Abstract
An atypical pestivirus (‘Hobi’-like pestivirus, putative bovine viral diarrhoea 3, BVDV-3) was identified firstly in contaminated foetal calf serum batches and isolated subsequently from an outbreak of respiratory disease in a cattle herd in Italy. The isolation of the novel pestivirus from animals affected clinically posed concerns about the validity of BVDV eradication programs, considering that ‘Hobi’-like pestivirus (BVDV-3) is undetected or mistyped by the molecular diagnostic tools currently employed. In this paper, the development of a nested PCR (nPCR) assay for unambiguous typing of all bovine pestiviruses is reported. The assay consisted of a first-round amplification using an oligonucleotide pair which binds to conserved sequences located in the 5′ untranslated region and capsid gene, followed by a heminested PCR using virus-specific forward primers. The assay performances were evaluated analytically, showing good sensitivity and specificity. By analysis of 100 BVDV-positive samples typed using a nPCR assay discriminating ruminant pestiviruses, five samples recognised previously as BVDV-2 were not typed when submitted to the new assay (n = 2) or reacted as ‘Hobi’-like pestivirus BVDV-3 (n = 3). Sequence analysis of the first-round amplification products showed that the untyped strains were border disease viruses, whereas the other three strains were true ‘Hobi’-like viruses. The development of a molecular assay able to identify simultaneously all bovine pestiviruses known currently will help warrant biosafety of live vaccines and other biological products and assess the molecular epidemiology of ‘Hobi’-like pestivirus, thus leading to the improvement of the eradication programs through unambiguous typing of pestiviruses infecting cattle.
Keywords: Cattle, Bovine pestiviruses, ‘Hobi’-like strains (BVDV-3), Diagnosis, Nested PCR
1. Introduction
Bovine viral diarrhoea virus (BVDV) belongs to the genus Pestivirus of the family Flaviviridae, together with classical swine fever virus (CSFV), border disease virus (BDV) and other pestiviruses isolated from wild ruminants [1], [2], [3]. BVDV is an enveloped, single-strand positive-sense RNA virus responsible for polymorphic clinical signs [4], which includes two separate species, termed BVDV-1 and BVDV-2 [5], [6], and several subtypes within each species [7], [8], [9], [10], [11], [12], [13], [14], [15]. The BVDV genomic RNA encodes for a polyprotein (NH2-Npro-C-Erns-E1-E2-p7-NS2-3-NS4A-NS4B-NS5A-NS5B-COOH), which is processed by viral and cellular proteases, thus generating structural and non-structural proteins. The single large open reading frame is flanked by the 5′ and 3′ untranslated regions (UTRs) [4].
An atypical pestivirus was isolated from a contaminated batch of foetal calf serum (FCS) originating from Brazil [16]. This virus, named D32/00_‘Hobi’, was proposed as prototype of a new pestivirus species, BVDV-3 [17]. ‘Hobi’-like pestiviruses were detected in different parts of the world in FCS batches of southern American origin [18], [19], [20] or in animals with natural infection but without overt disease [18], [21]. ‘Hobi’-like sequences were detected in aborted bovine foetuses in Brazil, thus suggesting direct clinical implications [22]. More recently, a ‘Hobi’-like strain was associated to severe respiratory disease and abortion in a cattle herd in Italy [23], [24], which posed some issues about the real efficacy of BVDV control or eradication programs. In fact, the existing methods employed commonly in BVDV surveillance programs either do not detect at all [16], [20], [21] or have a low sensitivity in detecting the new pestivirus, which is in addition mistyped as BVDV-2 [25]. A real-time RT-PCR assay has been established for identification of ‘Hobi’-like strains [26], but this method is not able to detect simultaneously BVDV-1/BVDV-2 which should be required for extensive use in eradication programs.
With the aim to overcome the limitations of existing diagnostic methods, a nested PCR (nPCR) assay was developed which is able to detect and type all bovine pestiviruses, including the new species (putative BVDV-3).
