Abstract
The spike (S) protein of the infectious bronchitis virus (IBV) plays a central role in the pathogenicity, the immune antibody production, serotype and the tissue tropism. In this study, we generate 11 monoclonal antibodies (mAbs) against S1 subunit of IBV Sczy3 strain, and two mAbs 1D5 and 6A12 were positive in indirect ELISA against both His-S1 protein and the purified whole viral antigen. MAb 6A12 and 1D5 could recognized by other 10 IBV strains (IBVs) from five different genotypes, except that 1D5 had a relatively low reaction with two of the 10 tested IBVs. End-point neutralizing assay performed in chicken embro kidney (CEK) cells revealed that the neutralization titer of 6A12 and 1D5 against Sczy3 reached 1:44.7 and 1:40.6, respectively. After screening a phage display peptide library and peptide scanning, we identified two linear B-cell epitopes that were recognized by the mAbs 1D5 and 6A12, which corresponded to the amino acid sequences 87PPQGMAW93 and 412IQTRTEP418, respectively, in the IBV S1 subunit. Sequences comparison revealed that epitope 412IQTRTEP418 was conserved among IBVs, while the epitope 87PPQGMAW93 was relatively variable among IBVs. The novel mAbs and the epitopes identified will be useful for developing diagnostic assays for IBV infections.
Keywords: Infectious bronchitis virus (IBV), Monoclonal antibodies, Phage random peptide library, Neutralizing antigenic epitope
1. Introduction
Avian infectious bronchitis (IB) is a highly contagious, acute, and economically important disease of chickens. It can infect all ages of the chickens and replicate in many tissues, causing respiratory symptom, diarrhea, reduced weight gain and feed efficiency, and decline of egg production and quality (Cavanagh, 2007). The etiologic agent of IB is the infectious bronchitis virus (IBV), a member of the Coronaviridae family, subfamily coronavirinae, genus gamma-coronavirus that replicates primarily in the respiratory tract and also in some epithelial cells of the ileum, kidney, and oviduct (Abd et al., 2009, Benyeda et al., 2010). Currently, dozens of IBV serotypes and genotypes have been described. Although vaccines are now being used widely and extensively, outbreaks of IB still occur frequently because of infections with field strains that differ serologically from vaccine strains (Li et al., 2010, Zou et al., 2010, Mahmood et al., 2011).
IBV is an enveloped, non-segmented, single-stranded, positive-sense RNA virus with a genome of approximately 27.6 kb (Boursnell et al., 1987). The remaining one-third of the genome encodes four main structural proteins: the spike glycoprotein (S), the small membrane protein (E), the integral membrane protein (M), and the nucleocapsid protein (N) (Lai et al., 1981). The S glycoprotein is a large type I transmembrane glycoprotein that is responsible for receptor binding and membrane fusion (Cavanagh et al., 1986). It consists of the N-terminal S1 and C-terminal S2 subunits, which are generated during post-translational cleavage of S protein (Jackwood et al., 2001). The S1 subunit is the main protein to induce the protective antibodies, virus-neutralization antibodies, hemagglutination inhibition antibodies, cross-reactivity ELISA antibodies and cell-mediated immune response (Cavanagh et al., 1997, Wang et al., 2009, Zhang et al., 2014). The S1 protein is involved in cell attachment and carries virus-neutralizing and serotype-specific determinants. It also plays an important role in tissue tropism and the degree of virulence of the virus (Casais et al., 2003).
To determine the antigenic epitopes is critical for the vaccine development and elucidation of the mechanism of virus-antibody interactions. It is also important to analyze the antigenic epitopes of prevalent strains of IBV for better understanding the antigenic change of IBV in genetic evolution. The antigenic epitopes of many viruses have been defined, and there are few reports on the antigenic epitope analysis of IBV proteins which have focused mainly on the S, N and M proteins (Boots et al., 1991, Hu et al., 2007, Seah et al., 2000, Xing et al., 2009, Yu et al., 2010). In the present work, mAbs against the S1 subunit derived from China isolate of an QX-like IBV strain Sczy3 was produced and the neutralizing activity of each mAbs was analyzed by neutralizing test on chicken embryonated kidney (CEK) cells. The neutralizing antibodies were then further used for antigenic epitopes analysis by using phage display peptide library, and the linear epitopes were analyzed.
