Abstract
Live vaccines can generate false-positive results on common influenza assays including reverse transcriptase-PCR (RT-PCR), culture and antigen tests. This threatens the integrity of epidemiological data and may misdirect treatment and control efforts. We report the development of RT-PCR tests that distinguish live FluMist™ vaccine (FMV) strains from circulating influenza strains in clinical samples. Primers were validated using influenza-positive samples from unvaccinated patients, packaged FMV, and one PCR-positive asymptomatic vaccine. Furthermore, the assay was used to experimentally test our lab's collection of influenza-positive samples from the 2004–05 and 2005–06 influenza seasons and several 2005 preseason isolates to determine the rate of vaccine-derived false-positive results under differing epidemiological conditions. Analytical and clinical validations show that the assay is both sensitive and specific. Experimental results demonstrate that 51 out of 51 influenza-positive samples collected during influenza season from ill, previously-vaccinated military personnel represent real infections with circulating strains. Finally, the assay shows that four preseason influenza-positive samples were false positives resulting from vaccine shedding. The vaccine-discriminatory RT-PCR methods described here provide the first test designed to distinguish FMV strains from circulating strains. The results show that the test is effective, and demonstrate the importance of such tests in the age of live vaccines.
Keywords: PCR, RT-PCR, LAIV, Vaccine shedding, FluMist
1. Introduction
1.1. Live attenuated influenza vaccination
The FMV is a live, attenuated, cold-adapted, trivalent influenza vaccine that is administered intranasally. It contains three influenza strains: B, A/H3N2 and A/H1N1. Each strain is a 6/2 reassortant in which the two genomic segments encoding the primary surface antigens, hemaggluttinin (H) and neuraminidase (N), are derived from antigenically relevant wild-type strains [1]. The products of these genes confer the ability to induce protective immunity against currently circulating influenzas [2] and are taken from isolates that are recommended annually for inclusion in vaccine formulations. The remaining six genomic segments are derived from master donor virus (MDV) strains which are themselves derived from the A/Ann Arbor/6/60 and B/Ann Arbor/1/66 strains, originally isolated in 1960 and 1966, respectively. The original isolates were evolved by serial passage at sequentially lower temperatures to confer the temperature-sensitive, cold-adapted, and attenuated phenotype [3]. Previous studies have shown that live vaccine strains can be recovered from vaccinated individuals for up to 21 days post-vaccination and could therefore yield false influenza-positive results on RT-PCR tests, rapid antigen assays and culture-based diagnostics [4], [5]. In one study, shedding was demonstrated in 50% of the vaccinated subjects 3 days post-vaccination, and in approximately 5% seven days post-vaccination [5]. Given that our study population (military recruits) is highly vaccinated and our samples are generally collected during training within approximately 10 weeks of vaccination, the risk of generating false-positive influenza results due to FMV shedding is significant.
1.2. Molecular diagnosis of influenza
The use of molecular diagnostics for detection of influenza is becoming more widespread due to low cost, high sensitivity, and increased speed relative to viral culture [6]. The Naval Health Research Center (NHRC) laboratory uses RT-PCR as the principal diagnostic method for detecting influenza A and B in clinical samples. The universal influenza primers used by NHRC (sets 1, 2 and 7 in Table 1 ) and many of the primers described in the literature [7] target conserved regions in the matrix gene and are incapable of discriminating between the MDVs and current circulating strains. To ensure that influenza-positive samples identified by routine influenza A and influenza B RT-PCR assays are community-acquired infections and not the result of shedding from recent vaccination, primers were developed that can discriminate between the MDVs and currently circulating influenza strains.
Table 1.
