Abstract
Stimulator of interferon gene (STING) plays an important role in the cyclic GMP-AMP synthase (cGAS)-mediated activation of type I IFN responses. In this study, we identified and cloned canine STING gene. Full-length STING encodes a 375 amino acid product that shares the highest similarity with feline STING. Highest levels of mRNA of canine STING were detected in the spleen and lungs while the lowest levels in the heart and muscle. Analysis of its cellular localization showed that STING is localizes to the endoplasmic reticulum. STING overexpression induced the IFN response via the IRF3 and NF-κB pathways and up-regulated the expression of ISG15 and viperin. However, knockdown of STING did not inhibit the IFN-β response triggered by poly(dA:dT), poly(I:C), or SeV. Finally, overexpression of STING significantly inhibited the replication of canine influenza virus H3N2. Collectively, our findings indicate that STING is involved in the regulation of the IFN-β pathway in canine.
Keywords: Canine, STING, Type I IFN, H3N2, CIV
Highlights
-
•
Here we first identified and cloned canine STING gene.
-
•
STING overexpression induced the IFN response via the IRF3 and NF-kB pathways.
-
•
Overexpression of STING inhibited the replication of H3N2 CIV.
1. Introduction
The innate immune response is an important defense against invading pathogens. Following infection, nucleic acids from microbes are pattern recognition receptors (PRRs) that subsequently trigger host innate immune responses [1]. Several cytosolic DNA sensors have been found, including cGAS [2], IFI16 (gamma interferon-inducible protein 16) [3], DAI (DNA-dependent activator of IFN-regulatory factors) [4], AIM2 (absent in melanoma 2) and RNA polymerase III [5]. The activation of DNA sensors transmit signals to adaptor proteins and activate signaling cascades that ultimately result in the production and secretion of interferons (IFNs), pro-inflammatory cytokines, and chemokines. Among the adaptor proteins, STING (stimulator of IFN genes), which resides in the endoplasmic reticulum (ER) or mitochondria-associated membranes, recruits TANK-binding kinase 1 (TBK1) and IRF3, and leads to the phosphorylation of IRF3 by TBK1 [6]. STING is also involved in modulating innate immune responses against RNA virus infections [7]. Human immunodeficiency virus (HIV), influenza A virus, Sendai virus (SeV), and vesicular stomatitis virus trigger STING signaling dependently and independently of DNA detection [8]. Additionally, STING may be associated with autoimmune diseases, lipid metabolism [8], and tumor development [9]. Upon infection with a DNA virus or retrovirus, cGAS recognizes cytosolic viral DNA and causes the production of type-I IFN and the expression of interferon stimulated genes (ISGs) [10]. However, some viruses have evolved the ability to inhibit the function of STING and evade the host antiviral defenses. Kaposi's sarcoma-associated herpesvirus (KSHV) encodes viral interferon regulatory factor 1 (vIRF1) that blocks STING by preventing its interaction with TANK binding kinase 1 (TBK1), thereby inhibiting STING's phosphorylation, resulting in an inhibition of the DNA sensing pathway [11]. HSV-1 UL46 protein binds to STING and blocks the interferon response triggered by 2′ 3’ -cGAMP [12]. Human T lymphotropic virus type 1 (HTLV-1) Tax and HBV polymerase reduce the K63-linked ubiquitination of STING and interfere with the interactions between STING and TBK1, which may promote the persistent infections [13], [14]. Except that some DNA viruses can regulate the cGAS-STING pathway, a recent study reported that enveloped RNA viruses, including influenza virus (IAV), are able to stimulate a cGAS-independent STING pathway, and IAV antagonizes the pathway through its conserved hemagglutinin fusion peptide (FP) interacting with STING [15].
STING orthologs have been characterized in many species including mouse, rat, pig, chicken, feline, and zebrafish [16], [17], [18], [19]. However, little is known about the characteristics and function of STING in canines. Here, we characterized canine STING and show that canine STING induces the activation of the IRF3, NF-κB pathways, and the expression of ISGs in MDCK cells. Knockdown of STING does not affect the production of IFN-β triggered by poly(dA:dT), poly(I:C), and SeV. Moreover, STING overexpression inhibits the replication of canine influenza virus (CIV) H3N2 in vitro.
2. Materials and methods
2.1. Tissues, cells, and viruses
Tissue samples from dogs were provided by the Experimental Animal Centre, Nanjing Agricultural University. MDCK cells, 293T cells and A549 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% antibiotics. Cells were cultured at 37 °C with 5% (v/v) CO2. SeV and CIV H3N2 were propagated in SPF chick embryos as previously described [20].
2.2. RNA preparation and cDNA synthesis
Total RNA was isolated from tissues and PBMCs using Trizol (pufei, China). The amount of DNA and RNA in each sample was quantified using a NanoDrop 2000 (Thermo). The preparation of cDNA from total RNA was performed using the RevertAid First Strand cDNA Synthesis Kit (Thermo).