2. Methods and materials
2.1. Cell line and viruses
The myeloma cell line SP2/0 was purchased from the ATCC and cultured in Dulbecco's modified eagle medium (DMEM, Gibco BRL, Paisley, UK) with 10% fetal calf serum (FCS, Gibco), and maintained at 37 °C and 5% CO2. IBV Sczy3 strain was isolated in 2009 from a broiler chickens in Sichuan Province (Zou et al., 2010). Other ten IBV strains from 5 different genotypes were selected for cross-reaction analysis (Table 1 , marked with c). Newcastle disease virus (NDV) and subtype H9 avian influenza virus (AIV) and subtype H5 AIV antigen were selected for specificity analysis. All viruses were isolated by us before and stored at −70 °C, except that the H5 AIV antigen was purchased from Qingdao Yebio Bioengineering Co., Ltd (Qingdao, China). IBV Sczy3 strain was purified using differential velocity centrifugation.
Table 1.
Background information of IBV strains used in the present study.
| IBV Strain | Countrya | Yearb | Type (Genotype) (Zou et al., 2010) | Genbank accession number |
|---|---|---|---|---|
| CK/CH/SCEM/09Ic | China | 2009 | LX4 | GU384207 |
| CK/CH/SCMS/10Ic | China | 2010 | LX4 | HM106334 |
| CK/CH/SCYA/10Ic | China | 2010 | Proventriculus-Type | HM363027 |
| CK/CH/SCMY/10Ic | China | 2010 | TW-I | HM363028 |
| 4/91c | Vaccine | 1992 | 4/91 | AF093794 |
| W93c | Vaccine | – | Mass | AY842862 |
| M41c | Vaccine | 1965 | Mass | DQ834384 |
| 28/86c | Vaccine | – | Mass | AY846750 |
| Ma5c | Vaccine | – | Mass | AY561713 |
| H120c | Vaccine | – | Mass | EU822341 |
| Beaudette C | USA | 1937 | Mass | M95169 |
| H52 | Vaccine | – | Mass | AF352315 |
| A2 | China | 1996 | CK/CH/LSC/99I-Type | EU526388 |
| SAIB14 | China | unknown | JP-Type | AF397527 |
| SAIBK | China | unknown | CK/CH/LSC/99I | DQ288927 |
| CK/CH/LSC/95I | China | 1995 | CK/CH/LSC/99I-Type | DQ167146 |
| tl/CH/LDT3/03 | China | 2003 | CK/CH/LSC/99I-Type | AY702975 |
| DY04 | China | 2004 | CK/CH/LSC/99I-Type | GQ265950 |
| DY05 | China | 2005 | LX4 | GQ265928 |
| CQ04-1 | China | 2004 | CK/CH/LSC/99I-Type | GQ265952 |
| CK/CH/LSD/07III | China | 2007 | LX4 | FJ345385 |
| CK/CH/LSD/08I | China | 2008 | LX4 | GQ258336 |
| CK/CH/LJS/08 | China | 2008 | LX4 | GQ258321 |
| HB08 | China | 2008 | LX4 | GQ265934 |
| LX4 | China | 1999 | LX4 | AY338732 |
| QXIBV | China | 1997 | LX4 | AF193423 |
| JP8127 | Japan | 1993 | JP-Type | AY296744 |
| 2575/98 | Taiwan | 1998 | TW-I | AY606314 |
| Italy02 | Italy | 1999 | Italy-02 | AM260962 |
| J2 | China | Before 1999 | Proventriculus-Type | AF286303 |
| Q1 | China | Before 1999 | Proventriculus-Type | AF286302 |
Country where the viruses were isolated.
Year when viruses were isolated.
Strains used in cross-reactivity test.
2.2. Prokaryotic expression and purification of the S1 recombinant protein
The S1 sequence was amplified from Sczy3 viral RNA by RT-PCR using forward primer S1F: 5′-GGATCCTCCATAGCTATGACAGCACCTC-3′ with BamHI site (underlined) comprising position 241-162 of S1 coding sequence and reverse primer S1R: 5′-CTCGAG TCACATTAATACTAAGGGCTCCGT-3′ with XhoI site (underlined) comprising position 1266-1246 of S1 coding sequence. The amplified S1 gene was ligated with the prokaryotic expression vector pET-32a (+) and transformed into Escherichia coli BL21 (DE3) competent cell. The transformation cells were induced with 1 mM Isopropyl β-d-1-Thiogalactopyranoside (IPTG), and the recombinant fusion proteins were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The recombinant proteins were then purified by HisTrap FF crude Columns (GE Healthcare Life Sciences, Piscataway, NJ, USA) as the manufacturer's introductions and named as pET-S1.