2. Material and methods
2.1. Primer design
The full-length genomes of BVDV-1, BVDV-2, ‘Hobi’-like pestivirus (BVDV-3), BDV and CSFV strains were retrieved from the GenBank database (http://www.ncbi.nlm.nih.gov/Genbank/index.html) and aligned using the BioEdit software package [27]. First-round amplification (RT-PCR) primers, amplifying a region encompassing the 5′ UTR, Npro and C genes, were designed using the Primer3 software, version 0.4.0 (http://frodo.wi.mit.edu/primer3/) in order to bind to bovine pestivirus conserved regions. Typing nPCR was carried out using the first-amplification reverse primer and a panel of three species-specific forward primers that were designed manually on bovine pestivirus discriminating regions through visual inspection of the aligned sequences.
Primers were synthesised by Primm srl (Milan, Italy). The position and sequence of the primers used for the assay are reported in Table 1 .
Table 1.
Oligonucleotides used in the nPCR assays for pestivirus typing.
| Reference | Assay | Target | Primer | Sequence 5′–3′ | Sense | Positiona | Specificity | Amplicon size (bp) |
|---|---|---|---|---|---|---|---|---|
| Ref. [35] | RT-PCR | Erns | P1 | AACAAACATGGTTGGTGCAACTGGT | + | 1424–1448 | Bovine pestiviruses, BDV, CSFV | 826 |
| P2b | CTTACACAGACATATTTGCCTAGGTTCCA | − | 2222–2250 | |||||
| nPCR | TS1 | TATATTATTTGGAGACAGTGAATGTAGTAG | + | 1684–1713 | BDV | 566 | ||
| TS2 | TGGTTAGGGAAGCAATTAGG | + | 1802–1821 | BVDV-2 | 448 | |||
| TS3 | GGGGGTCACTTGTCGGAGG | + | 2027–2045 | BVDV-1 | 223 | |||
| This study | RT-PCR | 5′ UTR, Npro, C | PanBVDVpcrF | CTCTGCTGTACATGGCACATG | + | 368–388 | Bovine pestiviruses, BDV, CSFV | 1013 |
| PanBVDVpcrRb | CGTCGAACCAGTGACGACT | − | 1364–1383 | |||||
| nPCR | BVDV-1 npcrF | TTTCAAGCTGCTCHGAYAC | + | 879–897 | BVDV-1 | 501 | ||
| BVDV-2 npcrF | ATCCTGACCAATGCTAGGTCC | + | 551–571 | BVDV-2 | 829 | |||
| BVDV-3 npcrF | TCCTGTGGCAACCGGTAGGT | + | 1173–1192 | ‘Hobi’-like | 210 |
Oligonucleotide position is referred to the genomic sequence of BVDV-1 strain NADL (GenBank accession no. M31182).
RT-PCR reverse primers were also used in nPCR assays.
2.2. RT-PCR
RT-PCR was carried out using SuperScript™ One-Step RT-PCR for Long Templates (Life Technologies, Invitrogen, Milan, Italy) and the following thermal protocol: reverse transcription at 50 °C for 30 min, inactivation of Superscript II RT at 94 °C for 2 min, 45 cycles of 94 °C for 30 s, 50 °C for 30 s, 68 °C for 1 min, with a final extension at 68 °C for 10 min. The PCR products were detected by electrophoresis through a 1.5% agarose gel and visualisation under UV light after bromide ethidium staining.
2.3. Nested PCR
Nested PCR was performed using AmpliTaq Gold (Applera Italia, Monza, Italy) The reaction was carried out in a total volume of 50 μl containing PCR buffer 1× (KCl 50 mM, Tris–HCl 10 mM, pH 8,3), MgCl2 2 mM, 200 μM of each deoxynucleotide (dATP, dCTP, dGTP, dTTP), 1 μmol l−1of the RT-PCR reverse primer and of each internal species-specific primer, 1 U of AmpliTaq Gold and 5 μl of a 1:100 dilution in distilled water of the primary PCR product. The thermal conditions consisted of activation of AmpliTaq Gold polymerase at 94 °C for 10 min and 25 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s and polymerisation at 72 °C for 1 min, followed by a final extension at 72 °C for 10 min. The PCR products were detected as for the first-round amplification.
