Abstract
Objective
Human infections caused by avian influenza virus A(H7N9) re-emerged in late 2013. We reported the first Hong Kong patient without risk factors for severe A(H7N9) disease.
Methods
Direct sequencing was performed on the endotracheal aspirate collected from a 36-year-old female with history of poultry contact. Bioinformatic analysis was performed to compare the current strain and previous A(H7N9) isolates.
Results
The influenza A/Hong Kong/470129/2013 virus strain was detected in a patient with acute respiratory distress syndrome, deranged liver function and coagulation profile, cytopenia, and rhabdomyolysis. The HA, NA and MP genes of A/Hong Kong/470129/2013 cluster with those of other human A(H7N9) strains. The PB1, PB2 and NS genes are most closely related to those of A/Guangdong/1/2013 strain identified in August 2013, but are distinct from those of other human and avian A(H7N9) strains. The other internal genes NP and PA genes are more closely related to those of non-A(H7N9) avian influenza A viruses. A unique PA L336M mutation, associated with increased polymerase activity, was found. The patient required salvage by extracorporeal membrane oxygenation.
Conclusions
The A/Hong Kong/470129/2013 virus is a novel reassortant derived from A/Guangdong/1/2013 virus. The unique mutation PA L336M may enhance viral replication and therefore disease severity.
Keywords: Avian influenza, H7N9, Phylogenetic tree, Evolution, L336M, PA
Introduction
Human infections by avian influenza viruses A(H5N1), A(H9N2), A(H7N7/3/2), A(H10N7), A(H7N9) and A(H6N1) have been reported.1, 2 However the influenza A(H7N9) virus has now become the most prevalent avian influenza virus affecting humans in China since its emergence in the spring of 2013.3, 4 The case-fatality rate of more than 30% is much higher than that of the 2009 pandemic influenza virus and the SARS coronavirus.5, 6, 7 Similar to other severe influenza virus infection, A(H7N9) typically causes severe pneumonia not responding to antibiotics covering for typical and atypical respiratory pathogens, and can lead to extrapulmonary complications.8 Seroepidemiological study found that 13.9% of poultry workers and 0.8% of the general population in the Zhejiang province of China had a haemagglutination inhibition antibody titre of 40 or more.9 Furthermore epidemiological and genetic studies have established the link between direct or indirect poultry contact and human A(H7N9) infection, with the possibility of limited person-to-person transmission.3, 10, 11
Similar to the seasonality for human A(H5N1) infections, the number of human A(H7N9) infections decreased during the summer with only four reported cases from May to September 2013.7, 12, 13 However, human A(H7N9) cases re-emerged since October 2013 with a total of eight cases reported as of 12th December 2013.12, 14 Five of these patients were from Zhejiang province, two patients from Hong Kong and one patient from Guangdong province. In Hong Kong, the first human case of A(H7N9) occurred in November 2013.15 In this study, we reported the clinical severity of this case and sequenced the viral genome directly from the endotracheal aspirate of the patient. We then performed phylogenetic analysis and compared the genetic differences between the virus from this Hong Kong patient and other human and avian A(H7N9) viruses.
Materials and methods
Patient and microbiological investigations
The clinical data and endotracheal aspirate specimen were collected from a 36-year-old female patient suffering from severe bilateral pneumonia on day 3 after hospitalization in Hong Kong. Acute respiratory distress syndrome was defined with standard criteria.16 The endotracheal aspirate was cultured in Madin Darby canine kidney cell line with trypsin for seven days. It was also tested by the multiplex polymerase chain reaction with ResPlex II v2.0 assay (Qiagen) for coinfection with respiratory syncytial virus, influenza B virus, parainfluenza viruses 1–4, human metapneumovirus, enteroviruses, rhinovirus, adenovirus, bocavirus, and coronaviruses NL63, HKU1, 229E, and OC43, in accordance with manufacturer's instructions. Nucleic acid extraction and sequencing were performed directly from this endotracheal aspirate specimen. Informed consent was obtained from the patient.
Real time reverse transcriptase-polymerase chain reaction (RT-PCR) for detection of A(H7N9) virus
Real time RT-PCR was performed as we described previously.8 Briefly, total nucleic acid was extracted using NucliSens easyMAG™ extraction system (BioMerieux, France). Taqman real-time RT-PCR was performed under standard thermocycling conditions to detect matrix protein (MP), H7, and N9 genes.
Sequencing of full virus genome from endotracheal aspirate
Sequencing was performed as we described previously using consensus primers for all eight gene segments of influenza A(H7N9) virus.3, 17 Direct sequencing of the PCR products was performed using an ABI Prism 3730 DNA analyzer.