2.3. Cloning and sequence analysis of canine STING
The full transcript of canine STING was obtained by RACE PCR (Invitrogen). Based on the results of 5′ and 3′ RACE, primers were designed (STING F1 and STING R1) to target the full coding region of canine STING (Table 1 ). PCR was performed using Phanta® Super-Fidelity DNA Polymerase (Vazyme). The PCR products were cloned into the pMD-18T vector (TaKaRa) and sequenced. The correct clone was named pMD-STING.
Table 1.
Primers used in this study.
| Primer | Sequence 5′-3′ |
|---|---|
| STING -F1 | ATGCTCCAGGCTAGCCTGCAC |
| STING-R1 | TCAGAAGATATCTGTGCGGAGTGGG |
| STING -pegfp-F | CCCTCGAGATGCTCCAGGCTAGCCTGCAC |
| STING -pegfp-R | CGGAATTCGGAAGATATCTGTGCGGAGTG |
| Flag-STING-F | CGGAATTCGGATGCTCCAGGCTAGCCTGCACCCAT |
| Flag-STING-R | CCCTCGAGTCAGAAGATATCTGTGCGGAGTGGG |
| q STING F | AACAACTGCCGCCTCATTG |
| q STING R | GCCCATAGTAACCTCCCTTTC |
| qGAPDH F | GGTCACCAGGGCTGCTTT |
| qGAPDH R | ATTTGATGTTGGCGGGAT |
| qVirepin F | AGATTAAAGCCCTGAACCC |
| qVirepin R | TCATCGCTGATAACAAACC |
| qISG 15-1 F | AGTATCGCCTACGAGGTCTG |
| qISG 15-1 R | ATGGGCTTCCCTTCAAAA |
| qCIV-m F | TAAGGCGACGATAAATACA |
| qCIV-m R | CCAGAAACGAATGGGAGT |
STING orthologs proteins were aligned using the Clustal X 2.0 program. A phylogenetic tree was constructed using the neighbor-joining method with 1000 bootstraps using the MEGA5 software. The functional domains, motifs, and features of canine STING were predicted using the Sample Modular Architecture Research Tool (SMART).
2.4. Plasmid construction
Canine STING was amplified from the plasmid pMD-STING and inserted into the pCMV-DYKDDDDK-N plasmid (N-terminal Flag tag; Flag-STING) and the pEGFP-N1 plasmid (C-terminal EGFP tag; STING-EGFP). All constructs were verified by sequencing.
2.5. Analysis of the expression levels of STING in different tissues and its subcellular localization
STING mRNA levels were evaluated in canine tissues using quantitative real-time PCR (qPCR). Individual transcripts from each sample were normalized to the mRNA level of glyceraldehyde 3-phosphate dehydrogenase (GADPH) (internal control). qPCR was performed using SYBR Green master Mix (Vazyme) with the primer pairs shown in Table 1.
To determine the subcellular localization of STING, MDCK cells were seeded in Glass Bottom Dishes (Nunc) at 50% confluence per dish. The plasmid pDsRed2-ER (Clontech), which encodes the DsRed2a fluoresce protein that localizes to the ER [17], was cotransfected with the STING-EGFP vector using the Lipofectamine 3000 (Invitrogen). 24 h post-transfection, the cells were washed three times with PBST and then fixed with 4% paraformaldehyde at −20 °C for 20 min. Following permeabilization with 0.2% Triton X-100 for 20 min at room temperature, the cells were stained using 1 μM DAPI (Sangon, China) for 10 min. A confocal laser scanning microscope (Nikon) was used to detect fluorescent signals.
2.6. Reagents
Poly(I:C) high molecular weight and poly(dA:dT) were purchased from InvivoGen. Cells were stimulated with either poly(I:C) or poly(dA:dT) using Lipofectamine 2000 (Invitrogen) at a 1:1 (wt/v) ratio of which was added to the cells at a concentration of 4 μg/mL.
2.7. Reporter plasmids and luciferase assays
Luciferase reporter plasmids for the evaluation of canine IFN-β promoter (IFN-Luc), NF-κB response element (3×PRDII-Luc), IRF3 response element (3×PRDIII/I-Luc), and ISRE promoter (ISRE-Luc) were constructed as previously described [21]. Cells grown in 48-well plates were co-transfected with 0.25 μg of luciferase reporter plasmids and 0.25 μg of various expression plasmids using the GenJet™ In Vitro DNA Transfection Reagent. As an internal control, the pRL-TK plasmid (Promega) (0.01 μg per well) was simultaneously transfected. After 24 h of transfection, cells were stimulated with SeV, poly(I:C), poly(dA:dT), or left untreated. Cells were lysed 12 h after stimulation. Luciferase activity was determined using a Dual-Luciferase Assay Kit according to the manufacturer's instructions (Promega). All reporter assays were performed in triplicate and repeated three independent times. The relative level of luciferase activity in each sample was normalized to Renilla luciferase activity. The graphs represent the average and standard deviations (SD).
