, as the free 6SL successfully bound and therefore blocked the binding sites of H1N1 influenza A virus. Therefore, the relative potential difference between 6SL and 3SL can be confidently attributed to specific binding of influenza A virus. The equilibrium dissociation constant (KD) range of lectin-carbohydrate binding is 10−2 to 10−5 M (Lis and Sharon, 1998), which is lower than that of antibody or aptamer. However, multisite binding of lectin-carbohydrate interactions can enhance the interaction strength by multiplier effect. In this experiment, the density of immobilized sialyllactose was greater than 1.46±0.33 molecules/nm2, as determined by QCM (Supporting Information, Fig. S3). Therefore, multisite binding was possible between the sensor surface and influenza virus A subtype H1N1, which is 93±23 nm in diameter on average (Fig. 2B). HA makes up 25–30% of all influenza virus proteins (Aitken and Hannoun, 1980), and this ratio may be sufficient to form multisite bonds on the HA-dense surface of the viral envelope.
Notably, the potential shifts were positive when the virus was bound to human SA moieties on the gold electrodes; this is in contrast to the zeta potential of H1N1 influenza A virus as −8.4 mV (Fig. 2B). The nanostructure on the surface of the viral particle may explain this. HA and other proteins are expressed on the virus surface as spike proteins projecting from the virus membrane. HA forms homotrimeric structures on the viral surface, with a molecular weight of around 80 kDa (Aitken and Hannoun, 1980). These spikes may have resulted in formation of a gap of more than several nanometers between the viral envelope and sensing surface when HA was bound to sialyllactose. As a result, the negatively charged viral membrane is considered to be outside the electrical double layer that suppresses the contribution of the negative charge to the potential changes. However, the human SA moiety on the SAM surface can interact strongly with the HA epitope on the virus surface. It has been reported that two basic amino acids (H183 and R229) and one acidic amino acid (E190) were close to SA when HA was bound to the SA immobilized surfaces (Sawada et al., 2010). It has also been reported that the potentials showed a positive shift when free HA was bound to human SA surfaces (Hideshima et al., 2013). According to these considerations, the positively charged epitope area of HA is considered to contribute to the positive potential response.
3.3. Label-free influenza HA detection using electrical measurements
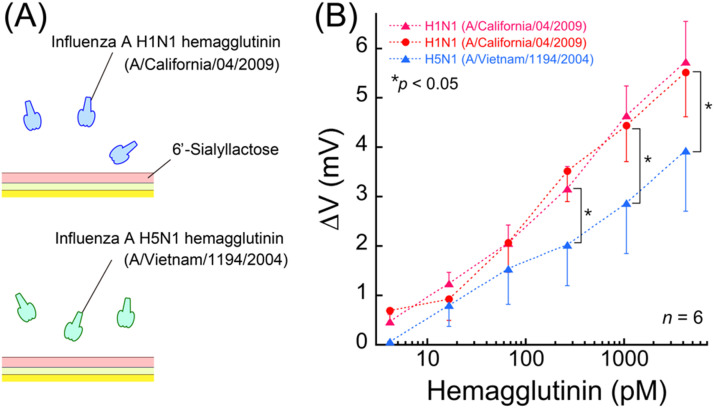
Indicators of virus titer such as HAU, transduction unit, plaque forming units and infectious units are calculated from the viewpoint of an infectivity scale, and as such, these values do not necessarily represent the absolute number of viruses. To understand the sensing performance more clearly on a physical scale, binding assays using commercially available recombinant HAs were carried out. In these experiments, the HA solutions derived from H1N1 (A/California/04/2009) or H5N1 (A/Vietnam/1194/2004) were applied to the 6SL immobilized gold surface ( Fig. 4A) and a significant difference of relative potentials between the two subtypes of HAs was observed from subnanomolar concentrations of the HA solutions (Fig. 4B). However, HA forms homotrimers (Wiley and Skehel, 1987) that can increase the affinity strength by multisite binding. It has been reported that the result of the KD between recombinant HA derived from influenza A virus subtype H5N3 (A/duck/Hong Kong/313/4/1978 (H5N3)) and GD1a (an avian SA containing structure) was 6.10×10−9 M according to QCM measurements (Takahashi et al., 2013a). Based on these factors, the relative difference observed was of the subnanomolar order. In this study, specific recognition was performed using human influenza virus only. However, selective binding between human influenza HA and avian influenza HA was confirmed. According to the previous report (Watanabe et al., 2015), it has already been demonstrated that each sialic acid receptor can distinguish each influenza virus using ICT test. On the basis of these results, we concluded that selective detection of human or avian influenza could be realize using chemically modified electrodes to improve the diagnostic performance.