Phylogenetic analysis
All sequences were assembled and edited with Lasergene 6.0 (DNASTAR, WN, USA). Bioedit 7 was used for alignment and analysis of amino acid residues. The phylogenetic trees of haemagglutinin (HA), neuraminidase (NA), acidic polymerase (PA), basic polymerases (PB1, PB2), MP, nucleoprotein (NP) and non-structural protein (NS) genes were constructed using neighbour-joining method with Tamura-Nei model of nucleotide substitution using MEGA software package, version 5.05. The bootstrap values from 1000 replicates were calculated to evaluate the reliability of the phylogenetic trees. Our gene sequences were deposited within the GenBank sequence database under accession numbers KF952506 to KF952513. All other gene sequences were obtained from NCBI Influenza Virus Resource database, except A/Anhui/1/2013 and A/Shanghai/1/2013 which were obtained from Global Initiative on Sharing All Influenza Data (GISAID) EpiFlu™ Database (Supplementary Table).
Amino acid sequence comparison
The amino acid sequence of A/Hong Kong/470129/2013 virus was compared to that of other human and avian A(H7N9) virus strains available in the NCBI Influenza Virus Resource database as of 10th December, 2013. H7 and human N9 numbering were used for denoting the amino acid positions in the HA and NA proteins, respectively.
Results
Patient
The patient was a 36-year-old female Indonesian domestic helper living in Hong Kong. Her past health was unremarkable except for some vitiligo over the neck and back which was treated with topical steroid. Though her antinuclear antibody titre was over 640, there is no evidence of autoimmune disease activity as evident by negative anti-double stranded DNA antibody and normal complement levels. On 17th November 2013, she travelled to Shenzhen, China, where she visited a live poultry market. On 21st November, she developed malaise. Subsequently, she developed fever since 23rd November, associated with cough, sputum production, sore throat and dyspnoea. She did not have vomiting or diarrhoea. She was hospitalized on 27th November. On admission, her body temperature was 40.0 °C. Her respiratory rate was 18 breaths per minute, and the oxygen saturation was 97% while breathing ambient air. The pulse rate was 116 beats per minute, and the blood pressure was 120/63 mmHg. Her chest radiograph showed right lower zone consolidation and blunting of right costophrenic angle. Her condition deteriorated rapidly despite intravenous amoxicillin-clavulanate 1.2 g every 8 h and oral azithromycin 500 mg every 24 h given from day 1–2 after hospitalization. She also received one dose of oral oseltamivir 75 mg on day 2 after hospitalization. Serial chest radiographs showed progression to bilateral lower zone consolidation. She subsequently required intubation and positive pressure ventilation on day 2 after hospitalization. With further deterioration and the development of acute respiratory distress syndrome, extracorporeal membrane oxygenation was started on day 3 after hospitalization. After the diagnosis of A(H7N9) was confirmed, she was started on intravenous zanamivir 600 mg every 12 h from day 5 after hospitalization. On day 3 after hospitalization, complete blood picture showed a lowish white cell count of 3.7 (normal >3.9) × 109/L (neutrophil 2.7 and lymphocyte 0.9[normal >1.1]) and a platelet count of 131 × 109/L (normal >150). The coagulation profile was deranged with a prothrombin time of 15.4 (normal <13.5) seconds and an activated partial thromboplastin time of 42 (normal <33.7) seconds. The plasma creatine kinase and lactate dehydrogenase peaked at 1976 (normal <161) and 990 (normal <218) U/L respectively. Urine for myoglobin was positive. Her hepatic parenchymal enzyme, alanine transaminase, peaked at 206 U/L (normal <36) on day 18 after symptom onset.
RT-PCR for influenza A virus MP gene and H7 gene were positive in the endotracheal aspirate sample collected on day 3 after hospitalization. No viral co-infection was detected by multiplex RT-PCR. Viral culture of this endotracheal aspirate in Madin Darby canine kidney cell line was positive for cytopathic effect on day three after inoculation. The presence of influenza A virus in positive cell culture was confirmed by immunostaining for NP protein and A(H7N9) by specific RT-PCR. Bacterial cultures were negative in blood, endotracheal aspirate and right pleural fluid.