Primers
| Set | Amplicon | Influenza A primers | Sequence (5′–3′) |
|---|---|---|---|
| 1 | 93 bp | FluA_Universal_1 F | AGCAAAAGCAGGTAGATRTT |
| FluA_Universal_1 R | TCGGCTTTGAGGGG | ||
| 2 | 239 bp | FluA_Universal_2 F | GACCIATCCTGTCACCTCTGAC |
| FluA_Universal_2 R | CATICAACTGGCIAGIGCAC | ||
| 2.5 | 239 bp | FluA_Universal_2.5 Fa | GACCRATYCTGTCACCTCTGAC |
| FluA_ Universal_2.5 R | CATYCAACTGGCAAGTGCAC | ||
| 3 | 647 bp | FMV_A (not H3N2) F | TACGTTCTCTCTATCATCCCG |
| FMV_A (not H3N2) R | GAGCTAGGATGAGTCCCAATAAC | ||
| 4 | 264 bp | Circ_H3N2 (not Vaccine) F | CTGTAACCACTGAAGTGGCA |
| Circ_H3N2 (not Vaccine) R | AGCTAGGATGAGTCCCAATGG | ||
| 5 | 264 bp | Circ_H1N1 (not Vaccine) Fb | CTGTRACCACCGAATCAGCA |
| 6 | 264 bp | Circ_H1N2 (not Vaccine) F | CTGTAACCACCGAAGTGGCA |
| Circ_H1N2 (not Vaccine) R | AACTAGGATGAGTCCCAACGG | ||
| Set | Amplicon | Influenza B primers | Sequence (5′–3′) |
| 7 | 91 bp | FluB_Universal F | GGATTAAACAAAAGCAAGCCTTCCT |
| FluB_Universal R | CAAGCTTCAAAGGTGTTTTC | ||
| 8 | 472 bp | FMV_B F | GCCTACCTGCTTTCACTA |
| FMV_B R | CTCTGCTCTGCTATGAGCTCTT | ||
| 9 | 472 bp | Circ_B F | GCCTACCTGCTTTCATTG |
| Circ_B R | CTCTGCTATGAGCCCTG | ||
All primers were developed by NHRC except the FluA_Universal_1 primers (developed by Luke Daum at Brooks Air Force Base, San Antonio, TX).
FluA_Universal_2.5 (set 2.5) is a current, drift-updated version of the FluA_Universal_2 primers. All FluA_Universal primers have been validated against culture results on more than 100 samples at NHRC and serve (or served) as part of our CAP-certified diagnostic RT-PCR for influenza A.
Circ_H1N1 F uses Circ_H3N2 R as a reverse primer.
1.3. Primer design
The mutations accrued by the MDVs during cold adaptation are too widely dispersed to allow distinguishing primers to be designed around them. Fortunately, the MDVs were derived from strains that were originally collected over 35 years ago, allowing enough time for natural divergence to create many suitable polymorphic regions for strain-specific primer design.
Within H/N subtypes, the molecular evolution of the matrix gene follows a pattern of serial replacement in which new variant alleles arise from recently circulating alleles. Hence, their change is characterized by the accumulation of mutations (drift) over time, forming a linear phylogenetic succession [8]. With this evolutionary pattern in mind, strain-specific discriminating primers were designed for the three MDVs and for circulating strains of the three dominant influenza A subtypes (H3N2, H1N1, and H1N2) and influenza B. Each set of primers exploits polymorphisms that arose in specific subtype lineages after the isolation of the relevant vaccine strain. Fig. 1 depicts our primer choice strategy in the context of a sequence alignment that illustrates the general pattern of influenza evolution, using the influenza A H3N2 matrix gene and the FMV_A (not H3N2) forward primer as an example.
Fig. 1.
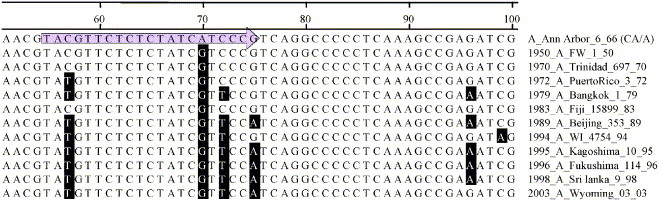
Sequence alignment of a temporal series of influenza A/H3N2 matrix gene sequences corresponding to the region targeted by the FMV_A (not H3N2) forward primer (shaded arrow).
Phylogenetically, the matrix gene of the influenza A MDV (an H2N2 strain) lies directly between the currently circulating H1N1 and H3N2 clades (current H1N2 strains are very close to H3N2 strains with respect to the matrix gene). This pattern represents the fact that current H1N1 and H3N2 matrix genes have diverged independently from the MDV sequence, and hence differ from the MDV in different ways. Thus, a single pair of PCR tests cannot differentiate between the entire spectrum of circulating strains of influenza A and the FMV influenza A MDV. The FMV_A (not H3N2) primers can distinguish the influenza A MDV from circulating H3N2 strains, but will also give a positive result for circulating H1N1 strains (all primers are shown in Table 1). The Circ_H1N1 (not vaccine) primers, designed to yield a positive result for all current H1N1 strains, will not give a positive result for the MDV but will yield a positive result for some circulating H3N2 strains. Hence, a definitive identification of the influenza A MDV requires three results: a positive with the FMV_A primer set, and negative results with both the Circ_H3N2 and Circ_H1N1 primer sets. An analysis matrix is presented in Table 2 for clarity. There are no such issues with influenza B typing because all currently circulating influenza Bs are in the same distinct clade with respect to the influenza B MDV.