2.8. RNA interference
MDCK cells were transfected with siRNA using Lipofectamine 2000. Knockdown efficiency was determined using real-time PCR and western blot analysis. Cells were co-transfected with 0.25 μg of luciferase reporter plasmid, 0.02 μg of pRL-TK plasmid, and 5 pmol of siRNA. After 24 h of transfection, cells were stimulated using SeV, poly(I:C), poly(dA:dT), or left untreated. After 12 h of stimulation, cells were lysed and luciferase activity determined. The following siRNA were used: siSTING1, TCCCTGTTGTCTTCCAGAA (75–93); siSTING2, CAGGCATTGCACAACAACA (657–675); and siSTING3, TGCACAACAACATGCTACA (664–682). The siRNAs for canine STING and the control siRNA were produced by RiboBIO (Guangzhou, China).
2.9. Western blot analysis
Briefly, cells were co-transfected with Flag-STING and siRNA. At 24 h or 36 h post-transfection, cells were lysed using NP-40 lysis buffer (Beyotime, China) containing 1 mM of PMSF. Lysates were then collected and centrifuged. A total of 30 μg of each protein sample was separated using 12% SDS-PAGE and then transferred into nitrocellulose membranes. Membranes were then blocked in 5% skimmed milk for 2 h at room temperature and incubated overnight at 4 °C or for 2 h at room temperature with specific mouse anti-Flag antibody (Huabio, China), mouse anti-NP antibody (eEnzyme, USA) or rabbit anti-GAPDH antibody (Goodhere Biotechnology, China). Membranes were then incubated at 37 °C for 1 h with hrp-conjugated anti-mouse IgG or anti-rabbit IgG (KPL) at a 1:10,000 dilution in 5% skimmed milk.
2.10. Statistics
The data are presented as the mean ± SD. Statistical significance was determined using unpaired student's t-tests and one-way ANOVA using the Prism 5.0 software (GraphPad Software). For all tests, a p < 0.05 was considered to indicate a significant difference.
3. Results
3.1. Cloning and sequence analysis
Based on the published canine genome and sequences of STING orthologous, specific primers were designed to clone full-length canine STING using 5′ and 3′ RACE from total RNA isolated from PBMCs. Sequence comparisons indicated that the coding sequence (CDS) of canine STING is 1128 bp long and encodes 376 amino acids (aa).
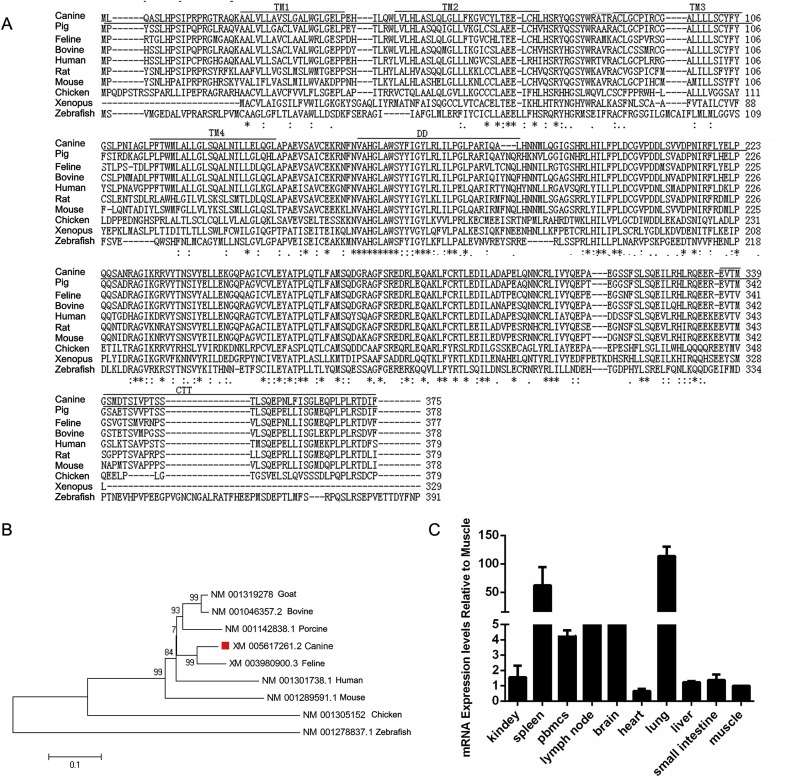
Comparison of the structural characteristics of STING orthologs revealed that canine STING has four transmembrane (TM) domains (21–139 aa) in its N-terminal region, a central c-di-GMP-binding domain (CBD, 155–335 aa), and a C-terminal tail (CTT, 336–375 aa) (Fig. 1 A). Moreover, canine STING has a dimerization domain (DD, 155–180 aa) within the CBD domain (Fig. 1A). All of these domains are highly conserved in mammals.