Fig. 4.
Influenza virus hemagglutinin detection using electrical measurement. (A) Schematic illustration of the influenza HA detection experiment. Hemagglutinin from H1N1 (A/California/04/2009) was used as the specific target while HA from H5N1 (A/Vietnam/1194/2004) was used as a control experiment. (B) The potential changes on the 6′-sialyllactose immobilized surfaces after application of the two kinds of HA solution.
3.4. The evaluation of validity for practical use
In the previous section, the sensing performance of virus binding assay was discussed. To understand the similarities, differences and advantages between the proposed detection system and existing methods, a comparison table was shown in Table 1. The important points for comparison are sensitivity in terms of required virus titer, detection time and cost. In our method, the specific response was observed at 1/16 HAU by QCM measurement (Fig. 3C). In the case of electrical measurement, the specific response was observed at 1/80 HAU (Fig. 3F). The total exposures in the virus solution are (1/32 HAU for 5 min)+(1/16 HAU for 5 min) for QCM measurement, and (1/1280 HAU for 10 min)+(1/320 HAU for 10 min)+(1/80 HAU for 10 min) for electrical measurement. According to these result, 1/16 HAU virus can be detected in 10 min with QCM, and at least 1/80 HAU can be detected in 30 min with the electrical measurement. In the Table 1, these values were defined as sensitivity in terms of required virus titer and detection time for comparison. ICT is widely used because it takes short time to diagnose from the sampling from patients. On the other hand, real-time PCR technique can detect the trace amount of virus precisely. According to the previous report (Takahashi et al., 2013b), detection of virus HA gene with real-time reverse-transcription-PCR showed high sensitivity compared with traditional solid-phase virus binding assay, TLC virus overlay assay and ICT (Table 1). However, gene detection methods need much time (several hours) compared with ICT because extraction of gene from virus and amplification process for the detection are required. If we use the virus detection in medical facilities such as a hospital and a physician's office, a result is desirable to be obtained before a patient leave for home. In the case of our method, the lower concentration of virus solution can be detected with both QCM and electrical measurements compared with existing techniques. However, a cost of QCM sensor chip was 4000 JPY (approximately 35 USD), which was almost four times higher than that of ICT. On the other hand, the cost of electrical detection system will be much lower than that of ICT. As shown in Fig. 1B, the sensor chip can be removable from the device and the price of this unit is 100 JPY (less than 1 USD). The total cost including the modification process of sialyllactose will be less than 2 USD. If the chips were mass-produced in an industrial production line, the price will become lower. Even though the initial cost for the electrical device is required, the cost of the sensor chip is reasonable range for diagnosis. In this study, we took sufficient time for the detection of virus because of the stabilization of electrode potential. However, as described earlier, it is possible to shorten the detection time if the optimum approximation scheme is adopted for the baseline correction.
Table 1.
Advantage and validity of the new detection system compared with existing technique.
| Type of detection system | Sensitivity in terms of virus titer (HAU) | Detection time (min) | Cost (USD) |
|---|---|---|---|
| Label-free influenza virus detection (QCM detection) | 2–4 | 10 | 35 |
| Label-free influenza virus detection (electrical detection) | 2–6 | 30 | 2 |
| Immunochromatographic technique (ICT) | 22–24 | 5–15 | 8–10 |
| Detection of HA gene with PCR | 20 | 240 | – |
| TLC virus overlay assay | 28–210 | – | – |
| Solid-phase virus binding assay | 25–210 | – | – |
*The detection performance and cost of ICT was calculated from (Watanabe et al., 2015, Yeo et al., 2014) and product specifications of commercially available ICT kit. The detection performance of PCR, TLC virus overlay assay and solid-phase virus binding assay were referred from (Takahashi et al., 2013b).