Table 2.
Analysis Matrix
| Template | Primer set |
|||||
|---|---|---|---|---|---|---|
| FMV_A | Circ_ H3N2 | Circ_H1N1 | Circ_H1N2 | Circ_B | FMV_B | |
| FluMist MDV (A) | + | — | — | — | — | — |
| Influenza A H3N2 | — | + | ± | ± | − | − |
| Influenza A H1N1 | + | ± | + | − | − | − |
| Influenza A H1N2 | − | ± | − | + | − | − |
| Influenza B | − | − | − | − | + | − |
| FluMist MDV (B) | − | − | − | − | − | + |
Expected results for different primer sets amplifying from different templates.
+ indicates expected amplification, − indicates expected negative, and ± indicates that primer may give either negative or positive results.
The H3N2 primers presented here will cover more than 90% of the influenza A strains currently circulating in the United States (and in most of the world) if the current pattern of infection (http://www.cdc.gov/flu/) holds true. If the pathogen has already been identified as influenza A, a paired test with the Circ_H3N2 primers and the FMV_A primers will usually definitively identify the strain as community-acquired by giving a positive result for the Circ_H3N2 primers only. However, if these tests yield a negative result with the Circ_H3N2 primers and a positive result with the FMV_A primers, then the Circ_H1N1 primers must be used to determine if the strain is vaccine or H1N1. These three PCRs should definitively discriminate between FMV shedding and circulating strains in 99% of influenza A cases (http://www.cdc.gov/flu/). Finally, rare H1N2 strains have also been identified in recent years, and some of these are slightly diverged from H3N2 at the Circ_H3N2 primer sites. Hence, we also present primers designed to catch these circulating strains.
Like most primers designed for surveillance of rapidly evolving RNA viruses, the primers for circulating strains will need to be replaced as strains continue to evolve. However, it is unlikely that circulating strains will back mutate to (or be replaced by) strains with target alleles identical to those of the vaccine strain, and hence it is improbable that the vaccine primers will ever require replacement. The inclusion of primer pairs for both the vaccine and circulating strains allows identification of genetic drift affecting recognition by one primer pair because such changes will yield strains that give negatives on all tests. When such results become common, circulating strains should be sequenced in the primer-targeted regions to address possible mismatches.
1.4. Test validation
Primers were initially tested for specificity against a panel of 84 negative controls consisting of common respiratory pathogens (both bacterial and viral, with the exception of influenza) and common environmental and commensal bacteria (like Escherichia coli) (see Table 6 for specific identities).
Table 6.
Negative controls used to assay specificity of vaccine discriminatory primers, shown as Species (# of strains tested)
| Acinetobacter baumannii (1) | Legionella pneumophila (1) | Staphylococcus epidermidis (1) |
| Bordetella holmesii (1) | Moraxella catarrhalis (1) | Streptococcus agalactiae (1) |
| Bordetella parapertussis (1) | Mycobacterium kansasii (1) | Streptococcus pneumoniae (2) |
| Bordetella pertussis (1) | Mycoplasma fermentans (1) | Human adenovirus (13) |
| Chlamydia muridarum (1) | Mycoplasma genitalium (1) | Coronavirus (2) |
| Chlamydia pneumoniae (1) | Mycoplasma hominis (1) | Coxsackievirus (4) |
| Chlamydia trachomatis (2) | Mycoplasma hyopneumoniae (1) | Rhinovirus (2) |
| Corynebacterium diptheriae (1) | Mycoplasma mobile (1) | Echovirus (4) |
| Enterococcus faecalis (1) | Mycoplasma penetrans (1) | Enterovirus (2) |
| Escherichia coli (1) | Mycoplasma pneumoniae (2) | Epstein-Barr virus (1) |
| Streptococcus pyogenes (4) | Mycoplasma pulmonis (1) | Herpes simplex virus (4) |
| Haemophilus influenzae (2) | Proteus mirabilis (1) | Mumps virus (1) |
| Haemophilus parainfluenzae (2) | Pseudomonas aeruginosa (1) | Human parainfluenza virus (6) |
| Klebsiella pneumoniae (2) | Staphylococcus aureus (2) | Respiratory syncytial virus (4) |
For FMV A and FMV B positive controls, a 10-fold dilution series of nucleic acid extracted from FluMist™ vaccine (MedImmune) was tested to evaluate the sensitivity of vaccine-specific primers (FMV_A and FMV_B) and the specificity of circulating strain primers (Circ_H3N2, Circ_H1N1, Circ_H1N2, and Circ_B) (Table 1). One sample, collected as a nasal swab from one of the authors (N.E.F.) approximately one hour after FMV vaccination, served as a positive clinical control sample for the live vaccine strains.