2.4. Test specificity and sensitivity
To rule out cross-reactivities between bovine pestiviruses and other viral pathogens, the specificity of the assay was evaluated by testing isolates of the following viruses: bovine coronavirus [28], bovine rotaviruses [29], bovine respiratory syncytial virus (vaccine strain BRSV/375, Cattle Master 4, Pfizer Italia srl), bovine parainfluenza virus (vaccine strain TS RLB 103, Cattle Master 4, Pfizer Italia srl), and bovine herpesvirus types 1 [30] and 4 [31]. A BDV isolate, strain BD91 [32], and the CSFV lapinised Chinese vaccine [33] were also submitted to the test.
Faecal, nasal and EDTA-blood samples collected from ten pestivirus-negative calves as well as sterile water were also included in the analysis as negative controls and blanks, respectively.
To evaluate the detection limit of the nPCR assay, tenfold dilutions in Dulbecco’s minimal essential medium of reference strains BVDV-1 NADL (courtesy of Dr Ferrari, Istituto Zooprofilattico Sperimentale di Lombardia ed Emilia Romagna, Brescia, Italy), BVDV-2 232/02 [34] and ‘Hobi’-like strain 1/10-1-Italy [23], having titres of 104.50, 104.50 and 103.50 TCID50/50 μl, respectively, as determined by an immunofluorescence test on infected Madin Darby bovine kidney (MDBK) cell cultures, were used. Each virus dilution was quantified three times separately.
2.5. Clinical samples
A total of 98 clinical bovine samples tested positive by an nPCR assay established previously for typing of ruminant pestiviruses [35] was submitted to the novel assay. These samples included nine tissue samples from aborted foetuses, 17 respiratory specimens from calves with respiratory disease, six fecal samples from calves with enteritis and 66 EDTA-blood samples from animals infected persistently. In addition, two caprine tissue samples previously typed as BVDV-2 were analysed.
2.6. Internal control
In order to verify the absence of RNA losses during the extraction step and the presence of RT-PCR inhibitors in the RNA templates, an internal control (IC), consisting of an RNA synthetic transcript containing the M gene of canine coronavirus (CCoV) type II [36], was added to the lysis buffer (AVL buffer, QIAGEN S.p.A.) at a concentration of 10,000 RNA copies ml−1 of buffer prior to nucleic acid extraction. RNA extracts were submitted in parallel to a CCoV RT-PCR assay [37] and samples from which the amplicons of the expected size were not obtained were excluded from the analysis.
3. Results
3.1. Performance of the nPCR assay for bovine pestivirus typing
The first-amplification (RT-PCR) yielded products of the expected size from all reference pestivirus strains, including BDV and CSFV. By the developed nPCR assay, BVDV-1, BVDV-2 and ‘Hobi’-like reference strains were typed as predicted on the basis of species-specific oligonucleotides and viral sequences, giving amplicons of 501, 829, and 210 bp, respectively. No cross-reactions were observed between different bovine pestiviruses (Fig. 1 ). BDV and CSFV did not react with any of the bovine pestivirus-specific primers. By using the old protocol for typing of ruminant pestiviruses [35], which targets the Erns gene, BVDV-1 and BVDV-2 were correctly typed, whereas ‘Hobi’-like pestivirus and BDV reference strains were both mistyped as BVDV-2, a finding that had been already observed [25], [38]. As expected from a previous study [35], CSFV was detected by the first-round amplification, but not typed by the Erns nPCR.
Fig. 1.
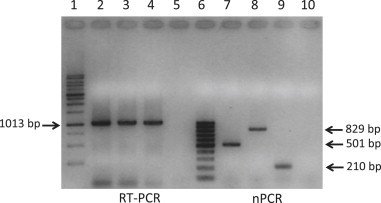
Gel electrophoresis of products obtained from RT-PCR (lines 2–5) and nPCR (lines 7–10) assays for detection and typing of bovine pestiviruses. Line 1, marker GeneRuler 100 bp DNA Ladder (MBI Fermentas GmbH, St. Leon-Rot, Germany); Line 6, marker GeneRuler 1 kb DNA Ladder (MBI Fermentas GmbH); lines 2, 7, BVDV-1 strain NADL; lines 3, 8, BVDV-2 strain 232/02; lines 4, 9, ‘Hobi’-like (BVDV-3) strain 1/10-1-Italy; lines 5, 10, negative control (blood from a pestivirus-negative calf).