4. Conclusion
In summary, we have demonstrated the highly sensitive and label-free detection of influenza A virus subtype H1N1 on densely immobilized human SA surfaces using QCM and electrical detection methods. The identification technique for the subtype of influenza A virus may be able to help reduce the risk of pandemic outbreaks of HPAI in future. Many kinds of viruses such as coronavirus, rhinovirus, adenovirus and dengue virus have specific binding sites for oligosaccharide receptors. These viruses can also be measured using the same method described in this report. That is, the multi-sensing system using a high-density array of oligosaccharide probes can be developed for highly sensitive and label-free multiple virus tests in the future.
Acknowledgements
This work was supported by the Impulsing Paradigm Change through Disruptive Technologies (ImPACT) Miyata Program on the “Ultra-high-speed multiplexed sensing system beyond evolution for the detection of extremely small amounts of substances” (launched in 2014 and funded by the Council for Science, Technology and Innovation headed by the Prime Minister of Japan) from the Japan Science and Technology (JST) Agency.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bios.2017.02.023.
Appendix A. Supporting information
Supplementary material
References
- Aitken A., Hannoun C. Eur. J. Biochem. 1980;107:51–56. doi: 10.1111/j.1432-1033.1980.tb04623.x. [DOI] [PubMed] [Google Scholar]
- Carrat F., Flahault A. Vaccine. 2007;25:6852–6862. doi: 10.1016/j.vaccine.2007.07.027. [DOI] [PubMed] [Google Scholar]
- Goda T., Miyahara Y. Anal. Chem. 2010;82:1803–1810. doi: 10.1021/ac902401y. [DOI] [PubMed] [Google Scholar]
- Goda T., Miyahara Y. Biosens. Bioelectron. 2013;45:89–94. doi: 10.1016/j.bios.2013.01.053. [DOI] [PubMed] [Google Scholar]
- Gopinath S.C.B., Kumar P.K.R. Acta Biomater. 2013;9:8932–8941. doi: 10.1016/j.actbio.2013.06.016. [DOI] [PubMed] [Google Scholar]
- Hamilton B.S., Whittaker G.R., Daniel S. Viruses. 2012;4:1144–1168. doi: 10.3390/v4071144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideshima S., Hinou H., Ebihara D., Sato R., Kuroiwa S., Nakanishi T., Nishimura S.-I., Osaka T. Anal. Chem. 2013;85:5641–5644. doi: 10.1021/ac401085c. [DOI] [PubMed] [Google Scholar]
- Horiguchi Y., Miyachi S., Nagasaki Y. Langmuir. 2013;29:7369–7376. doi: 10.1021/la304548m. [DOI] [PubMed] [Google Scholar]
- Horiguchi Y., Nakayama N., Kanayama N., Nagasaki Y. Biomater. Sci. 2014;2:819–826. doi: 10.1039/c3bm60212e. [DOI] [PubMed] [Google Scholar]
- Jeon S.H., Kayhan B., Ben-Yedidia T., Arnon R. J. Biol. Chem. 2004;279:48410–48419. doi: 10.1074/jbc.M409059200. [DOI] [PubMed] [Google Scholar]
- Kogure T., Suzuki T., Takahashi T., Miyamoto D., Hidari K.P.J., Guo C.-T., Ito T., Kawaoka Y., Suzuki Y. Glycoconj. J. 2006;23:101–106. doi: 10.1007/s10719-006-5442-z. [DOI] [PubMed] [Google Scholar]
- Lis H., Sharon N. Chem. Rev. 1998;98:637–674. doi: 10.1021/cr940413g. [DOI] [PubMed] [Google Scholar]
- Ostuni E., Chapman R.G., Liang M.N., Meluleni G., Pier G., Ingber D.E., Whitesides G.M. Langmuir. 2001;17:6336–6343. [Google Scholar]
- Park S., Yousaf M.N. Langmuir. 2008;24:6201–6207. doi: 10.1021/la8005663. [DOI] [PubMed] [Google Scholar]