For clinical circulating-strain controls, twelve unvaccinated patients’ throat swab (TS) samples which had previously tested positive for influenza A were tested with the new vaccine-discriminating primer sets (sets 3–5, Table 1). Similarly, 25 influenza A culture-positive/RT-PCR-positive TS samples from individuals treated with inactivated injectable vaccine were also tested, since injectable vaccine would not be expected to yield influenza-positive results by culture or RT-PCR. Both of these sample types served as positive controls for sensitivity of the circulating strain primer sets as well as negative controls for specificity of the vaccine primer sets (with the exception of samples identified as H1N1, which were expected to cross-react with the FMV_A primers).
For circulating influenza B-positive clinical controls, two TS samples collected from a generally unvaccinated border population and two samples from unvaccinated recruits, all of which had previously tested positive for influenza B by both culture/IF and RT-PCR, were tested with primer sets 7–9. Similarly, 14 influenza B culture-positive/RT-PCR-positive TS samples from recruits treated with inactivated injectable vaccine were also tested (Table 1).
As clinical circulating-strain negative controls, seven TS samples from asymptomatic recruits who had been recently vaccinated with FMV were tested. These samples had previously tested negative for influenza by culture/IF and RT-PCR (Universal primer sets 1 and 2, Table 1).
1.5. Experimental testing
The developed assay was used to test a collection of 55 influenza-positive clinical TS samples from symptomatic patients who had been vaccinated with FMV between 1 day and 4 months prior to sample collection. These samples could have originally tested positive due to either the vaccine strains or community-acquired wild-type strains. Forty-five of these had originally tested positive for influenza A by culture/IF and/or RT-PCR, and ten had tested positive for influenza B. Of the 45 influenza A samples chosen, six were culture-negative/RT-PCR-positive. These were specifically included because FMV is growth-limited at 37 °C and vaccine shedding might be expected to generate this type of discrepant result.
2. Materials and methods
2.1. Primer design
Independent sequence alignments of influenza A H3N2, A H1N1 and A H1N2 matrix genes were made using the Clustal W algorithm in the Lasergene MegAlign software (DNASTAR, Madison, WI). These represented a temporally diverse selection of human isolates collected between 1933 and 2003. Sequences were found using GenBank (http://www.ncbi.nlm.nih.gov/) and the Influenza Sequence Database (http://www.flu.lanl.gov/) [9]. Each alignment also included the matrix sequence of the FMV influenza A MDV, kindly provided by Jin Hong (MedImmune, Inc., Gaithersburg, MD). Vaccine-specific primers were chosen that would amplify sequences from the MDV and not from the primary current circulating subtype (H3N2), while circulating strain primers were chosen for each subtype that would amplify sequences from circulating strains and not from the vaccine strain (all primers shown in Table 1). The same methods were used to design primers that distinguish circulating influenza B from the influenza B MDV. Primers were obtained from Integrated DNA Technologies (Coralville, IA).
2.2. Extraction and RT-PCR
RNA was extracted from FMV and TS samples using the QIAamp 96 DNA Blood and Body Fluid Kit (Qiagen, Valencia, CA) following manufacturer's instructions. RT-PCR was performed using the One-Step RT-PCR kit (Qiagen) according to manufacturer's instructions (“Protocol Using Q-Solution”) with 600 nM final primer concentrations and the following modifications: Final reaction volumes were 25 μl, and 5 μl of sample was used as template. Reactions were performed on iCycler PCR machines (Bio-Rad, Hercules, CA). Cycling conditions included a 10-min final extension at 72 °C and a final hold at 4 °C. In regard to other parameters, reactions were cycled as shown in Table 3 . All PCRs appeared robust using annealing temperatures of up to 58 °C, but it is suggested that annealing temperatures be tuned for any new reagent set, sample type, or thermal cycling platform (specifically, machines with different ramp rates). RT-PCRs were analyzed by agarose gel electrophoresis with ethidium bromide visualization.