2.3. Preparation of monoclonal antibodies against IBV S1 subunit protein
SPF six- to eight-week-old female BALB/c mice were purchased from the Chengdu Dossy biological technology Co., Ltd (Chengdu, China). Each of five mice was immunized subcutaneously with 200 μg of the purified virus particles with an equal volume of adjuvant three times at 2-week intervals. Three days after the final booster immunizations with 100 μg purified virus, the mice were sacrificed, and spleen cells were harvested and fused with SP2/0 myeloma cells in the presence of polyethylene glycol (PGE1450, Sigma Chemical Co., St Louis, USA). The cell fusion mix was then seeded in 96-well plates with peritoneal macrophages from non-immunized Kunming mice and cultured in DMEM medium contained with 20% FBS and 1×hypoxanthine- aminopterin- thymidine (HAT; Sigma, St Louis, USA) for 7-12 days. The hybridoma culture supernatants of primary clones were screened by whole viral antigen–based indirect ELISA, and the positive clones were further subcloned by limiting dilution. After three rounds of subcloning, the positive hybrydoma clones were further tested by pET-S1-based ELISA and Western blot. The mAbs reacting positively with both whole viral antigen and recombinant S1 protein in indirect ELISA (hereinafter referred to as anti-S1 MAbs) were used for further characterization. The hybridomas secreting anti-S1 mAbs were intraperitoneal injected into the 10-week-old BALB/c female mice, and the ascite fluids were collected and purified by Protein L Resin (Genscript Co., Nanjing, China). The mAb classes and subclasses were determined by using a mouse monoclonal antibody isotyping reagents kit (Sigma, St Louis, USA). The specificity and cross reaction of anti-S1 mAbs were analyzed by indirect ELISA as the method described before by using other avian viruses (NDV, AIV H5 and AIV H9) and other genotypes of IBVs (Table 1, marked with c) as coating antigen, respectively. Two mAbs, designated as 1D5 and 6A12, were identified and used for further fine-level epitope mapping.
2.4. Virus neutralization assay
IBV Sczy3 strain was passaged in 9-11-day-old SPF embryonated chicken eggs for six generations. The kidney of 18-day-old chicken embryos were dispersed with 0.25% trypsin and seeded in 24-well plates (Corning Inc., Corning, NY). The chicken embryonated kidney (CEK) cells were maintained in MEM supplemented with 10% fetal bovine serum (FBS). The allantoic fluids containing IBV-Sczy3 strain was used to infect CEK cells at the confluency of 70–80% and the culture medium was replaced by MEM supplemented with 2% FBS. The supernatant was harvested 40 h post-inoculation and passaged in CEK cells for 20 generations until the characteristic cytopathic effect (CPE) such as syncytia was observed. Determination of the TCID50 of the 20th generation of Sczy3 virus in CEK cells was conducted as the method of Reed and Muench. For the end-point neutralizing assay of the anti-S1 mAbs, equal volumes of 100 TCID50 of the 20th generations of Sczy3 and serial 2-fold dilutions of purified anti-S1 mAbs were mixed and kept at 37°C for 1 h. 0.2 mL of the virus-antibody mix and 0.2 mL of DMEM with 2% FBS were then transferred into CEK cells cultures in 24-well plates (6 wells for each dilutions). The plates were incubated until CPE developed in virus control group, and the 50% end-point neutralizing titers were calculated by the method of Reed and Muench.