Fig. 1.
Characterization of canine STING. (A) Alignment of the amino acid (aa) sequence of STING orthologs was performed using Clustal X. The numbers indicate the aa positions. Asterisks or dots indicate identical or similar aa residues. Gaps were added to optimize the alignment. The canine STING sequence is underlined. (B) Neighbor joining tree based on protein sequences is shown to describe the relationships between canine STING and other STING orthologs. Scale bar = 0.1. (C) Quantitative analysis of the distribution of canine STING transcripts in healthy canine organs. STING mRNA levels in different organs are presented relative to mRNA expression in muscle. The error bars indicate standard deviations.
Nucleotide and protein alignments of STING orthologs revealed that canine STING shares the highest similarity with feline STING (90.1% at the nucleotide and 84.8% at the protein level). The levels of protein identity with goat, bovine, porcine, human, mouse, chicken and zebrafish were 88.2%, 87.3%, 85.9%, 78.3%, 76.4%, 57.9% and 48.0% respectively. Phylogenic analysis showed that canine STING belongs to the group containing feline STING (Fig. 1B).
3.2. Organ-specific expression analysis of canine STING
Next, the expression profiles of STING in canine organs were evaluated using qPCR. Canine STING mRNA was expressed widely in all examined organs. Moreover, spleen and lungs displayed the most abundant expression of STING (Fig. 1C). This indicates that STING is expressed in both immune and non-immune cells.
3.3. Subcellular localization analysis of canine STING
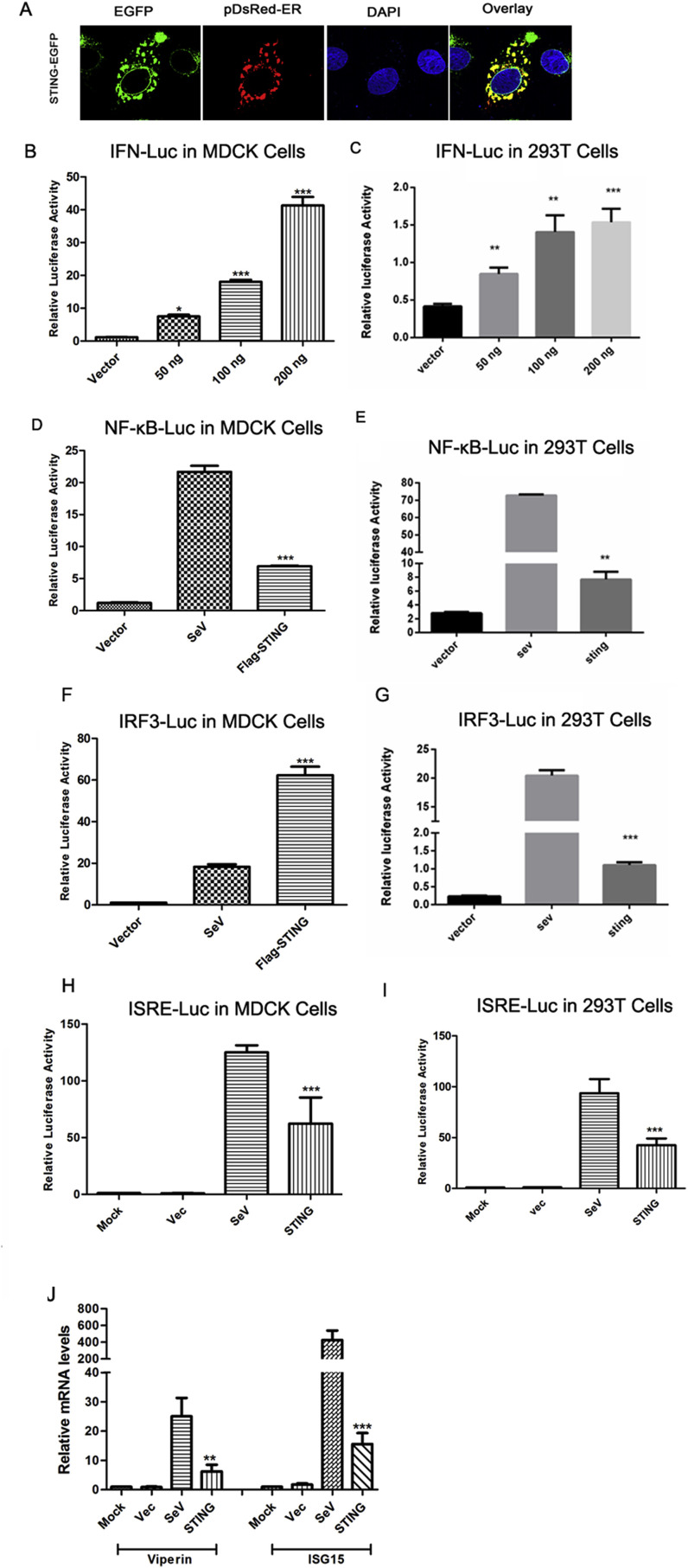
Inspection of the sequence of canine STING revealed the presence of a N-terminal RPR motif at aa positions 12–14 (Fig. 1A). Motifs such as KDEL, KKXX, and RXR are necessary for ER retention of proteins [22]. To determine the subcellular localization of canine STING, a STING-EGFP expression vector was constructed. Confocal microscopy indicated that the STING-EGFP fusion protein co-localized with the DsRed-ER marker indicating that canine STING localizes to ER membrane (Fig. 2 A).