Table 3.
Cycling conditions
| PCR step | Cycles | Flu_A_Universal_1 | Flu_A_Universal_2, 2.5 | Circ_H1N1 | FMV_A and B Circ_H3N2, H1N2, B |
|---|---|---|---|---|---|
| RT | 1 | 45:00 (50 °C) | 45:00 (50 °C) | 45:00 (50 °C) | 45:00 (50 °C) |
| Activation | 1 | 15:00 (95 °C) | 15:00 (95 °C) | 15:00 (95 °C) | 15:00 (95 °C) |
| Denaturation | 40a | 0:30 (94 °C) | 0:30 (94 °C) | 0:30 (94 °C) | 0:30 (94 °C) |
| Annealing | 1:00 (50 °C) | 1:00 (58 °C) | 1:30 (58 °C) | 1:30 (55 °C) | |
| Extension | 1:00 (72 °C) | 1:00 (72 °C) | 2:00 (72 °C) | 2:00 (72 °C) |
All conditions are shown as time (temperature), with time shown as minutes:seconds.
FluA_Universal primer sets are run for 35 cycles.
2.3. Clinical sample collection
All clinical samples from symptomatic individuals were collected as part of NHRC's ongoing respiratory surveillance efforts in which TS samples were taken with consent from military recruits meeting the case definition of a temperature at or above 38 °C and a respiratory symptom. Swabs were placed in Viral Transport Medium (Remel, Lenexa, KS), frozen at −80 °C, and sent on dry ice to NHRC. Samples were then pre-aliquotted and stored at −80 °C in monitored freezers. Samples were tested by NHRC's College of American Pathologists (CAP)-certified influenza A and B culture/immunofluorescence (culture/IF) and universal influenza A RT-PCR tests (Sets 1–2.5, Table 1). The NHRC influenza A surveillance RT-PCR involves two independent amplifications with different primer sets, and positives represent samples that yielded amplicons for both primer sets. Influenza B identifications were initially achieved at NHRC by either culture/IF or with a single universal influenza B real-time PCR (Set 7, Table 1). From this surveillance collection, we chose 104 influenza-positive TS samples collected during the 2004/2005 and 2005/2006 influenza seasons, between December and March. Four similar samples were chosen that had been collected during the intervening non-influenza season, from September to November 2005. Additionally, seven TS samples were collected from healthy individuals at Lackland Air Force Base and processed in a similar fashion. These samples were from individuals who had received FMV within one week of sample collection and had previously tested negative for influenza by both culture and RT-PCR. Two influenza B-positive TS samples were collected from symptomatic individuals from a US/Mexico border population with very low vaccination rates (assumed to be unvaccinated), and one nasal swab sample was collected from one of the authors (N.E.F.) who had just received FMV.
2.4. Negative control panel
All primer pairs were tested against a negative control panel representing 84 potential confounders, including respiratory bacteria and viruses that might cause similar symptoms, and commensals and environmentally common species that might incidentally be present in specimens. These were obtained from either the American Type Culture Collection or NHRC's College of American Pathologists (CAP) certified bacteriology and virology labs, and were tested at titers between 103 and 108/ml, suspended in distilled water. These tests were performed with appropriate positive controls in 96-well PCR plates.
3. Results
3.1. Clinical control samples
Representative PCR results are shown in Fig. 2 .
Fig. 2.
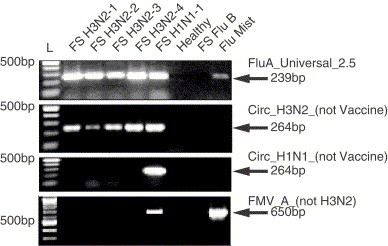
Representative RT-PCR results from described protocols. All gels feature 100 bp ladders (L) with the 500 bp band indicated. Seven field strain (FS) isolates and an extraction of the FluMist vaccine, listed on the top of the gel, were tested with described primers as indicated on the right. Arrows indicate location and size of expected RT-PCR products. Influenza types indicated in the name of the isolates were originally determined by culture and immunofluorescent methods.
The sensitivities of both the FMV_A and FMV_B primer sets were found to be between 0.158 and 0.0158 Tissue culture infectious doses (TCID50). This result was calculated from a TCID50 of 106.5−7.5 (FMV package insert) per 0.5 ml dose. All FMV dilutions also tested negative by all four circulating strain primer sets (see Table 4 ).
Table 4.