- Peiris J.S.M., de Jong M.D., Guan Y. Clin. Microbiol. Rev. 2007;20:243–267. doi: 10.1128/CMR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier G.E., Tarlov M.J., Rushmeier H.E. Langmuir. 1994;10:3383–3386. [Google Scholar]
- Rowe T., Abernathy R.A., Hu-Primmer J., Thompson W.W., Lu X., Lim W., Fukuda K., Cox N.J., Katz J.M. J. Clin. Microbiol. 1999;37:937–943. doi: 10.1128/jcm.37.4.937-943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada T., Fedorov D.G., Kitaura K. J. Am. Chem. Soc. 2010;132:16862–16872. doi: 10.1021/ja105051e. [DOI] [PubMed] [Google Scholar]
- Skehel J.J., Wiley D.C. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Subbarao K., Klimov A., Katz J., Regnery H., Lim W., Hall H., Perdue M., Swayne D., Bender C., Huang J., Hemphill M., Rowe T., Shaw M., Xu X., Fukuda K., Cox N. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- Takahashi T., Kawagishi S., Masuda M., Suzuki T. Glycoconj. J. 2013;30:709–716. doi: 10.1007/s10719-013-9477-7. [DOI] [PubMed] [Google Scholar]
- Takahashi T., Kawakami T., Mizuno T., Minami A., Uchida Y., Saito T., Matsui S., Ogata M., Usui T., Sriwilaijaroen N., Hiramatsu H., Suzuki Y., Suzuki T. PLoS One. 2013;8:e78125. doi: 10.1371/journal.pone.0078125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zhao J., Jiang T., Kwon Y.M., Lu H., Jiao P., Liao M., Li Y. J. Virol. Methods. 2013;189:362–369. doi: 10.1016/j.jviromet.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Ito T., Ibrahim M.S., Arai Y., Hotta K., Phuong H.V.M., Hang N.L.K., Mai L.Q., Soda K., Yamaoka M., Poetranto E.D., Wulandari L., Hiramatsu H., Daidoji T., Kubota-Koketsu R., Sriwilaijaroen N., Nakaya T., Okuno Y., Takahashi T., Suzuki T., Ito T., Hotta H., Yamashiro T., Hayashi T., Morita K., Ikuta K., Suzuki Y. Biosens. Bioelectron. 2015;65:211–219. doi: 10.1016/j.bios.2014.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley D.C., Skehel J.J. Annu. Rev. Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- Wu Z., Liang H., Lu J. Macromolecules. 2010;43:5699–5705. [Google Scholar]
- Xiong X., Tuzikov A., Coombs P.J., Martin S.R., Walker P.A., Gamblin S.J., Bovin N., Skehel J.J. Virus Res. 2013;178:12–14. doi: 10.1016/j.virusres.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Yamada S., Suzuki Y., Suzuki T., Le M.Q., Nidom C.A., Sakai-Tagawa Y., Muramoto Y., Ito M., Kiso M., Horimoto T., Shinya K., Sawada T., Kiso M., Usui T., Murata T., Lin Y., Hay A., Haire L.F., Stevens D.J., Russell R.J., Gamblin S.J., Skehel J.J., Kawaoka Y. Nature. 2006;444:378–382. doi: 10.1038/nature05264. [DOI] [PubMed] [Google Scholar]
- Yao L., Korteweg C., Hsueh W., Gu J. FASEB J. 2008;22:733–740. doi: 10.1096/fj.06-7880com. [DOI] [PubMed] [Google Scholar]
- Yeo S.-J., Huong D.T., Hong N.N., Li C.-Y., Choi K., Yu K., Choi D.-Y., Chong C.-K., Choi H.S., Mallik S.K., Kim H.S., Sung H.W., Park H. Theranostics. 2014;4:1239–1249. doi: 10.7150/thno.10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen K.Y., Chan P.K.S., Peiris M., Tsang D.N.C., Que T.L., Shortridge K.F., Cheung P.T., To W.K., Ho E.T.F., Sung R., Cheng A.F.B. Lancet. 1998;351:467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material