Phylogenetic analysis
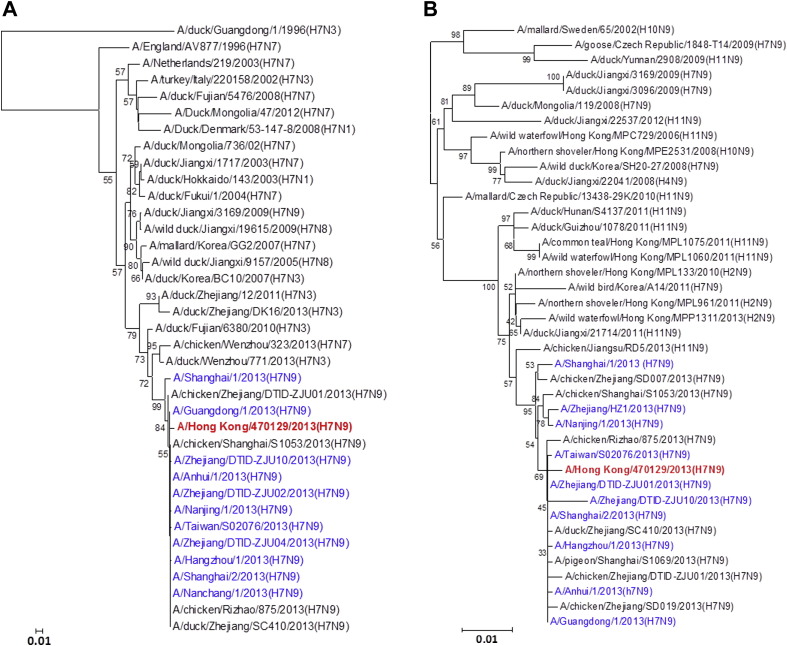
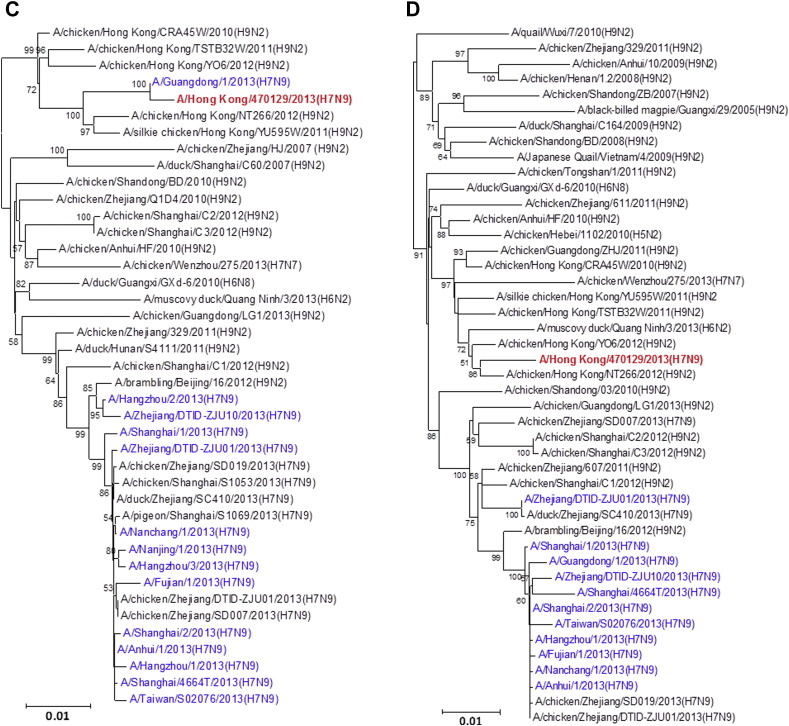
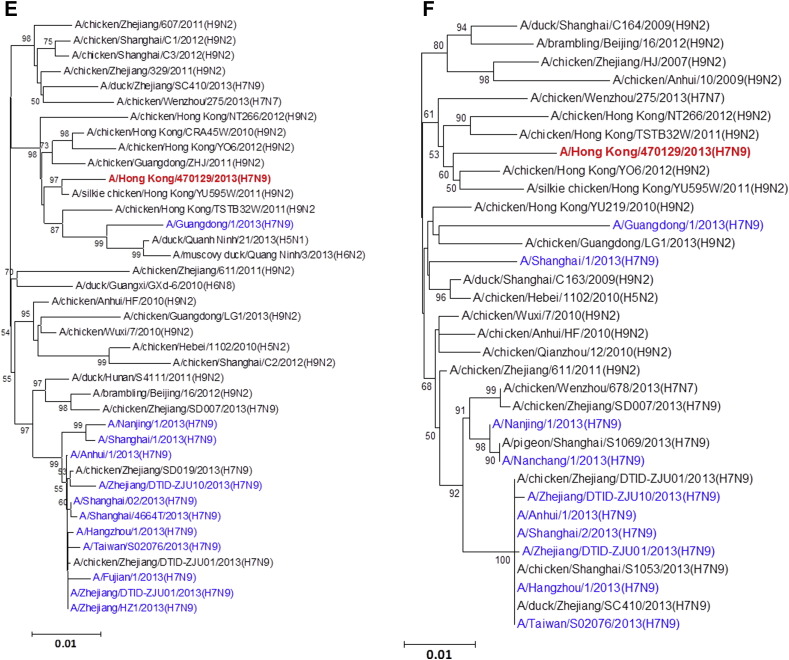
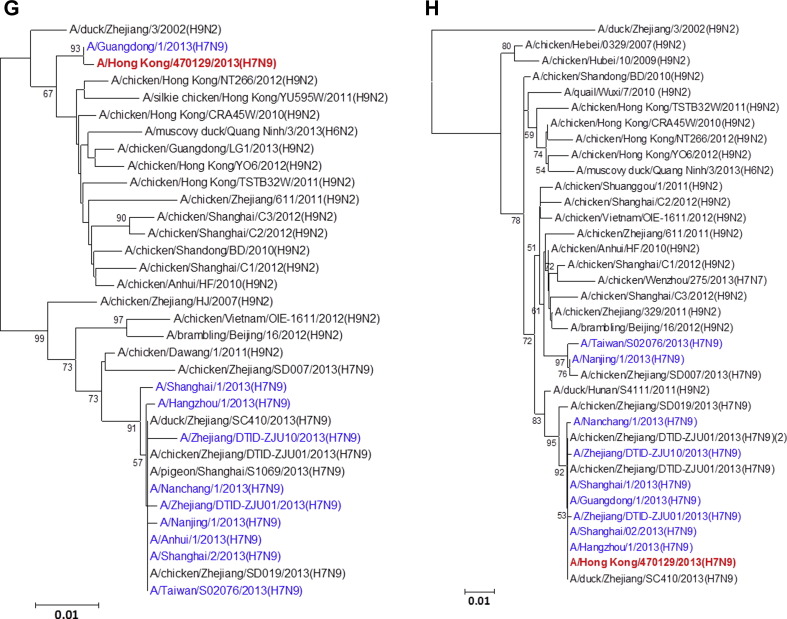
All eight gene segments of this A/Hong Kong/470129/2013 virus were sequenced directly from the endotracheal aspirate sample (Fig. 1 ). Manual inspection of the chromatogram did not show double peaks suggesting of a mixed population. The HA1, NA and MP genes cluster with other human A(H7N9) isolates characterized in the 2013 spring outbreak while the NP appears to be coming from additional donor closely related to A(H9N2) strains found in poultries from China. The PB2, PB1 and NS genes form a separate cluster with the human A(H7N9) isolate A/Guangdong/1/2013 which was identified in the Guangdong province adjacent to Hong Kong. Notably, the PA gene of A/Hong Kong/470129/2013 cluster with other avian A(H9N2), A(H6N2) and A(H7N7) viruses but not with those PA of other previously characterized A(H7N9) viruses including the A/Guangdong/1/2013, suggesting that the A(H7N9) virus is continuously reassorting with other avian viruses in the field.
Figure 1.
Phylogenetic trees showing the relationship of A/Hong Kong/470129/2013 to other A(H7N9) and non-A(H7N9) avian influenza viruses. (A) HA1. (B) NA. (C) PB2. (D) PA. (E) PB1. (F) NP. (G) NS. (H) MP. The phylogenetic trees were constructed using neighbour-joining method with Tamura-Nei model of nucleotide substitution using MEGA software package, version 5.05. The bootstrap values from 1000 replicates were performed to evaluate the reliability of the phylogenetic trees. The A/Hong Kong/470129/2013 was highlighted in bold font in red, while other human A(H7N9) strains were highlighted in blue. Sequences not determined in this study were obtained from NCBI Influenza Virus Resource database, except A/Anhui/1/2013 and A/Shanghai/1/2013 which were obtained from Global Initiative on Sharing All Influenza Data (GISAID) EpiFlu™ Database (Supplementary Table).