The template controls, pestivirus-negative specimens and the other tested viral pathogens of cattle did not yield any detectable PCR product by first- and second-round amplifications of the novel assay, thus confirming that both RT-PCR and nPCR assays are highly specific for pestiviruses.
The detection limits of the old and novel protocols for bovine pestivirus identification and typing were calculated (Table 2 ), showing that the assay established newly is more sensitive for BVDV-2 and ‘Hobi’-like pestivirus detection, whereas BVDV-1 is more sensitively detected by the Erns nPCR.
Table 2.
Evaluation of the sensitivity of the old and new protocols for bovine pestivirus detection and typing.a
| Reference | Assay | Pestiviral titre (TCID50 50 μl−1) |
||
|---|---|---|---|---|
| BVDV-1 | BVDV-2 | ‘Hobi’-likea | ||
| Ref. [35] | RT-PCR | 102.50 | 101.50 | 100.50 |
| nPCR | 10−2.50 | 10−0.50 | 100.50 | |
| This study | RT-PCR | 101.50 | 100.50 | 101.50 |
| nPCR | 10−0.50 | 10−1.50 | 10−2.50 | |
‘Hobi’-like pestivirus is mistyped as BVDV-2 by the old protocol [35].
3.2. Analysis of clinical samples
All clinical samples tested positive by the Sullivan and Akkina’s protocol were confirmed to contain pestiviral RNA by using the novel assay (Table 3 ). Totally, 95 samples were in agreement by both nPCR assays, but five samples previously typed as BVDV-2 gave contrasting results. Using the developed assay, three of these samples reacted with the ‘Hobi’-like pestivirus primer, as shown by the 210-bp product observed after gel electrophoresis. The remaining two samples, albeit positive by the panpestivirus RT-PCR assay, were not typed by nPCR. Sequence analysis of the first-round amplification products from the three samples confirmed the pestivirus specificity predicted by the novel assay for the ‘Hobi’-like pestivirus positive samples, whereas the two untyped caprine strains were recognised as true BDVs (Table 3).
Table 3.
Results of typing of field pestivirus strains by two nPCR assays.
| Method | Reference | BVDV-1 | BVDV-2 | ‘Hobi’-like | BDV | Not typed |
|---|---|---|---|---|---|---|
| nPCR (Erns) | Ref. [35] | 80 | 20 | NA | 0 | 0 |
| nPCR (5’ UTR, Npro, C) | This study | 80 | 15 | 3 | NA | 2b |
| Sequence analysisa | NA | ND | ND | 3 | 2b | NA |
NA, not applicable; ND, not done.
Sequence analysis was carried out only on the five pestivirus strains that were differently typed by the two nPCR assays.
Strains detected in tissue samples from dead kids, that were erroneously typed as BVDV-2 by the Erns nPCR and found to be true BDVs by sequence analysis.
3.3. Internal control detection
The IC was detected in all the examined samples, thus ruling out relevant RNA losses during nucleic acid extraction or DNA polymerase inhibition during amplifications.
4. Discussion
To date, at least twelve ‘Hobi’-like strains have been obtained worldwide, most of which were of Brazilian origin, having been detected in contaminated bovine serum batches [16], [18], [19], [20], in a buffalo [18] or in bovine aborted foetuses [22]. Only three viruses were recovered outside South America, one from a bovine serum in Thailand [21] and the other two from an Italian cattle herd [23], [24]. At the moment, the Italian viruses are the unique ‘Hobi’-like strains that were associated to overt disease in cattle.