Fig. 2.
Subcellular localization and function analysis of canine STING. (A) MDCK cells were co-transfected with C-terminal EGFP-tagged STING (STING-EGFP) and pDsRed2-ER. Cells were stained with DAPI 24 h post-transfection and analyzed using confocal microscopy. (B, C) MDCK Cells (B) and 293T cells (C) were co-transfected with 50–200 ng of the plasmid Flag-STING and IFN-Luc along with 0.01 μg of pRL-TK. (D–G) MDCK Cells (D, F, H) and 293T cells (E, G, I) were co-transfected with 0.25 μg of one of reporter plasmids-NF-κB-Luc (D, E), IRF3-Luc (F,G) or ISRE-Luc (H, I) along with 0.02 μg of pRL-TK and 0.25 μg of the Flag-STING or empty vector. SeV stimulation was used as a positive control. 24 h after transfection luciferase activity was measured. (J) MDCK cells were transfected with 0.25 μg of the indicated expression plasmid, Flag-STING, or empty vector. SeV infection was used as a positive control. 24 h post-transfection the mRNA levels of ISG15 and viperin were examined using qPCR. ISGs levels were normalized to the level of GAPDH. * represent differences between experimental and control groups (p < 0.05).
3.4. Overexpression of canine STING leads to activation of the IFN-β promoter via both the NF-κB and IRF3 pathways
As a signaling adaptor, STING plays a pivotal role in the activation of type I IFN responses during infection with DNA viruses, some RNA viruses, and bacteria. To identify whether canine STING is involved in the induction of type I IFN, a IFN-β-luciferase reporter vector was used. As shown in Fig. 2B and C, overexpression of canine STING led to a significant increase in IFN-β promoter activity compared to empty controls in a dose-dependent manner on the MDCK cells and 293T cells.
STING could induce the activation of the transcription factors NF-κB and IRF3 [23], [24]. To further investigate whether canine STING is also involved in both pathways, the canine STING expression vector was co-transfected with NF-κB-Luc or IRF3-Luc. As shown in Fig. 2D–G, canine STING significantly activated the NF-κB and IRF3 pathways on the MDCK cells and 293T cells.
Previous studies demonstrated that STING indirectly activates the JAK/STAT1 pathway by elevating the expression of type I IFNs [24], [25]. To examine the role of canine STING in regulating antiviral signaling, we examined the effect of STING expression on the activation of the ISRE promoter. As shown in Fig. 2H and I, expression of canine STING led to a significant induction of the ISRE promoter on the MDCK cells and 293T cells. Moreover, expression of STING increased the mRNA levels of ISG15 and viperin (Fig. 2J), indicating that canine STING can activate JAK/STAT1 pathway. Collectively, these results suggest that canine STING is a potent stimulator in the IFN response.
3.5. Knockdown of STING does not affect the activation of the IFN-β promoter mediated by poly(dA:dT), poly(I:C), and SeV
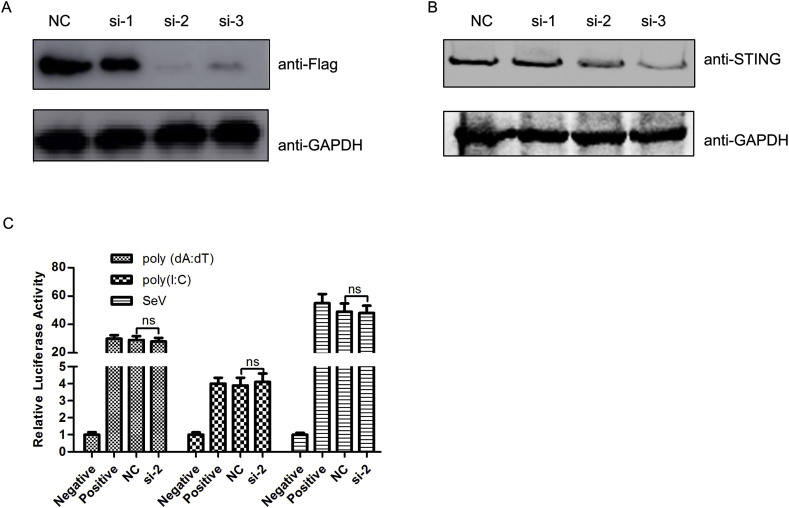
To further understand the role of canine STING in innate immune responses, we analyzed the effect of decreased STING expression on the IFN response in MDCK cells using RNAi. As shown in Fig. 3 A and B, both the levels of exogenous and endogenous STING were reduced by the transfection of siRNAs (si-2 and si-3).
