Abstract
Capsid proteins are structural components of virus particles. They are nucleic acid-binding proteins whose main recognized function is to package viral genomes into protective structures called nucleocapsids. Research over the last 10 years indicates that in addition to their role as genome guardians, viral capsid proteins modulate host cell signaling networks. Disruption or alteration of intracellular signaling pathways by viral capsids may benefit replication of the virus by affecting innate immunity and in some cases, may underlie disease progression. In this review, we describe how the capsid proteins from medically relevant RNA viruses interact with host cell signaling pathways.
Keywords: RNA virus, Capsid, Apoptosis, Signaling proteins, Flaviviruses, SARS, Rubella virus
1. Introduction
RNA viruses cause the majority of virus-associated acute and chronic diseases in humans including some of the most devastating epidemics over the last few centuries. For example, it is estimated that more people (> 40 million) were killed by orthomyxoviruses as a result of the 1918 influenza pandemic than in World War I [1]. Other simple RNA viruses including hepatitis C and dengue viruses have infected more than 400 million people worldwide.
Without exception, all viruses that infect mammalian cells must replicate before the host immune system defenses shutdown virus production and/or destroys the infected cell. It is well documented that large DNA viruses such as pox and herpes viruses express regulatory proteins that interfere with the host defense systems thereby providing a window of opportunity for the virus to replicate and/or disseminate [2]. By comparison, RNA viruses are genetically simple, often encoding less than 10 proteins many of which are involved in viral assembly. However, they too face the same challenges as genetically complex viruses in that they must at least temporarily evade the immune response until viral replication and egress take place.
It has been known for quite some time that RNA viruses produce structural proteins in vast excess to what is needed or used for assembly of virions. Moreover, even though they may be the first viral proteins synthesized in infected cells, structural proteins are not necessarily incorporated into virions immediately after synthesis [3]. This suggests that there are large pools of viral structural proteins present within infected cells that are available for “nonstructural” or “ambassadorial” functions [4]. Given their relatively small genome sizes, it is certainly beneficial for RNA viruses to encode multifunctional proteins. In the following review, we discuss the evidence that in addition to serving as building blocks of virions, RNA virus capsid proteins function as important mediators of virus–host interactions (Fig. 1 ). Unlike other viral structural antigens such as envelope glycoproteins which are largely sequestered within membranes of the central vacuolar system, capsid proteins are exposed to a wide variety of host cell proteins in the cytoplasm and nuclei. In addition, they are often produced in such large quantities that they can in theory, overwhelm signaling circuits by engaging in stoichiometric interactions with pathway components. As such, they are prime candidates for mediating virus–host interactions at the cellular level. For the sake of brevity, our discussion of capsids is limited to those of RNA viruses that cause serious human diseases.
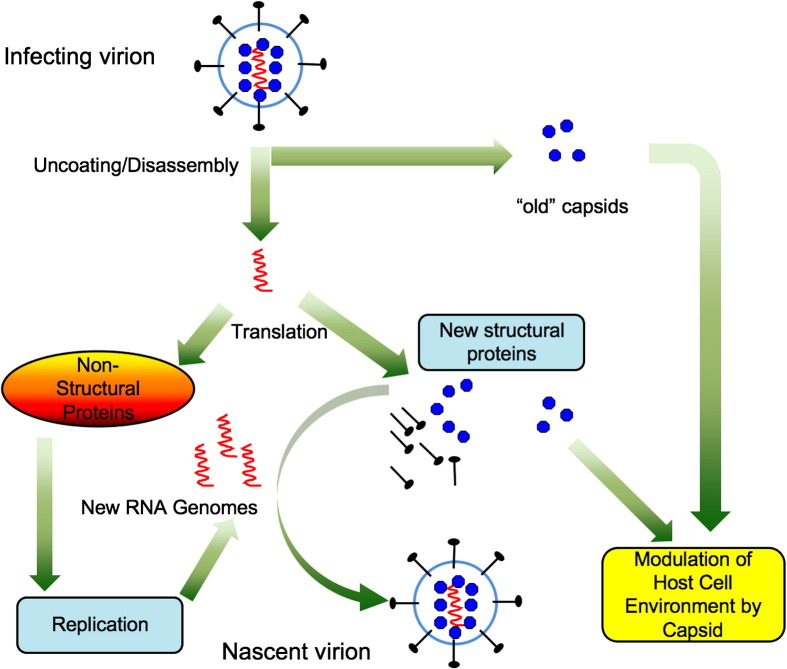
Fig. 1.
Multifunctionality of RNA virus capsid proteins. A generic RNA virus is used as the model. Virions consist of a host-derived lipid envelope containing virus-encoded membrane glycoproteins. The interior of the virion consists of genomic RNA bound to capsid proteins. Upon entry, deposition of nucleocapid into the cytoplasm provides the first opportunity for capsids to interact with signaling proteins. The viral genome is translated to produce structural and nonstructural proteins. The nonstructural proteins function in replication of the genome whereas the structural proteins function in assembly of new virions. Recent evidence suggests that nascent viral capsids also interface with cellular proteins to alter the host cell environment so that it is more permissive for viral replication.