The detection of the novel virus in different parts of the world posed some concerns about the ability of commonly used molecular assays to detect ‘Hobi’-like strains [16], [19], [20], [25]. The panpestivirus RT-PCR developed by Vilcek et al. [39], which is commonly used for BVDV molecular screening, does not detect ‘Hobi’-like sequences due to the presence of a mismatch at the 3′ end of primer 324 that prevents the correct primer annealing. Other conventional and real-time RT-PCR protocols are able to detect ‘Hobi’-like pestivirus but do not provide any virus typing, which is helpful to assess virus epidemiology [40], [41], [42], [43]. A TaqMan assay that was claimed to be specific for the new pestivirus (putative BVDV-3) was recently developed [26], but this assay could not type simultaneously BVDV-1 and BVDV-2 and showed a limited cross-reaction with high-titre BVDV-2 samples (N. Decaro, personal observation). If current diagnostic tests based on nucleic acid detection may miss ‘Hobi’-like strains, there may be antigen-detection kits that do detect the virus. These assays may have a certain value for pestivirus surveillance in cattle herds, but are likely to be less useful for eradication programs that require virus typing to determine the source of infection and evaluate lack of protection by using available vaccines.
In the present study, an nPCR assay able to detect and type simultaneously all bovine pestivirus species was developed and compared to a PCR protocol established previously for ruminant pestivirus typing targeting the Erns gene [35]. Although alternative PCR protocols have been established for BVDV-1/BVDV-2 typing [44], [45], the Sullivan and Akkina’s protocol was chosen for comparison with the novel assay, as it is employed routinely in our lab and all clinical samples processed in this study had been already analysed by that assay. Processing of field samples resulted in the identification of three additional ‘Hobi’-like strains in southern Italy, which had been mistyped as BVDV-2 by using the Erns protocol. Two of these strains were associated to abortion in the same cattle herd [24], whereas the third strain was recovered from an outbreak of respiratory disease (N. Decaro, unpublished). Although the novel assay was set up using only one ‘Hobi’-like reference strain (Italy-1/10-1), detection of these three additional strains in field samples accounts for its ability to correctly identify and type the new bovine pestivirus. Indeed, sequence analysis of the most divergent ‘Hobi’-like strain currently known, Th/04_KhonKaen [46], showed no point mutations in the specific-oligonucleotide binding regions, thus supporting the hypothesis that diverse ‘Hobi’-like strains are likely detected by the established nPCR assay. Nevertheless, ongoing testing would be needed to fully validate the test particularly with the limited ‘Hobi’-like strains tested.
Considering that most ‘Hobi’-like strains were detected in contaminated bovine serum batches, the established assay will warrant simultaneous screening of live vaccines and other biological products for the presence of all bovine pestiviruses.
Although nPCR protocols are inherently difficult to utilise in large scale testing without contamination of the facility, the established assay may overcome the limitation of existing methods. Presently, at least two separate real-time PCR assays should be run for typing all bovine pestiviruses and real-time PCR technology is not handled in all laboratories. In addition, lacking specific serological assays that are complicated by the cross-reactivity existing among different pestiviral species [16], [21], this molecular assay represents the unique tool currently available for unambiguous virus detection and typing during pestivirus surveillance activities in cattle.
Continuous epidemiological surveillance by using specific molecular methods will help assess to which extent ‘Hobi’-like pestivirus is widespread in cattle population worldwide and impacts on animal productions, thus requiring specific prophylactic measures.