Fig. 3.
Knockdown of canine STING does not block poly(dA:dT)-, poly(I:C)- and SeV-mediated activation of the IFN-β promoter. (A) MDCK cells were co-transfected with 0.25 μg of Flag-STING and 5 pmol of siRNA or a negative control siRNA. 24 h post-transfection, the expression of exogenous STING was examined by western blot. (B) Cells were transfected with 5 pmol of siRNA or a negative control siRNA (si-Negative). 24 h post-transfection, the expression of endogenous STING was examined using western blot. (C) MDCK cells were co-transfected with 5 pmol of siRNA or a negative control siRNA in addition to 0.01 μg of pRL-TK and 0.25 μg of IFN-Luc. 24 h post-transfection, cells were transfected with 4.0 μg of poly(dA:dT) or poly(I:C), or infected with SeV. After 12 h, cells were lysed and luciferase activity quantified. * represent differences between experimental and control groups (p < 0.05).
To investigate whether canine STING is involved in the type I IFN response triggered by dsDNA poly(dA:dT) and dsRNA poly(I:C), MDCK cells were co-transfected with an IFN-β reporter plasmid and si-2 siRNA. Cells were transfected with poly(dA:dT) or poly(I:C) or infected with SeV at 24 h post-transfection. After 24 h post-stimulation, the levels of luciferase activity were determined. As shown in Fig. 3C, poly(dA:dT), poly(I:C), and SeV were able to activate the IFN-β promoter however, knockdown of STING did not block this activation. Taken together, these data indicate that canine STING may be not necessary for the type I IFN response triggered by poly(dA:dT), poly(I:C), and SeV.
3.6. Overexpression of STING inhibits the replication of canine influenza virus H3N2
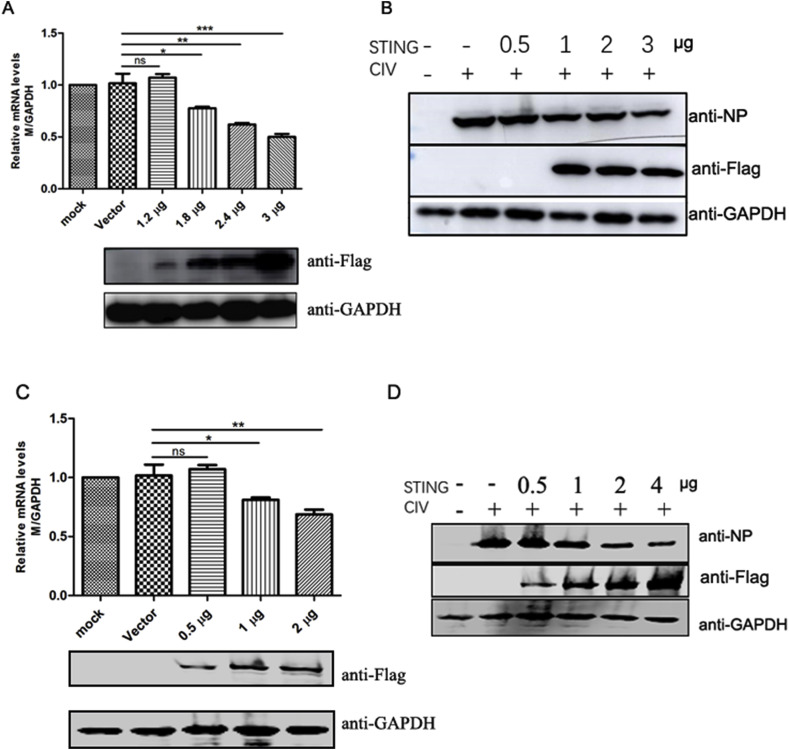
To investigate the role of STING in inhibiting virus replication, we analyzed the effect of canine STING over-expression on CIV H3N2 replication. MDCK cells were transfected with canine STING-expressing plasmid and 24 h after infected with CIV H3N2. Virus RNA levels were determined 24 h post infection. As shown in Fig. 4 A, the virus RNA levels decreased between 25% and 50% in a dose-dependent manner with canine STING expression and expression of viral NP protein was also decreased (Fig. 4B). Moreover, we confirmed the anti-CIV activity of canine STING on A549 cells (Fig. 4C and D). Taken together, these data indicate that canine STING can inhibit CIV H3N2 replication.