FluMist™ (FMV) controls
| FMV Dilution | Primer set |
|||||
|---|---|---|---|---|---|---|
| Circ_B | FMV_B | FMV_A | Circ_H1N2 | Circ_H1N1 | Circ_H3N2 | |
| 10−8–10−9 | Negative | Negative | Negative | Negative | Negative | Negative |
| 100–10−7 | Negative | Positive | Positive | Negative | Negative | Negative |
| Negative | Negative | Negative | Negative | Negative | Negative | Negative |
| FMV same daya | Negative | Positive | Positive | Negative | Negative | Negative |
Pooled results indicate range of 10-fold dilution series over which the reported results were obtained.
FMV same day is a nasal swab taken from an author (N.E.F.) immediately after FMV vaccination.
The 18 influenza B circulating-strain control samples (four from symptomatic, unvaccinated patients and 14 from symptomatic patients vaccinated with injectable vaccine) tested negative with the FMV_B primers and positive with the Circ_B primers (see Table 5 ). There were no cross-reactions between influenza A and influenza B tests for any control samples.
Table 5.
Clinical control samples
| N | Flu_A_Universala | Flu_B_Universal | Viral Culture/IF | Vaccine type | FMV_A | Circ_H3N2 | Circ_H1N1 | Circ_H1N2 | Circ_B | FMV_B |
|---|---|---|---|---|---|---|---|---|---|---|
| 12 | Positive | Negative | A H3N2 | None | Negative | Positive | Negative | Positive | Negative | Negative |
| 21 | Positive | Negative | A H3N2 | Injected | Negative | Positive | Negative | Positive | Negative | Negative |
| 1 | Positive | Negative | A H3N2 | Injected | Negative | Negative | Negative | Positive | Negative | Negative |
| 3 | Positive | Negative | A H1N1 | Injected | Positive | Negative | Positive | Negative | Negative | Negative |
| 7 | Negative | Negative | Negative | FMV | Negative | Negative | Negative | Negative | Negative | Negative |
| 4 | Negative | Positive | Influenza B | None | Negative | Negative | Negative | Negative | Positive | Negative |
| 14 | Negative | Positive | Influenza B | Injected | Negative | Negative | Negative | Negative | Positive | Negative |
N indicates the number of samples with the indicated pattern of test results and vaccination state. All vaccinations were between 2 days and 3 months prior to the collection date.
A positive for Flu_A_Universal indicates a dual-positive for both Flu_A_Universal_1 and Flu_A_Universal_2 primer sets.
All 34 influenza A H3N2-positive control samples from symptomatic recruits who had not received FMV (that is, who were unvaccinated or received injectable vaccine) tested negative with the FMV_A primers. Thirty-three of the 34 tested positive with the Circ_H3N2 primers and the remaining sample was positive only with the Circ_H1N2 primers (see Table 5). Within this sample set, the specificity of the FMV_A primers was 100%, and the sensitivity of the Circ_H3N2 primers was 97% with respect to culture and immunofluorescence and with respect to the Universal FluA primers.
Both the Circ_H1N1 and the FMV_A primers gave positive results for the three H1N1 samples, as expected (see Table 5, also see Introduction: Primer Design for explanation and Table 2 for specificity/analysis matrix). The Circ_H1N1 primers gave negative results for the 34 control H3N2 samples and all FMV dilutions, yielding 100% sensitivity and specificity in this sample set.
The Circ_H1N2 primers could not be validated for sensitivity here because we had no access to H1N2 samples, but they were tested on the other samples and, as expected, cross-reacted strongly with H3N2 strains but not with FMV.
3.2. Analytical control samples
None of the six novel discriminatory primer sets (sets 3, 4, 5, 6, 8 and 9 in Table 1) yielded positive results when tested against 84 negative control samples. The identities of all negative control isolates are listed in Table 6 .
3.3. Experimental samples
The seven influenza A RT-PCR-positive/culture-negative samples collected from symptomatic FMV-vaccinated recruits during the 2004/2005 influenza season all tested positive for one or more of the circulating strains and negative for the vaccine. All ten influenza B RT-PCR-positive/culture-positive samples collected from symptomatic FMV-vaccinated recruits tested positive for circulating influenza B and negative for the vaccine. Thirty of the 34 RT-PCR-positive/culture-positive influenza A samples collected from symptomatic FMV-vaccinated recruits during the influenza season tested negative with FMV_A primers and positive with Circ_H3N2 primers. The remaining four samples tested positive for both vaccine and circulating influenza A strains (all experimental results are shown in Table 7 ). In summary, all 51 influenza-season experimental samples appear to contain community-acquired influenza strains.