2.5. Epitope prediction and identification
A commercially available 12-mer, M13 gene 3-based random peptide phage display library was purchased from New England Biolabs inc (Ipswich, MA). This dodecapeptide library contained 2.7 × 109 electroporated sequences (1.5 × 109 pfu/ml). Three successive rounds of affinity selection biopanning were carried out according to the manufacturer's instruction. Briefly, each 96-well microtiter plates were coated with mAb (10 μg/well) in coating buffer (0.06 M NaHCO3, pH 8.6) for 10 wells at 4 °C for overnight, followed by blocking with blocking buffer (0.1 M NaHCO3 pH 8.6, 5 mg/ml BSA, 0.02% NaN3) for 1 h at 37 °C. 2 × 1011 phages (10 μL from the original library) were added to a well and incubated at room temperature for 1 h. The unbound phage particles were removed by ten successive washes with 0.1% Tween 20 in TBS. The specifically bound phage particles were eluted by general elution buffer (0.1 M Glycine-HCl pH 2.2, 1 mg/ml BSA) and rocked gently for 20 min at room temperature. The eluted phages were amplified in Escherichia coli ER2738 for 4 h and partially purified by adding PEG 8000 (Sigma, St Louis, USA). After three rounds of peptide selection, ten individual phage clones against each mAb were selected and assayed for target binding using an sandwich ELISA as the method above except that the bound phages were reacted with horseradish peroxidase (HRP) –conjugated anti-M13 monoclonal (GE Healthcare, USA). The positive phage clones were sent for sequencing with sequencing primer by 5′-TGA GCG GAT AAC AAT TTC AC-3′.
Two gene fragments containing the predicted S1 gene epitopes were amplified by RT-PCR with primer pairs Se1F: 5′-GGATC CAAAGTGCGGCTTCCATA G-3′ with BamHI site corresponding to 229-247 of S1 coding sequence and Se1R: 5′-CTCGAGTCATTCAGAAAAGTTACAGTGTGCAC-3′ with XhoI site corresponding to 299-321 of S1 coding sequence, and primer pairs Se2F: 5′-GGATCCGGATTGCTGGTTTA TGTTACTA-3′ with BamHI site corresponding to 1195-1216 of S1 coding sequence and Se2R: 5′-CTCGAGTCA GTGTTGCATTAATACTAAGGGCT-3′ with XhoI site corresponding to 1250-1272 of S1 coding sequence respectively. The amplified fragments were linked with pET-32a (+) to generate two recombinant plasmids pET-32a-Se1 and pET-32a-Se2, and the fusion proteins pET-se1 and pET-se2 were induced with 1 mM IPTG, and analyzed by SDS-PAGE. The fusion proteins were purified by HisTrap FF crude Columns, and used for further ELISA and Western-blot analysis, as previously described (Han et al., 2013).
The conservation of the epitope-containing sequences of IBV strains of different genotypes was further analyzed. The S1 subunit protein genes from the 32 IBV strains were retrieved from GenBank (Table 1). Phylogenetic analysis of the nucleotide sequences showed that the analyzed strains belong to eight different genotypes. Analysis of the epitope-containing sequences of IBVs were performed using the MegAlign application in the Lasergene software package.
3. Results and discussion
3.1. Expression and purification the recombinant fusion protein pET-S1
The result of SDS-PAGE analysis demonstrated that the recombinant S1 subunit protein pET-S1 was successfully expressed in BL21 strains. The purified recombinant protein pET-S1 could react with anti-Sczy3 mouse polyclonal serum in Western blot. The size of the recombinant fusion protein was estimated to be approximately 57.5 kDa including the amino acids encoded by the vector. (data not shown).
3.2. Cultivation of CEK-adapted IBV Sczy3 strain
The allantoic fluids containing IBV Sczy3 strain was used to infect CEK cells, and the supernatant of CEK cells was harvested 40 h post-inoculation and passaged blindly. After being serially passaged in CEK cultures for 5 generations, specific CPE (syncytium) of IBV infection could be observed at 18 h post infection in the CEK monolayers cells, suggesting the CEK-adapted strain of Sczy3 was generated successfully. As the passaging continues, the time when syncytia appeared became earlier, and more and more syncytia were observed. The TCID50 of the 20th generation of Sczy3 strain was calculated as 10−5.64/0.2 ml (data not shown).