References
- 1.Avalos-Ramirez R., Orlich M., Thiel H.-J., Becher P. Evidence for the presence of two novel pestivirus species. Virology. 2001;286:456–465. doi: 10.1006/viro.2001.1001. [DOI] [PubMed] [Google Scholar]
- 2.Becher P., Avalos Ramirez R., Orlich M., Cedillo Rosales S., König M., Schweizer M. Genetic and antigenic characterization of novel pestivirus genotypes: implications for classification. Virology. 2003;311:96–104. doi: 10.1016/s0042-6822(03)00192-2. [DOI] [PubMed] [Google Scholar]
- 3.Thiel H.-J., Collett M.S., Gould E.A., Heinz F.X., Houghton M., Meyers G. Family Flaviviridae. In: Fauquet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball L.A., editors. Virus taxonomy. Eighth report of the international committee on taxonomy of viruses. Elsevier/Academic Press; , London: 2005. pp. 978–987. [Google Scholar]
- 4.Thiel H.-J., Plagemann P.G.W., Moenning V. Pestiviruses. In: Fields B.N., Knipe D.M., Howley P.M., editors. Fields virology. 3rd ed. Lippincott-Raven; Philadelphia: 1996. pp. 1059–1073. [Google Scholar]
- 5.Pellerin C., van den Hurk J., Lecomte J., Tussen P. Identification of a new group of bovine viral diarrhea virus strains associated with severe outbreaks and high mortalities. Virology. 1994;203:260–268. doi: 10.1006/viro.1994.1483. [DOI] [PubMed] [Google Scholar]
- 6.Ridpath J.F., Bolin S.R., Dubovi E.J. Segregation of bovine viral diarrhea virus into genotypes. Virology. 1994;205:66–74. doi: 10.1006/viro.1994.1620. [DOI] [PubMed] [Google Scholar]
- 7.Baule C., van Vuuren M., Lowings J.P., Belák S. Genetic heterogeneity of bovine viral diarrhoea viruses isolated in Southern Africa. Virus Res. 1997;52:205–220. doi: 10.1016/s0168-1702(97)00119-6. [DOI] [PubMed] [Google Scholar]
- 8.Becher P., Orlich M., König M., Thiel H.J. Nonhomologous RNA recombination in bovine viral diarrhea virus: molecular characterization of a variety of subgenomic RNAs isolated during an outbreak of fatal mucosal disease. J Virol. 1999;73:5646–5653. doi: 10.1128/jvi.73.7.5646-5653.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couvreur B., Letellier C., Collard A., Quenon P., Dehan P., Hamers C. Genetic and antigenic variability in bovine viral diarrhea virus (BVDV) isolates from Belgium. Virus Res. 2002;85:17–28. doi: 10.1016/s0168-1702(02)00014-x. [DOI] [PubMed] [Google Scholar]
- 10.Flores E.F., Ridpath J.F., Weiblen R., Vogel F.S., Gil L.H. Phylogenetic analysis of Brazilian bovine viral diarrhea virus type 2 (BVDV-2) isolates: evidence for a subgenotype within BVDV-2. Virus Res. 2002;87:51–60. doi: 10.1016/s0168-1702(02)00080-1. [DOI] [PubMed] [Google Scholar]
- 11.Tajima M. Bovine viral diarrhea virus 1 is classified into different subgenotypes depending on the analyzed region within the viral genome. Vet Microbiol. 2004;99:131–138. doi: 10.1016/j.vetmic.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 12.Vilcek S., Alenius S., Paton D.J., Mittelholzer C., Belák S. Genetic clustering of bovine viral diarrhoea viruses in cattle farms: genetic identification and analysis of viruses directly from cattle sera. Vet J. 1999;158:33–38. doi: 10.1053/tvjl.1999.0363. [DOI] [PubMed] [Google Scholar]
- 13.Vilcek S., Drew T.W., McGoldrick A., Paton D.J. Genetic typing of bovine pestiviruses from England and Wales. Vet Microbiol. 1999;69:227–237. doi: 10.1016/s0378-1135(99)00111-x. [DOI] [PubMed] [Google Scholar]
- 14.Vilcek S., Paton D., Lowings P., Björklund H., Nettleton P., Belák S. Genetic analysis of pestiviruses at the 3′ end of the genome. Virus Genes. 1999;18:107–114. doi: 10.1023/a:1008000231604. [DOI] [PubMed] [Google Scholar]
- 15.Wolfmeyer A., Wolf G., Beer M., Strube W., Hehnen H.R., Schmeer N. Genomic (5′ UTR) and serological differences among German BVDV field isolates. Arch Virol. 1997;142:2049–2057. doi: 10.1007/s007050050222. [DOI] [PubMed] [Google Scholar]