Table 7.
Experimental clinical samples
| Influenza season? | N | Flu_A_Universal | Flu_B_Universal | Viral culture | Vaccine type | FMV_A | Circ_H3N2 | Circ_H1N1 | Circ_H1N2 | Circ_B | FMV_B |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Yes | 26 | Positive | Negative | A H3N2 | FMV | Negative | Positive | Negative | Positive | Negative | Negative |
| Yes | 3 | Positive | Negative | A H3N2 | FMV | Negative | Positive | Positive | Positive | Negative | Negative |
| Yes | 1 | Positive | Negative | A H3N2 | FMV | Negative | Positive | Negative | Negative | Negative | Negative |
| Yes | 4 | Positive | Negative | A H3N2 | FMV | Positive | Positive | Negative | Positive | Negative | Negative |
| Yes | 6 | Positive | Negative | Negative | FMV | Negative | Positive | Negative | Positive | Negative | Negative |
| Yes | 1 | Positive | Negative | Negative | FMV | Negative | Negative | Negative | Positive | Negative | Negative |
| Yes | 10 | Negative | Positive | B | FMV | Negative | Negative | Negative | Negative | Positive | Negative |
| No | 4 | Positive | Negative | Negative | FMV | Positive | Negative | Negative | Negative | Negative | Negative |
N indicates the number of samples with the indicated pattern of test results and vaccination state. All vaccinations were between 2 days and 3 months prior to the collection date.
The four RT-PCR-positive samples collected from symptomatic FMV-vaccinated recruits before the beginning of the 2005/2006 influenza season all tested negative for circulating strains and positive for the vaccine (FMV A). These appear to all be cases of false-positive test results related to vaccine shedding.
Many of the primer sets tested here cross-react with strains outside the scope of their designed discriminatory capacity (as expected; see Section 1.3 and Table 2). It must be reiterated here that the purpose of this test is to distinguish FMV shedding from legitimate community-acquired influenza infection, not to determine subtype.
4. Discussion and conclusions
Together, the results demonstrate the sensitivity and specificity of the designed tests. The results also strongly suggest that, when used on samples collected during the influenza season, our CAP-certified influenza culture and RT-PCR tests are primarily yielding positive results from patients with legitimate community-acquired pathogenic influenza as opposed to false positives resulting from vaccine strain shedding.
On the other hand, several results from our work suggest that general RT-PCR methods can detect FMV, and hence reiterate the consensus from the literature that live intranasal vaccination can confound tests designed to identify community-acquired pathogens [4], [5]. Supporting results include the four samples from FMV-vaccinated recruits who tested positive for both vaccine and circulating influenza A strains and the positive results from the control nasal swab sample of an author taken immediately after FMV vaccination. Most importantly, this fall our lab identified what appeared to be four very early-season influenza A infections at recruit training centers using our primary RT-PCR method, discoveries that would have been of epidemiological concern given the time of year. However, all four of these samples tested positive by FMV_A primers and negative by Circ_H3N2 and Circ_H1N1 primers (see Table 7), suggesting that the original FluA_Universal positive results were due to shedding of vaccine strains. This suggestion was substantiated by the facts that these recruits had been vaccinated with FMV within a few days of sampling and these samples did not yield culturable virus at 37°C. The results show that influenza-positive RT-PCR results from surveillance of vaccinated populations during times when pathogenic influenza is not widely circulating are likely to be false-positives from FMV strain shedding, strongly supporting the epidemiological and clinical necessity of tests that can discriminate between vaccine strains and circulating pathogenic strains. This is particularly critical at a time when public health agencies are increasing off-season influenza surveillance in an effort to protect the public from the threat of an influenza pandemic.
News reports indicate that the military alone has contracted to purchase between 850,000 and 1.9 million doses of FluMist for the 2006/2007 influenza season (Baltimore Business Journal, June 19, 2006). In the past two vaccination seasons, MedImmune has sold between 450,000 and three million doses of the drug worldwide. The expected introduction of a refrigerator-stable version is likely to increase sales, as is increased public confidence and familiarity (FluMist was just introduced in the 2003/2004 influenza season, and is the first inhaled influenza vaccine).