3.3. Two MAbs with neutralizing activity against S1 subunit of IBV Sczy3 strain were generated
11 hybridoma cell lines producing antibodies against the Sczy3 virus were established. Among them, two hybrydoma clones reacted positively with both whole viral antigen and recombinant S1 protein in indirect ELISA and Western blot, and were designated as 1D5 and 6A12. 1D5 and 6A12 were injected into BABL/c mice to generate ascites. Isotyping revealed that the mAbs 1D5 and 6A12 were of the IgM class. In addition, these two mAbs could specifically react with IBV but not with NDV, AIV H5 or H9. The cross-react analysis revealed that the mAb 6A12 and 1D5 could cross-react with other 10 IBV strains in five different genotypes, except that mAb 1D5 had a relatively low cross-reaction with CK/CH/SCYA/10I and CK/CH/SCMY/10I, indicating that these two MAbs especially 6A12 could be used for the establishment of diagnostic assays such as indirect immunofluorescence assay (IFA) and antigen capture ELISA (AC –ELISA) against IBVs. End-point neutralizing assay of anti-S1 mAbs showed that the neutralization titer of 1D5 and 6A12 were 1:40.6 and 1:44.7, respectively (data not shown), indicating that these mAbs had the potential for immunotherapy.
3.4. Epitope prediction and identification
To identify the epitopes recognized by the neutralizing mAbs 1D5 and 6A12, biopanning of a 12-mer random peptide library was performed using affinity purified MAbs. After three rounds of biopanning, 10 individual positive phage clones of each mAbs were isolated. Their reactivities were assessed with mAbs by phage ELISA, the results showed that 9 single phage displayed peptides bound to mAb 1D5 and 10 to mAb 6A12 (OD450 > 0.47). The BSA control wells were no interaction (OD450 < 0.15). The positive phage clones against 1D5 and 6A12 were selected for sequencing of single-stranded DNA inserts. For 1D5, sequence alignment showed that seven phage clones shared the same sequence: ALWPPQGHAWVP, one clone had the sequence HGENPPSGMAWV, and the final clone had the sequence HGVNPPSGMAWV. For 6A12, 8 clones shared the same sequence: HIQTRLEPQWKS, and one clone had the sequence: HGIQTATEPQWS, the last one clones had the sequence: HTLQTRTPPQSP. Comparison of these sequences with the sequence of S1 protein of Sczy3 (GenBank Accession No: GU384206) showed that homology of 1D5 occurred at amino acids 87PPQGMAW93, and 6A12 occurred at amino acids: 412IQTRTEP418 (Fig. 1, Fig. 2 ).
Fig. 1.
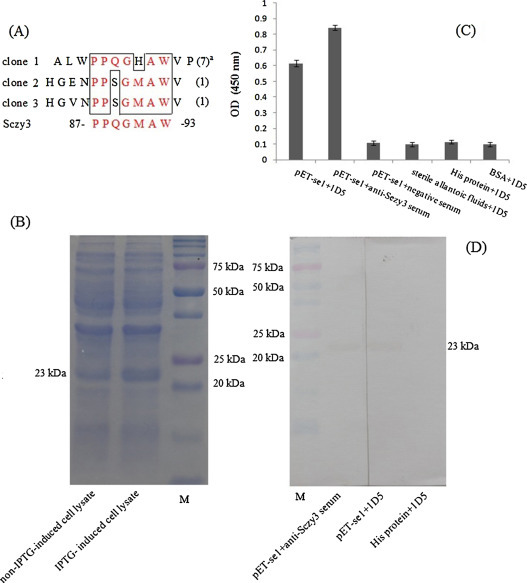
Fine mapping of the epitope for MAb 1D5 in S1 protein of Sczy3. The sequences of 12 peptides displayed by the selected phage clones were shown. A consensus sequence, 87PPQGMAW93, displayed by the 9 phages had a good match with the S1 protein of Sczy3 at amino acids 87-93 (A). The reactivity of MAb 1D5 with a truncated recombinant S1 protein pET-se1 (B) by indirect ELISA (C) and Western blot were illustrated (D).
a Times of the clone sequences occurred.
Fig. 2.