Fig. 4.
Overexpression of canine STING inhibits the replication of CIV H3N2. (A, B) MDCK cells were transfected with different doses of Flag-STING or empty vector. 24 h after transfection, cells pre-inoculated in 12 well plate were infected with 1000 TCID50 of CIV H3N2. The RNA levels of viral M gene (A) and the expression of viral NP gene (B) were determined by qPCR and Western blotting, respectively, 24 h after infection. * represent differences between experimental and control groups (p < 0.05). (C, D) A549 cells were transfected with different doses of Flag-STING or empty vector. 24 h after transfection, cells pre-inoculated in 12 well plate were infected with 10000 TCID50 of CIV H3N2. The RNA levels of viral M gene (C) and the expression of viral NP gene (D) were determined by qPCR and Western blotting, respectively, 24 h after infection.
4. Discussion
There are a variety of infection diseases which may have a major threat to public health among dogs, such as canine parvovirus, rabies virus, influenza virus, coronavirus and so on [26], [27], [28], [29], [30], [31], so the research of canine innate immunity should be gave more attention in future study. STING is an important signaling adaptor in the activation of type I IFN responses during infection with DNA and RNA viruses and bacteria. However, it has emerged as a target that is recognized and antagonized by viruses including DENV, HCV, HCoV-NL63, PEDV, and SARS-CoV to ultimately block the IFN pathway [7]. In this study, we characterized canine STING and found that its overexpression leads to the induction of the IFN response via IRF3 and NF-κB. However, knockdown of STING did not inhibit the IFN-β response triggered by poly(dA:dT), poly(I:C), or SeV. Finally, overexpression of STING significantly inhibited the replication of canine influenza virus H3N2. Collectively, our findings indicate that STING is involved in the regulation of the IFN-β pathway in canine.
A previous study indicated that feline and canine MAVS share the highest similarity [32]. Here, we found that canine STING is most similar at the sequence level to feline STING, which may be due to the closed relationship between canines and felines.
Structure analysis indicated that canine STING contains TM, DD, and CTT domains, similarly to human, mouse, and other vertebrates [33]. The TM domain of human STING interacts with other adaptor molecules and it is necessary for its localization to the ER membrane or mitochondria [34]. In this region, some conserved motifs, such as KDEL, KKXX, and RXR are necessary for ER retention of proteins [22]. In this study, we found that canine STING contains an RPR motif at positions 12–14 aa and localizes to ER membrane.
STING is a key adaptor molecule of the immune system and is involved in antiviral responses against intracellular bacteria and viruses as well as anti-tumor responses [35]. STING overexpression activates type I IFN through the IRF3 and NF-κB pathways [24]. In this study, canine STING induced the activation of the IFN-β promoter via the IRF3 and NF-κB pathways as well. We also found that canine STING activated the JAK/STAT1 pathway. These data indicate that the STING-mediated innate immune response is conserved in canines. However, knockdown of canine STING did not inhibit the activation of the IFN-β promoter, which so far has not been reported for other species. We speculate that there is an alternative molecule that has a similar function to STING in canines.
The STING pathway is more commonly used by the host to inhibit the replication of viruses such as HSV-1. In the absence of STING, viruses replicate more efficiently, demonstrating a protective antiviral role of the STING pathway [36]. To check whether canine STING is involved in an antiviral response, we analyzed its ability to inhibit the replication of CIV H3N2 in vitro. We found that overexpression of canine STING inhibited CIV H3N2 replication in a dose-dependent manner.
Conflict of interest
The authors declare no financial or commercial conflicts of interest.
Acknowledgements
This work was funded by the Fundamental Research funds for the central Universities. (Grant No. KYZ201633; KYZ201730).