Important amino acid substitutions associated with antiviral resistance, mammalian adaptation and viral replication in the A/Hong Kong/470129/2013 virus were examined and summarized in Table 1 . The M2 protein contains the S31N mutation which confers resistance to adamantanes.5 However the NA does not contain the R289K substitution which confers resistance to neuraminidase inhibitors.4 In the HA, G177V and Q217L substitutions which were reported to confer increased binding to α2,6-linked sialic acid receptors present in the upper respiratory tract of humans, are found.4 In the PB2 polymerase subunit, A/Hong Kong/470129/2013 contains L89V and E627K substitutions which are associated with enhanced viral polymerase activity in mammalian cells.4 However, the PB2 T271A and D701N substitutions associated with enhanced polymerase activity and mammalian adaptation were not found. The PB1 protein contains I368V, which enables droplet transmission in ferrets.18 Other mutations present only in A/Hong Kong/470129/2013 and/or A/Guangdong/1/2013 but not among other human or avian A(H7N9) isolates available in NCBI Influenza Virus Resource database were summarized in Table 2 . Surprisingly, the PA L336M substitution of the polymerase subunit, which was reported in one study to be associated with enhanced polymerase activity,19 is found in the A/Hong Kong/470129/2013 but not other human or avian A(H7N9) isolates (Table 2 and Table 3 ). The mutations V236I, K261E, N322T, N341D, and K460R of the NA gene, that were first identified in the A/Zhejiang/22/2013 and A/Zhejiang/DTID_ZJU10/2013 strain isolated from patients in October 2013,14 are not found in A/Hong Kong/470129/2013.
Table 1.
Amino acid substitutions well known to be associated with mammalian adaptation, viral replication and antiviral resistance.
| Date of collection | Significance | Site | Virus strain |
||
|---|---|---|---|---|---|
| A/Hong Kong/470129/2013 |
A/Guangdong/1/2013 |
A/Zhejiang/DTID_ZJU10/2013 |
|||
| 30 November 2013 | 10 August 2013 | 14 October 2013 | |||
| HA (H7 numbering) | Mammalian adaptation | 177 | V | V | V |
| Increased binding to α2,6-linked sialic acid receptor | 217 | L | L | L | |
| NA (human N9 numbering) | Resistant to neuraminidase inhibitors | 289 | R | R | R |
| M2 | Resistant to adamantanes | 31 | N | N | N |
| PB1 | Enables droplet transmission in ferrets | 368 | V | V | V |
| PB2 | Enhanced polymerase activity | 89 | V | V | V |
| Enhanced polymerase activity | 271 | T | T | T | |
| Improved viral replication at 33 °C | 627 | K | K | E | |
| Mammalian adaptation | 701 | D | D | D | |
Table 2.
Unique amino acid substitutions present in the A/Hong Kong/470129/2013 and/or A/Guangdong/1/2013.
| Protein | Site | A/Hong Kong/470129/2013 | A/Guangdong/1/2013 | Other human and avian A(H7N9) virusesa |
|---|---|---|---|---|
| HA (H7 numbering) | 122 | A | A | T |
| NA (human N9 numbering) | 259 | V | I | I |
| PB2 | 139 | I | I | V |
| 283 | I | M | M | |
| 286 | G | S | S | |
| 569 | A | T | T | |
| 676 | V | V | M or T | |
| PB1 | 178 | D | E | E |
| 682 | V | I | I | |
| PA | 336 | Mb | L | L |
| NP | 313 | L | F | F |
| 359 | A | S | S | |
| 433 | A | T | T | |
| NS1 | 20 | R | K | K |
Only virus isolates available on NCBI Influenza Virus Resource database before 10 December 2013 were analysed. The number of sequences available (except A/Guangdong/1/2013) was: HA, 123; NA, 118; PB2, 104; PB1, 99; PA, 98, NP, 100; NS1, 101.
Associated with enhanced polymerase activity in one study.23
Table 3.
Differences between A/Hong Kong/470129/2013 and A/Guangdong/1/2013.