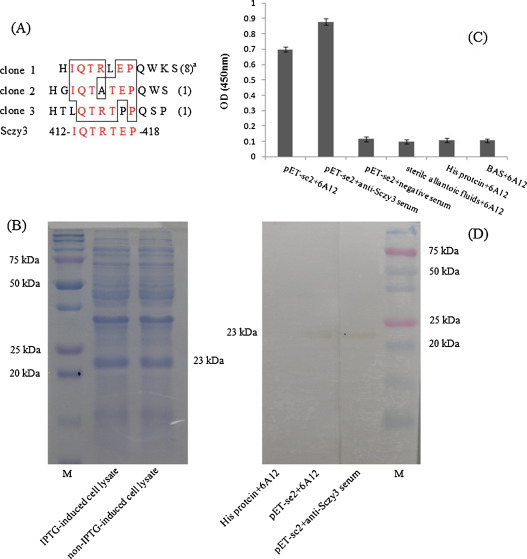
Fine mapping of the epitope for MAb 6A12 in S1 protein of Sczy3. The sequences of 12 peptides displayed by the selected phage clones were shown. A consensus sequence, 412IQTRTEP418, displayed by the 10 phages had a good match with the S1 protein of Sczy3 at amino acids 412-418 (A). The reactivity of MAb 6A12 with a truncated recombinant S1 protein pET-se2 (B) by indirect ELISA (C) and Western blot were illustrated (D).
a: times of the clone sequences occurred.
Fragment containing the predicted epitopes of S1 protein were then expressed and purified (Fig. 1, Fig. 2). Indirect ELISA and Western blot analysis confirmed that purified recombinant proteins pET-se1 and pET-se2 could interact with mAb 1D5 and mAb 6A12, respectively, and also could interact with mice hyperimmune sera of Sczy3 ((Fig. 1, Fig. 2).
Alignment of the sequences coding for the motif (87PPQGMAW93 and 412IQTRTEP418) on the S1 protein of IBV strains of other genotypes showed that all positions in the motif 412IQTRTEP418were conserved among most IBV strains, but the positions in the motif 87PPQGMAW93 were relatively variable, especially among the genotypes of CK/CH/LSC/99 I-type, proventriculus-type and the TW-type (Fig. 3 ). This sequence alignment result was consistent with the result of cross-reaction test which showed that mAb 6A12 had a better reaction with other 10 IBV strains from five different genotypes than mAb 1D5.
Fig. 3.
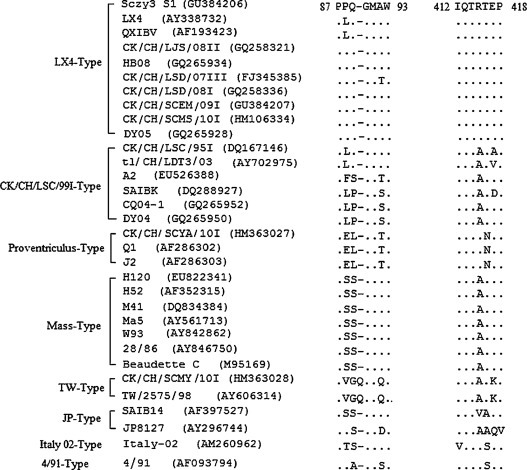
Alignment of the sequences of 32 IBV strains surrounding the epitope-coding region on the S1 protein. The GenBank accession numbers of the IBV strains used are indicated in parentheses. The genotypes of the viruses were listed on the left side.
The identification of antigenic site for the IBV structural S1 subunit was limited. In the early studies, sequence analysis was commonly used to predict the antigenic site, and diverse methods were conducted to identify the antigenic site, but most of the identified antigenic sites were not finely mapped (Hu et al., 2007, Ignjatovic and Sapats, 2005, Kant et al., 1992, Parr and Collissor, 1993, Wang et al., 1995). Recently, RPPD libraries were used to map the antigenic site more elaborately. Han, (2013) identified two novel linear B-cell epitopes 242FGPRTK247 and 195DLIARAAKI203 of IBV nucleocapsid protein by using a Ph.D.12TM Phage Display Peptide Library and two mAbs (Han et al., 2013). To update, there was no report of antigenic site mapping in IBV S1 subunit by RPPD library screening. In this study, we generated 2 mAbs against the S1 subunit protein of IBV and we finely mapped two neutralizing epitopes 87 PPQGMAW 93 and 412 IQTRTEP 418. To the best of our knowledge, these 2 epitopes are the first finely mapped neutralizing epitopes for the IBV S1 subunit protein. These two epitopes were not included in the regions of the previous studies. The reason of this discrepancy may be different viruses were used as the antigens for generating mAbs, and the production of mAbs was random. The strain Sczy3 is classified into LX-4 genotype, and recombinant analysis of Sczy3 genome demonstrated that Sczy3 was a chimeric strain derived from QX and H120 (Zhao et al., 2013).