References
- 1.Brubaker S.W. Innate immune pattern recognition: a cell biological perspective. Annu. Rev. Immunol. 2015;33:257–290. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun L.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Unterholzner L. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010;11(11) doi: 10.1038/ni.1932. 997-U42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takaoka A. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448(7152):501. doi: 10.1038/nature06013. -U14. [DOI] [PubMed] [Google Scholar]
- 5.Chiu Y.H., MacMillan J.B., Chen Z.J.J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138(3):576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka Y., Chen Z.J. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 2012;5(214) doi: 10.1126/scisignal.2002521. p. ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maringer K., Fernandez-Sesma A. Message in a bottle: lessons learned from antagonism of STING signalling during RNA virus infection. Cytokine Growth Factor Rev. 2014;25(6):669–679. doi: 10.1016/j.cytogfr.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y., Wilson H.L., Kiss-Toth E. Regulating STING in health and disease. J. Inflamm. (Lond) 2017;14:11. doi: 10.1186/s12950-017-0159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He L. STING signaling in tumorigenesis and cancer therapy: a friend or foe? Cancer Lett. 2017;402:203–212. doi: 10.1016/j.canlet.2017.05.026. [DOI] [PubMed] [Google Scholar]
- 10.Sun L. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Z. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc. Natl. Acad. Sci. U. S. A. 2015;112(31) doi: 10.1073/pnas.1503831112. p. E4306-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deschamps T., Kalamvoki M. Evasion of the STING DNA sensing pathway by the VP11/12 of herpes simplex virus type 1. J. Virol. 2017;91(16) doi: 10.1128/JVI.00535-17. pii: e00535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J. HTLV-1 Tax impairs K63-linked ubiquitination of STING to evade host innate immunity. Virus Res. 2017;232:13–21. doi: 10.1016/j.virusres.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y. Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways. J. Virol. 2015;89(4):2287–2300. doi: 10.1128/JVI.02760-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm C.K. Influenza A virus targets a cGAS-independent STING pathway that controls enveloped RNA viruses. Nat. Commun. 2016;7:10680. doi: 10.1038/ncomms10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie L. Molecular cloning and functional characterization of porcine stimulator of interferon genes (STING) Dev. Comp. Immunol. 2010;34(8):847–854. doi: 10.1016/j.dci.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Cheng Y. Chicken STING mediates activation of the IFN gene independently of the RIG-I gene. J. Immunol. 2015;195(8):3922–3936. doi: 10.4049/jimmunol.1500638. [DOI] [PubMed] [Google Scholar]
- 18.Ge R. Conservation of the STING-mediated cytosolic DNA sensing pathway in zebrafish. J. Virol. 2015;89(15):7696–7706. doi: 10.1128/JVI.01049-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X. The function of feline stimulator of interferon gene (STING) is evolutionarily conserved. Vet. Immunol. Immunopathol. 2016;169:54–62. doi: 10.1016/j.vetimm.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Su S. Identification of the IFN-beta response in H3N2 canine influenza virus infection. J. Gen. Virol. 2016;97(1):18–26. doi: 10.1099/jgv.0.000322. [DOI] [PubMed] [Google Scholar]
- 21.Tian J. Assessment of the IFN-beta response to four feline caliciviruses: infection in CRFK cells. Infect. Genet. Evol. 2015;34:352–360. doi: 10.1016/j.meegid.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Sun W. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. U. S. A. 2009;106(21):8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abe T., Barber G.N. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-kappaB activation through TBK1. J. Virol. 2014;88(10):5328–5341. doi: 10.1128/JVI.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burdette D.L., Vance R.E. STING and the innate immune response to nucleic acids in the cytosol. Nat. Immunol. 2013;14(1):19–26. doi: 10.1038/ni.2491. [DOI] [PubMed] [Google Scholar]
- 26.Su S. Epidemiology, evolution, and recent outbreaks of avian influenza virus in China. J. Virol. 2015;89:8671–8676. doi: 10.1128/JVI.01034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He W. Codon usage bias in the N gene of rabies virus. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2017;54:458. doi: 10.1016/j.meegid.2017.08.012. 54. [DOI] [PubMed] [Google Scholar]
- 28.Li G. Evolutionary and genetic analysis of the VP2 gene of canine parvovirus. BMC Genom. 2017;18(1):534. doi: 10.1186/s12864-017-3935-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su S. Epidemiology, evolution, and pathogenesis of H7N9 influenza viruses in five epidemic waves since 2013 in China. Trends Microbiol. 2017;25(9):713. doi: 10.1016/j.tim.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Su S. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan J. One health strategies for rabies control in rural areas of china. Lancet Infect. Dis. 2017;17(4):365. doi: 10.1016/S1473-3099(17)30116-0. [DOI] [PubMed] [Google Scholar]
- 32.Wu H. Molecular cloning and functional characterization of feline MAVS. Immunol. Res. 2015;64(1):82–92. doi: 10.1007/s12026-015-8682-9. [DOI] [PubMed] [Google Scholar]
- 33.Xie L.L. Molecular cloning and functional characterization of porcine stimulator of interferon genes (STING) Dev. Comp. Immunol. 2010;34(8):847–854. doi: 10.1016/j.dci.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 34.Zhong B. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29(4):538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 35.He L. Nucleic acid sensing pattern recognition receptors in the development of colorectal cancer and colitis. Cell Mol. Life Sci. 2017;74(13):2395–2411. doi: 10.1007/s00018-017-2477-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X. STING requires the adaptor TRIF to trigger innate immune responses to microbial infection. Cell Host Microbe. 2017;21(6):788. doi: 10.1016/j.chom.2017.05.007. [DOI] [PubMed] [Google Scholar]