| Protein | Site | A/Hong Kong/470129/2013 | A/Guangdong/1/2013 |
|---|---|---|---|
| HA (H7 numbering) | 47 | R | K |
| 267 | N | D | |
| 282 | S | N | |
| NA (human H9 numbering) | 259 | V | I |
| PB2 | 283 | I | M |
| 286 | G | S | |
| 292 | I | V | |
| 569 | A | T | |
| PB1 | 105 | H | N |
| 171 | M | V | |
| 178 | D | E | |
| 374 | A | V | |
| 397 | I | M | |
| 682 | V | I | |
| 694 | N | S | |
| PA | 100 | V | A |
| 336 | M | L | |
| 343 | A | T | |
| 356 | K | R | |
| 394 | D | N | |
| 519 | N | T | |
| NP | 313 | L | F |
| 359 | A | S | |
| 371 | M | I | |
| 375 | D | E | |
| 433 | A | T | |
| 482 | S | N | |
| NS1 | 20 | R | K |
| M1, M2, NS2 | No difference | ||
Discussion
We reported the first case of severe A(H7N9) infection in a Hong Kong patient returning from Shenzhen, China, who had contact with poultry. She developed acute respiratory distress syndrome and rhabdomyolysis which were known complications of A(H7N9) influenza.8 She also developed elevated level of alanine transaminase, cytopenia, and derangement in clotting profile. From our previous clinical and histopathological studies of fatal cases, these may be related to hepatic steatosis, disseminated intravascular coagulation and reactive haemophagocytosis.3, 8 If not for the support with extracorporeal membrane oxygenation, the patient may have developed other extrapulmonary complications such as renal tubular dysfunction leading to renal failure. Unlike most other previously reported fatal cases who were elderly or had active underlying chronic illnesses, our patient is young with no active autoimmune process and not on systemic immunosuppressives. While two out of 18 cases of A(H5N1) infections in Hong Kong in 1997 were domestic helpers with history of visiting live poultry market,20 the Hong Kong A(H7N9) patient was also a domestic home helper with a history of visiting live poultry market in Shenzhen, slaughtering and cooking of chicken.
Although influenza virus A(H7N9) has been isolated from avian species for many years, it was not until March 2013 when the first human case was reported.21 Phylogenetic studies using A(H7N9) viruses isolated from spring and early summer of 2013 showed that the human A(H7N9) virus was a reassortant of different avian influenza A viruses.4, 22, 23 The HA gene was most closely related to A(H7N3) viruses and A(H7N9) viruses in wild birds. The NA gene was most closely related to influenza A(H2N9) and A(H11N9) viruses of wild birds from Hong Kong and mainland China. The internal genes MP, NP, NS, PA, PB1 and PB2 were most closely related to avian A(H9N2) viruses circulating in the domestic poultry of China. In earlier studies comparing human A(H7N9) isolates obtained from patients in spring 2013, it was found that the internal genes appeared to come from multiple sources.21, 24 The PB2 gene of the human A(H7N9) strains isolated during spring and early summer of 2013 clusters closely together.22, 23 To monitor the evolution of the A(H7N9) virus, we have analysed the genomic sequence of the A(H7N9) virus (A/Hong Kong/470129/2013) causing severe pneumonia in a 36-year-old lady from Hong Kong in November 2013. We have shown that while the HA, NA, and MP gene of A/Hong Kong/470129/2013 cluster with other human A(H7N9) isolates, the PB2, PB1, and NS genes are closely related to the A/Guangdong/1/2013 isolated in August 2013 but distinct from other human and avian A(H7N9) viruses detected during the spring outbreak in China. The PA gene of A/Hong Kong/470129/2013 does not cluster with any human or avian A(H7N9) strains but clusters with avian A(H9N2), A(H6N2) and A(H7N7) viruses. Taken together, the phylogenetic analysis indicates that the A/Hong Kong/470129/2013 is a novel reassortant presumably originated from reassortment event involving A/Guangdong/1/2013 (H7N9) and other avian influenza A viruses in the field.
Many of the amino acid differences between the A/Hong Kong/470129/2013 and A/Guangdong/1/2013 isolates lie in the polymerase proteins which may affect the polymerase function and virus replication efficiency in mammalian hosts including humans (Table 3).25 In the PA protein, the L336M was only present in the A/Hong Kong/470129/2013 but not A/Guangdong/1/2013 strains or other human or avian A(H7N9) strains. In fact, L336M substitution is conserved among the 2009 pandemic A(H1N1)pdm09 virus isolates but rarely found in other human or avian influenza viruses.19 PA L336M substitution has been shown to increase the polymerase activity. Mice challenged with recombinant A(H1N1) virus containing PA L336M substitution had more body weight loss and higher lung viral titres than those infected with the wild type virus, suggesting that PA L336M substitution is associated with higher pathogenicity in mammals.19 This may have important clinical significance because our patient had no risk factors for severe influenza but she developed very severe pneumonia with rapid progression to acute respiratory distress syndrome which finally required respiratory support by extracorporeal membrane oxygen. Moreover she also had multi-organ dysfunction as evident by impaired liver function and coagulation profile, cytopenia and rhadomyolysis. Another substitution, PB2 V139I, was identified only in the A/Hong Kong/470129/2013 and A/Guangdong/1/2013 strains, but not in other human or avian A(H7N9) strains. A previous study has also shown that V139I was enriched in human A(H7N7) viruses when compared to avian A(H7N7) viruses.26 Further studies should be performed to ascertain the importance of these substitutions in A(H7N9) in terms of viral polymerase activity, viral load and cytotoxicity in infected cell lines or chick embryo, and virulence in animal challenge.