S1 gene is now commonly used as a marker for classification of IBV. Although highly variable, it remains the first choice for developing subunit vaccine against IB. Identification of the antigenic site accurately on S1 gene is critical for the development of subunit vaccine. In addition, S1 subunit anchors to the external surface of viral particles, making it to be the antigen more easily be recognized by IBV-specific antibody than other IBV antigens. As there are still relatively conserved region or epitopes in S1 subunit, S1 could also be used as targeted antigens in the development of diagnostic agents.
The availability of the mAbs and their corresponding epitope containing sequences identified in this study will facilitate the development of immunological assays for IBVs, although these 2 mAb can not differentiate field isolates with vaccine strains. Because of the IBVs available in our laboratory is limited, we only tested the reactivity between these two mAbs with 11 heterogeneous IBVs. In the future study, with the isolation of more and more field isolates of different genotypes or serotypes, the suitability of these two mAbs especially 6A12 for the development of immunological assays for IBVs will be tested.
4. Conflict of interest
The authors have no conflict of interest related to this research to report.
Acknowledgment
The work was financially supported by Program for Changjiang Scholars and Innovative Research Team in University “PCSIRT”(Grant No: IRTO848)
References:
- Abd E.R.S., El-Kenawy A.A., Neumann U., Herrler G., Winter C. Comparative analysis of the sialic acid binding activity and the tropism for the respiratory epithelium of four different strains of avian infectious bronchitis virus. Avian Pathol. 2009;38:41–45. doi: 10.1080/03079450802632049. [DOI] [PubMed] [Google Scholar]
- Benyeda Z., Szeredi L., Mato T., Suveges T., Balka G., Abonyi-Toth Z., Rusvai M., Palya V. Comparative histopathology and immunohistochemistry of QX-like Massachusetts and 793/B serotypes of infectious bronchitis virus infection in chickens. J. Comp. Pathol. 2010;143:276–283. doi: 10.1016/j.jcpa.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boots A.M., Kusters J.G., van Noort J.M., Zwaagstra K.A., Rijke E., van der Zeijst B.A., Hensen E.J. Localization of a T-cell epitope within the nucleocapsid protein of avian coronavirus. Immunology. 1991;74:8–13. [PMC free article] [PubMed] [Google Scholar]
- Boursnell M.E., Brown T.D., Foulds I.J., Green P.F., Tomley F.M., Binns M.M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 1987;68(Pt 1):57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- Casais R., Dove B., Cavanagh D., Britton P. Recombinant avian infectious bronchitis virus expressing a heterologous spike gene demonstrates that the spike protein is a determinant of cell tropism. J. Virol. 2003;77:9084–9089. doi: 10.1128/JVI.77.16.9084-9089.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Darbyshire J.H., Peters R.W. Coronavirus IBV: virus retaining spike glycopolypeptide S2 but not S1 is unable to induce virus-neutralizing or haemagglutination-inhibiting antibody, or induce chicken tracheal protection. J. Gen. Virol. 1986;67(Pt 7):1435–1442. doi: 10.1099/0022-1317-67-7-1435. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Elus M.M., Cook J.K. Relationship between sequence variation in the S1 spike protein of infectious bronchitis virus and the extent of cross-protection in vivo. Avian Pathol. 1997;26:63–74. doi: 10.1080/03079459708419194. [DOI] [PubMed] [Google Scholar]
- Han Z., Zhao F., Shao Y., Liu X., Kong X., Song Y., Liu S. Fine level epitope mapping and conservation analysis of two novel linear B-cell epitopes of the avian infectious bronchitis coronavirus nucleocapsid protein. Virus Res. 2013;171:54–64. doi: 10.1016/j.virusres.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J.Q., Li Y.F., Guo J.Q., Shen H.G., Zhang D.Y., Zhou J.Y. Production and characterization of monoclonal antibodies to (poly100)S1 protein of avian infectious bronchitis virus. Zoonoses Public Health. 2007;54:69–77. doi: 10.1111/j.1863-2378.2007.01030.x. [DOI] [PubMed] [Google Scholar]