The four symptomatic, influenza-positive individuals who tested positive for both circulating and vaccine strains of influenza A had received the FMV vaccine between 1 and 6 days prior to sample collection. Vaccine-induced immunity to influenza A, on average, takes up to 14 days to confer immunity. Hence, these results are consistent with the patients’ history, and suggest that these patients were simultaneously infected with a community-acquired influenza A strain and shedding FMV influenza A. It is interesting to note that these infected patients shed detectable levels of the vaccine strain while none of the seven asymptomatic patients sampled within six days of FMV vaccination gave positive results for the vaccine-specific tests (these individuals are among the asymptomatic controls shown in Table 5). This raises the possibility that vaccine strain survival and/or replication is supported by coinfection with community-acquired strains.
MedImmune, Inc. is currently expanding the line of FMVs to include avian influenzas. The MDVs used for this effort will be the same as those used in the current human influenza FMV, with avian H and N gene segments taking the place of the human influenza equivalents. The FMV primers presented here should be able to distinguish these vaccines from real avian influenza as effectively as they can discriminate human influenza FMV from community-acquired influenzas.
Acknowledgements
This work represents NHRC report No. 06–03, supported by the Department of Defense Global Emerging Infections Surveillance and Response System (GEIS) under research work unit 60501. The views expressed in this article are those of the authors and do not reflect the official policy or position of the Department of the Navy, Department of Defense nor the US Government. Approved for public release; distribution is unlimited. This research has been conducted in compliance with all applicable federal and international regulations governing the protection of human subjects in research, as documented in DoD Protocols NHRC.1999.0002.31271 (NHRC) and FWH20020124 H (Lackland). Informed consent was obtained from all subjects. All authors declare that there is no conflict of interest related to this work. The authors would like to thank Jin Hong of MedImmune Incorporated, the maker of FluMist™, for Master Donor Virus sequence data.
References
- 1.Cha T.A., Kao K., Zhao J., Fast P.E., Mendelman P.M., Arvin A. Genotypic stability of cold-adapted influenza virus vaccine in an efficacy clinical trial. J Clin Microbiol. 2000;38:839–845. doi: 10.1128/jcm.38.2.839-845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maassab H.F., Bryant M.L. The development of live attenuated cold-adapted influenza virus vaccine for humans. Rev Med Virol. 1999;9:237–244. doi: 10.1002/(sici)1099-1654(199910/12)9:4<237::aid-rmv252>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 3.Jin H., Lu B., Zhou H., Ma C., Zhao J., Yang C.F. Multiple amino acid residues confer temperature sensitivity to human influenza virus vaccine strains (FluMist™) derived from cold-adapted A/Ann Arbor/6/60. Virology. 2003;306:18–24. doi: 10.1016/s0042-6822(02)00035-1. [DOI] [PubMed] [Google Scholar]
- 4.Ali T., Scott N., Kallas W., Halliwell M.E., Savino C., Rosenberg E. Detection of influenza antigen with rapid antibody-based tests after intranasal influenza vaccination (FluMist™) Clin Infect Dis. 2004;38:760–762. doi: 10.1086/382887. [DOI] [PubMed] [Google Scholar]
- 5.Talbot T.R., Crocker D.D., Peters J., Doersam J.K., Ikizler M.R., Sannella E. Duration of virus shedding after trivalent intranasal live attenuated influenza vaccination in adults. Infect Control Hosp Epidemiol. 2005;26:494–500. doi: 10.1086/502574. [DOI] [PubMed] [Google Scholar]
- 6.Pregliasco F., Mensi C., Camorali L., Anselmi G. Comparison of RT-PCR with other diagnostic assays for rapid detection of influenza viruses. J Med Virol. 1998;56:168–173. doi: 10.1002/(sici)1096-9071(199810)56:2<168::aid-jmv11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 7.Fouchier R.A.M., Bestebroer T.M., Herfst S., Van Der Kemp L., Rimmelzwaan G.F., Osterhaus A.D.M.E. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J Clin Microbiol. 2000;38:4096–4101. doi: 10.1128/jcm.38.11.4096-4101.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferguson N.M., Galvani A.P., Bush R.M. . Ecological and immunological determinants of influenza evolution, Nature. 2003;422:428–433. doi: 10.1038/nature01509. [DOI] [PubMed] [Google Scholar]
- 9.Macken C., Lu H., Goodman J., Boykin L. The value of a database in surveillance and vaccine selection. In: Osterhaus A.D.M.E., Cox N., Hampson A.W., editors. Options for the control of influenza IV. Elsevier Science; Amsterdam: 2001. pp. 103–106. [Google Scholar]