Monitoring for mutations associated with antiviral resistance is important for treatment decisions. Previous studies have shown that R289K mutant, which confers reduced susceptibility to neuraminidase inhibitors, could emerge quickly after treatment with neuraminidase inhibitors.27, 28 However, R289K mutation was not found in the A/Hong Kong/470129/2013 strain, suggesting that this strain remain susceptible to neuraminidase inhibitors. We have performed direct sequencing from the endotracheal aspirate sample to decrease the chance of mutations being enriched during viral replication in the cell or chicken egg culture.29 We have also manually inspected the sequence chromatogram to observe for double peaks which signify the presence of quasispecies in the original specimen. Although we did not observe any double peaks, we cannot exclude the possibility of a mutant quasispecies population present in low frequency.
Since our study analysed gene sequences only from publicly accessible database, one limitation is that there may exist other human or poultry A(H7N9) strains that is closely related to our present isolate. In summary, our results showed that the A(H7N9) virus continues to evolve in nature and novel reassortants are likely to emerge in the future. Further studies are necessary to understand the virulence and transmissibility of this novel reassortant.
Acknowledgement
We are grateful for the generous support of Hui Hoy, and Hui Ming in the genomic sequencing platform. This work was partly supported by Providence Foundation Limited in memory of the late Lui Hac Minh, the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Disease for the HKSAR Department of Health, and Research Grants Council of the HKSAR (7620/10M and 7629/13M) . We acknowledge Yuelong Shu for permitting us to use the data he deposited in Global Initiative on Sharing All Influenza Data (GISAID) EpiFlu™ Database.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jinf.2014.02.012.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Chan J.F., To K.K., Tse H., Jin D.Y., Yuen K.Y. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013 Jun 13 doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuan J., Zhang L., Kan X., Jiang L., Yang J., Guo Z. Origin and molecular characteristics of a novel 2013 avian influenza A(H6N1) virus causing human infection in Taiwan. Clin Infect Dis. 2013 Nov;57(9):1367–1368. doi: 10.1093/cid/cit479. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y., Liang W., Yang S., Wu N., Gao H., Sheng J. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet. 2013 Jun 1;381(9881):1916–1925. doi: 10.1016/S0140-6736(13)60903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.To K.K., Chan J.F., Chen H., Li L., Yuen K.Y. The emergence of influenza A H7N9 in human beings 16 years after influenza A H5N1: a tale of two cities. Lancet Infect Dis. 2013;13:809–821. doi: 10.1016/S1473-3099(13)70167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng V.C., To K.K., Tse H., Hung I.F., Yuen K.Y. Two years after pandemic influenza A/2009/H1N1: what have we learned? Clin Microbiol Rev. 2012 Apr;25(2):223–263. doi: 10.1128/CMR.05012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007 Oct;20(4):660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization. Number of confirmed human cases of avian influenza A(H7N9) reported to WHO. Available at: http://www.who.int/influenza/human_animal_interface/influenza_h7n9/Data_Reports/en/index.html [accessed 10.12.13].
- 8.Yu L., Wang Z., Chen Y., Ding W., Jia H., Chan J.F. Clinical, virological, and histopathological manifestations of fatal human infections by avian influenza A(H7N9) virus. Clin Infect Dis. 2013 Nov;57(10):1449–1457. doi: 10.1093/cid/cit541. [DOI] [PubMed] [Google Scholar]
- 9.Yang S., Chen Y., Cui D., Yao H., Lou J., Huo Z. Avian-origin influenza A(H7N9) infection in influenza A(H7N9)-affected areas of China: a serological study. J Infect Dis. 2014 Jan 15;209(2):265–269. doi: 10.1093/infdis/jit430. [DOI] [PubMed] [Google Scholar]
- 10.Gao H.N., Lu H.Z., Cao B., Du B., Shang H., Gan J.H. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med. 2013 Jun 13;368(24):2277–2285. doi: 10.1056/NEJMoa1305584. [DOI] [PubMed] [Google Scholar]
- 11.Qi X., Qian Y.H., Bao C.J., Guo X.L., Cui L.B., Tang F.Y. Probable person to person transmission of novel avian influenza A (H7N9) virus in Eastern China, 2013: epidemiological investigation. BMJ. 2013;347:f4752. doi: 10.1136/bmj.f4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cowling B.J., Jin L., Lau E.H., Liao Q., Wu P., Jiang H. Comparative epidemiology of human infections with avian influenza A H7N9 and H5N1 viruses in China: a population-based study of laboratory-confirmed cases. Lancet. 2013 Jul 13;382(9887):129–137. doi: 10.1016/S0140-6736(13)61171-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.To K.K., Ng K.H., Que T.L., Chan J.M., Tsang K.Y., Tsang A.K. Avian influenza A H5N1 virus: a continuous threat to humans. Emerg Microbes Infect. 2012;1:e25. doi: 10.1038/emi.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen E., Chen Y., Fu L., Chen Z., Gong Z., Mao H. Human infection with avian influenza A(H7N9) virus re-emerges in China in winter 2013. Euro Surveill. 2013;18(43) doi: 10.2807/1560-7917.es2013.18.43.20616. [DOI] [PubMed] [Google Scholar]
- 15.The Government of the Hong Kong Special Administrative Region. SFH on confirmed human case of avian influenza A(H7N9). Available at: http://www.info.gov.hk/gia/general/201312/03/P201312030033.htm [accessed on 03.10.13].