- 16.Schirrmeier H., Strebelow G., Depner K., Hoffmann B., Beer M. Genetic and antigenic characterization of an atypical pestivirus isolate, a putative member of a novel pestivirus species. J Gen Virol. 2004;85:3647–3652. doi: 10.1099/vir.0.80238-0. [DOI] [PubMed] [Google Scholar]
- 17.Liu L., Xia H., Wahlberg N., Belák S., Baule C. Phylogeny, classification and evolutionary insights into pestiviruses. Virology. 2009;385:351–357. doi: 10.1016/j.virol.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Stalder H.P., Meier P., Pfaffen G., Wageck-Canal C., Rüfenacht J., Schaller P. Genetic heterogeneity of pestiviruses of ruminants in Switzerland. Prev Vet Med. 2005;72:37–41. doi: 10.1016/j.prevetmed.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 19.Peletto S., Zuccon F., Pitti M., Gobbi E., Marco L.D., Caramelli M. Detection and phylogenetic analysis of an atypical pestivirus, strain IZSPLV_To. Res Vet Sci. 2010 doi: 10.1016/j.rvsc.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 20.Ståhl K., Beer M., Schirrmeier H., Hoffmann B., Belák S., Alenius S. Atypical ‘HoBi’-like pestiviruses – recent findings and implications thereof. Vet Microbiol. 2010;142:90–93. doi: 10.1016/j.vetmic.2009.09.048. [DOI] [PubMed] [Google Scholar]
- 21.Ståhl K., Kampa J., Alenius S., Persson Wadman A., Baule C., Aiumlamai S. Natural infection of cattle with an atypical ‘HoBi’-like pestivirus—implications for BVD control and for the safety of biological products. Vet Res. 2007;38:517–523. doi: 10.1051/vetres:2007012. [DOI] [PubMed] [Google Scholar]
- 22.Cortez A., Heinemann M.B., De Castro M.G., Soares R.M., Pinto A.M., Alfieri A.A. Genetic characterization of Brazilian bovine viral diarrhea virus isolates by partial nucleotide sequencing of the 5′-UTR region. Pesquisa Veterinaria Brasileira. 2006;26:211–216. [Google Scholar]
- 23.Decaro N., Lucente M.S., Mari V., Cirone F., Cordioli P., Camero M. Atypical pestivirus and severe respiratory disease in calves, Europe. Emerg Infect Dis. 2011;17:1549–1552. doi: 10.3201/eid1708.101447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Decaro N, Lucente MS, Mari V, Sciarretta R, Pinto P, Buonavoglia D, et al. ‘Hobi’-like pestivirus in aborted bovine fetuses. J Clin Microbiol, in press. [DOI] [PMC free article] [PubMed]
- 25.Decaro N, Mari V, Lucente MS, Colaianni ML, Cirone F, Losurdo M, et al. Virus della diarrea virale bovina tipo 3 associato a malattia respiratoria. In: Atti XII Congresso Nazionale della Società Italiana di Diagnostica di Laboratorio Veterinaria (S.I.Di.L.V.), Genova; 27–29 Ottobre 2010. p. 96–97.
- 26.Liu L., Xia H., Belák S., Baule C. A TaqMan real-time RT-PCR assay for selective detection of atypical bovine pestiviruses in clinical samples and biological products. J Virol Methods. 2008;154:82–85. doi: 10.1016/j.jviromet.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Hall T.A. BioEdit: a user-friendly biological sequence alignment and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 28.Decaro N., Mari V., Desario C., Campolo M., Elia G., Martella V. Severe outbreak of bovine coronavirus infection in dairy cattle during the warmer season. Vet Microbiol. 2008;126:30–39. doi: 10.1016/j.vetmic.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pratelli A., Martella V., Tempesta M., Buonavoglia C. Characterization by polymerase chain reaction of ruminant rotaviruses isolated in Italy. New Microbiol. 1999;22:105–109. [PubMed] [Google Scholar]
- 30.Thiry J., Tempesta M., Camero M., Tarsitano E., Bellacicco A.L., Thiry E. A live attenuated glycoprotein E negative bovine herpesvirus 1 vaccine induces a partial cross-protection against caprine herpesvirus 1 infection in goats. Vet Microbiol. 2006;113:303–308. doi: 10.1016/j.vetmic.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Tempesta M., Marsilio F., Tiscar P.G., Buonavoglia D., Abdi Farah A., Vitellozzi G. Evaluation of the pathogenicity of a bovine herpesvirus-4 (BHV-4) strain in pregnant rabbits. Eur J Vet Pathol. 1996;2:83–85. [Google Scholar]