- Ignjatovic J., Sapats S. Identification of previously unknown antigenic epitopes on the S and N proteins of avian infectious bronchitis virus. Arch. Virol. 2005;150:1813–1831. doi: 10.1007/s00705-005-0541-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W., Hilt D.A., Callison S.A., Lee C.W., Plaza H., Wade E. Spike glycoprotein cleavage recognition site analysis of infectious bronchitis virus. Avian Dis. 2001;45:366–372. [PubMed] [Google Scholar]
- Kant A., Koch G., van Roozelaar D.J., Kusters J.G., Poelwijk F.A., van der Zeijst B.A. Location of antigenic sites defined by neutralizing monoclonal antibodies on the S1 avian infectious bronchitis virus glycopolypeptide. J. Gen. Virol. 1992;73(Pt 3):591–596. doi: 10.1099/0022-1317-73-3-591. [DOI] [PubMed] [Google Scholar]
- Lai M.M., Brayton P.R., Armen R.C., Patton C.D., Pugh C., Stohlman S.A. Mouse hepatitis virus A59: mRNA structure and genetic localization of the sequence divergence from hepatotropic strain MHV-3. J. Virol. 1981;39:823–834. doi: 10.1128/jvi.39.3.823-834.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Xue C., Chen F., Qin J., Xie Q., Bi Y., Cao Y. Isolation and genetic analysis revealed no predominant new strains of avian infectious bronchitis virus circulating in South China during 2004-2008. Vet. Microbiol. 2010;143:145–154. doi: 10.1016/j.vetmic.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood Z.H., Sleman R.R., Uthman A.U. Isolation and molecular characterization of Sul/01/09 avian infectious bronchitis virus, indicates the emergence of a new genotype in the Middle East. Vet. Microbiol. 2011;150:21–27. doi: 10.1016/j.vetmic.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr R.L., Collissor E.W. Epitopes on the spike protein of a nephropathogenic strain of infectious bronchitis virus. Arch. Virol. 1993;133:369–383. doi: 10.1007/BF01313776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seah J.N., Yu L., Kwang J. Localization of linear B-cell epitopes on infectious bronchitis virus nucleocapsid protein. Vet. Microbiol. 2000;75:11–16. doi: 10.1016/s0378-1135(00)00202-9. [DOI] [PubMed] [Google Scholar]
- Wang L., Parr R.L., King D.J., Collisson E.W. A highly conserved epitope on the spike protein of infectious bronchitis virus. Arch. Virol. 1995;140:2201–2213. doi: 10.1007/BF01323240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.F., Sun Y.K., Tian Z.C., Shi X.M., Tong G.Z., Liu S.W., Zhi H.D., Kong X.G., Wang M. Protection of chickens against infectious bronchitis by a recombinant fowlpox virus co-expressing IBV-S1 and chicken IFNgamma. Vaccine. 2009;27:7046–7052. doi: 10.1016/j.vaccine.2009.09.065. [DOI] [PubMed] [Google Scholar]
- Xing J., Liu S., Han Z., Shao Y., Li H., Kong X. Identification of a novel linear B-cell epitope in the M protein of avian infectious bronchitis coronaviruses. J. Microbiol. 2009;47:589–599. doi: 10.1007/s12275-009-0104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Han Z., Xu J., Shao Y., Li H., Kong X., Liu S. A novel B-cell epitope of avian infectious bronchitis virus N protein. Viral Immunol. 2010;23:189–199. doi: 10.1089/vim.2009.0094. [DOI] [PubMed] [Google Scholar]
- Zhang J., Chen X.W., Tong T.Z., Ye Y., Liao M., Fan H.Y. BacMam virus-based surface display of the infectious bronchitis virus (IBV) S1 glycoprotein confers strong protection against virulent IBV challenge in chickens. Vaccine. 2014;32:664–670. doi: 10.1016/j.vaccine.2013.12.006. [DOI] [PubMed] [Google Scholar]
- Zhao F., Zou N., Wang F., Guo M., Liu P., Wen X., Cao S., Huang Y. Analysis of a QX-like avian infectious bronchitis virus genome identified recombination in the region containing the ORF 5a ORF 5b, and nucleocapsid protein gene sequences. Virus Genes. 2013;46:454–464. doi: 10.1007/s11262-013-0884-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou N.L., Zhao F.F., Wang Y.P., Liu P., Cao S.J., Wen X.T., Huang Y. Genetic analysis revealed LX4 genotype strains of avian infectious bronchitis virus became predominant in recent years in Sichuan area China. Virus Genes. 2010;41:202–209. doi: 10.1007/s11262-010-0500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]