- 16.Bernard G.R., Artigas A., Brigham K.L., Carlet J., Falke K., Hudson L. The American-European Consensus Conference on ARDS. Definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med. 1994 Mar;149(3 Pt 1):818–824. doi: 10.1164/ajrccm.149.3.7509706. [DOI] [PubMed] [Google Scholar]
- 17.Chan K.H., To K.K., Chan B.W., Li C.P., Chiu S.S., Yuen K.Y. Comparison of pyrosequencing, Sanger sequencing, and melting curve analysis for detection of low-frequency macrolide-resistant mycoplasma pneumoniae quasispecies in respiratory specimens. J Clin Microbiol. 2013 Aug;51(8):2592–2598. doi: 10.1128/JCM.00785-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herfst S., Schrauwen E.J., Linster M., Chutinimitkul S., de Wit E., Munster V.J. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012 Jun 22;336(6088):1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bussey K.A., Desmet E.A., Mattiacio J.L., Hamilton A., Bradel-Tretheway B., Bussey H.E. PA residues in the 2009 H1N1 pandemic influenza virus enhance avian influenza virus polymerase activity in mammalian cells. J Virol. 2011 Jul;85(14):7020–7028. doi: 10.1128/JVI.00522-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuen K.Y., Chan P.K., Peiris M., Tsang D.N., Que T.L., Shortridge K.F. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998 Feb 14;351(9101):467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 21.Gao R., Cao B., Hu Y., Feng Z., Wang D., Hu W. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013 May 16;368(20):1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 22.Lam T.T., Wang J., Shen Y., Zhou B., Duan L., Cheung C.L. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature. 2013 Oct 10;502(7470):241–244. doi: 10.1038/nature12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu D., Shi W., Shi Y., Wang D., Xiao H., Li W. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013 Jun 1;381(9881):1926–1932. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L., Zhang Z., Weng Z. Rapid reassortment of internal genes in avian influenza A(H7N9) virus. Clin Infect Dis. 2013 Oct;57(7):1059–1061. doi: 10.1093/cid/cit414. [DOI] [PubMed] [Google Scholar]
- 25.Manz B., Schwemmle M., Brunotte L. Adaptation of avian influenza A virus polymerase in mammals to overcome the host species barrier. J Virol. 2013 Jul;87(13):7200–7209. doi: 10.1128/JVI.00980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jonges M., Bataille A., Enserink R., Meijer A., Fouchier R.A., Stegeman A. Comparative analysis of avian influenza virus diversity in poultry and humans during a highly pathogenic avian influenza A (H7N7) virus outbreak. J Virol. 2011 Oct;85(20):10598–10604. doi: 10.1128/JVI.05369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Y., Lu S., Song Z., Wang W., Hao P., Li J. Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet. 2013 Jun 29;381(9885):2273–2279. doi: 10.1016/S0140-6736(13)61125-3. [DOI] [PubMed] [Google Scholar]
- 28.Lin P.H., Chao T.L., Kuo S.W., Wang J.T., Hung C.C., Lin H.C. Virological, serological, and antiviral studies in an imported human case of avian influenza A(H7N9) virus in Taiwan. Clin Infect Dis. 2014 Jan;58(2):242–246. doi: 10.1093/cid/cit638. [DOI] [PubMed] [Google Scholar]
- 29.Ren X., Yang F., Hu Y., Zhang T., Liu L., Dong J. Full genome of influenza A (H7N9) virus derived by direct sequencing without culture. Emerg Infect Dis. 2013 Nov;19(11) doi: 10.3201/eid1911.130664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.