- 32.Buonavoglia C., Marsilio F., Tempesta M., Buonavoglia D., Cavalli A. Persistent pestivirus infection in sheep in Apulia (southern Italy) New Microbiol. 1994;17:163–165. [PubMed] [Google Scholar]
- 33.Buonavoglia C., Falcone E., Pestalozza S., Iovane G., Rivero V.B. Susceptibility of a minipig kidney cell line (MPK) to hog cholera virus. Microbiologica. 1988;11:263–264. [PubMed] [Google Scholar]
- 34.Decaro N., Camero M., Elia G., Martella V., Pratelli A., Gargano P. Malattia delle mucose da BVDV tipo 2: descrizione di un focolaio in Puglia. Large Anim Rev. 2004;Anno 10(2):29–34. [Google Scholar]
- 35.Sullivan D.G., Akkina R.K. A nested polymerase chain reaction assay to differentiate pestiviruses. Virus Res. 1995;38:231–239. doi: 10.1016/0168-1702(95)00065-x. [DOI] [PubMed] [Google Scholar]
- 36.Decaro N., Martella V., Ricci D., Elia G., Desario C., Campolo M. Genotype-specific fluorogenic RT-PCR assays for the detection and quantitation of canine coronavirus type I and type II RNA in faecal samples of dogs. J Virol Methods. 2005;130:72–78. doi: 10.1016/j.jviromet.2005.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pratelli A., Tempesta M., Greco G., Martella V., Buonavoglia C. Development of a nested PCR assay for the detection of canine coronavirus. J Virol Methods. 1999;80:11–15. doi: 10.1016/S0166-0934(99)00017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pratelli A., Martella V., Cirone F., Buonavoglia D., Elia G., Tempesta M. Genomic characterization of pestiviruses isolated from lambs and kids in southern Italy. J Virol Methods. 2001;94:81–85. doi: 10.1016/s0166-0934(01)00277-4. [DOI] [PubMed] [Google Scholar]
- 39.Vilcek S., Herring A.J., Herring J.A., Nettleton P.F., Lowings J.P., Paton D.J. Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch Virol. 1994;136:309–323. doi: 10.1007/BF01321060. [DOI] [PubMed] [Google Scholar]
- 40.Elvander M., Baule C., Persson M., Egyed L., Ballagi-Pordány A., Belák S. An experimental study of a concurrent primary infection with bovine respiratory syncytial virus (BRSV) and bovine viral diarrhoea virus (BVDV) in calves. Acta Vet Scand. 1998;39:251–264. doi: 10.1186/BF03547797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letellier C., Kerkhofs P., Wellemans G., Vanopdenbosch E. Detection and genotyping of bovine diarrhea virus by reverse transcription-polymerase chain amplification of the 5′ untranslated region. Vet Microbiol. 1999;64:155–167. doi: 10.1016/S0378-1135(98)00267-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaede W., Reiting R., Schirrmeier H., Depner K.R., Beer M. Detection and species-specific differentiation of pestiviruses using real-time RT-PCR. Berl Munch Tierarztl Wochenschr. 2005;118:113–120. [PubMed] [Google Scholar]
- 43.Hoffmann B., Depner K., Schirrmeier H., Beer M. A universal heterologous internal control system for duplex real-time RT-PCR assays used in a detection system for pestiviruses. J Virol Methods. 2006;136:200–209. doi: 10.1016/j.jviromet.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 44.Ridpath J.F., Bolin S.R. Differentiation of types 1a, 1b and 2 bovine viral diarrhoea virus (BVDV) by PCR. Mol Cell Probes. 1998;12:101–106. doi: 10.1006/mcpr.1998.0158. [DOI] [PubMed] [Google Scholar]
- 45.Gilbert S.A., Burton K.M., Prins S.E., Deregt D. Typing of bovine viral diarrhea viruses directly from blood of persistently infected cattle by multiplex PCR. J Clin Microbiol. 1999;37:2020–2023. doi: 10.1128/jcm.37.6.2020-2023.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu L., Kampa J., Belák S., Baule C. Virus recovery and full-length sequence analysis of atypical bovine pestivirus Th/04_KhonKaen. Vet Microbiol. 2009;138:62–68. doi: 10.1016/j.vetmic.2009.03.006. [DOI] [PubMed] [Google Scholar]