2. Flaviviruses
The family Flaviviridae is comprised of a large group (> 70 different types) of enveloped single strand RNA viruses that replicate in the cytoplasm of infected cells (for recent review [5]). Many flaviviruses are human pathogens and together, they are responsible for hundreds of millions of acute and chronic infections: The most serious of which include hemorrhagic fevers, encephalitis and hepatitis. Virions consist of a host cell-derived envelope that contains two virus-encoded glycoproteins that function in virus entry. Capsid proteins are internal structural components whose primary function is to package viral genomic RNA into nucleocapsids. A great deal of evidence suggests that in addition to their well-defined roles in virus assembly, flavivirus capsid proteins have important nonstructural functions. For instance, even though flavivirus replication is confined to the cytoplasm of host cells and assembly of nascent virions occurs on the endoplasmic reticulum (ER), large pools of flavivirus capsid proteins are localized to the nuclei of infected cells (Fig. 2 ). As there is no obvious role for nuclear localized capsid in virus assembly, the nuclear cohort of capsid is thought to engage in nonstructural functions, presumably in modifying the host cell environment in such a way that virus replication and/or dissemination is benefited.
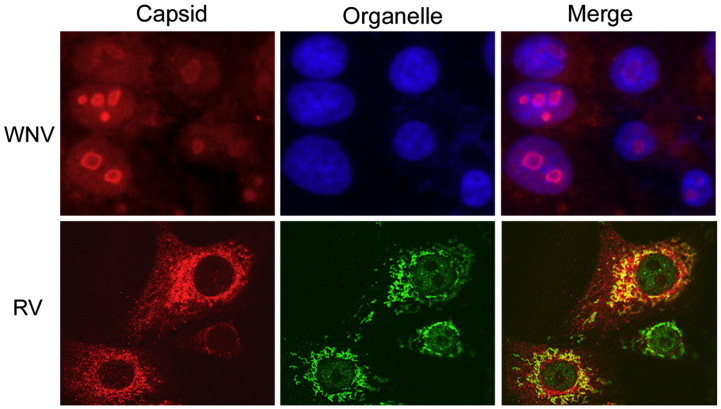
Fig. 2.
Capsid proteins localize to intracellular sites that are not related to virus assembly. West Nile virus (WNV) and Rubella virus (RV) both replicate in the cytoplasm and assemble on membranes of the endoplasmic reticulum and Golgi complex respectively. However, large pools of their capsid proteins localize to the nucleous (WNV) or mitochondria (RV). Cells were infected with WNV (upper panels) or RV (lower panels) and then fixed and processed for indirect immunofluorescence. Nuclei were stained with DAPI and mitochondria were detected with antibodies specific for the matrix protein p32.
2.1. Hepatitis C virus capsid/core protein
Hepatitis C virus (HCV) is the major etiological agent of non-A, non-B hepatitis. From a global and economic perspective, it is an extremely important pathogen and current estimates suggest that over 300 million individuals or 5% of the world population is infected with HCV (reviewed in [6], [7]). In approximately 85% of cases, infection becomes chronic in which case the virus can persist for decades. Patients chronically infected with HCV account for approximately 30–40% of all liver transplants. As such, HCV has been the focus of much research in an attempt to understand the mechanisms by which the virus causes and maintains a disease state.
The HCV capsid, also known as core protein, is by far the best studied of the flavivirus capsids. It has long been implicated in disease development through its roles in apoptotic pathways [8], [9] as well as regulating the innate immune response [10]. Data from studies over the last 10 years indicate that the HCV core protein interacts with at least 20 different host cell proteins ([11] and Table 1 ). In addition, microarray analyses revealed that it directly and/or indirectly regulates the expression of more than 400 human genes [12], [13]. Core is the first viral protein synthesized in an infected cell, however it is not required as a structural component until late in the assembly pathway. Furthermore, during viral entry it is the first viral protein to contact host cell proteins in the cytoplasm. Temporally and spatially, this places the HCV core protein in a strategic position to act as a modulator of the host cell environment.
Table 1.
Known host cell proteins that have been reported to interact with selected RNA virus capsid proteins
| Virus | Host cell protein | Function | Reference(s) |
|---|---|---|---|
| West Nile virus | I2PPA | Signal transduction | [61] |
| Jab1 | Transcriptional control, regulation of protein stability | [114] | |
| HDM2 | Cell cycle, regulation of protein stability | [63] | |
| Hepatitis C virus | Apolipoprotein AII | Lipid metabolism | [115], [116] |
| Cap-Rf | RNA helicase | [34] | |
| Complement Receptor gC1qR | T-cell response | [117] | |
| Cyclin dependent kinase 7 | Cell cycle regulation | [118] | |
| DEAD-box protein DBX | RNA helicase | [119] | |
| DEAD-box protein 3 | RNA helicase | [120] | |
| Heterogeneous nuclear ribonucleoprotein K | Transcriptional control | [121] | |
| JAK1/2 | Signal transduction | [122] | |
| Lymphotoxin-β receptor | Apoptosis | [123] | |
| p53 | Transcriptional control | [124] | |
| p73 | Transcriptional control | [125] | |
| Proteasome activator PA28γ | Protein degradation | [126] | |
| Retinoid X receptor α | Transcriptional control | [127] | |
| Smad3 | Transcriptional control | [128] | |
| sp110b | Transcriptional control | [129] | |
| STAT3 | Cell transformation | [130] | |
| TAFII28 | Transcriptional control | [124] | |
| Tumor necrosis factor receptor I | Apoptosis | [131] | |
| 14-3-3 protein | Regulate activity and transport of various cellular proteins | [132] | |
| Rubella virus | Par-4 | Apoptosis | [101] |
| p32 | Mitochondrial function, apoptosis | [98], [101] | |
| Hantavirus | Daxx | Apoptosis | [133] |
| SARS coronavirus | Ubc9 | Protein sumoylation | [134] |
| hnRNPA1 | mRNA processing and export | [113] | |
| Cyclophilin | Protein folding/maturation | [112] | |
| 14-3-3 proteins | Regulate activity and transport of wide variety of cellular proteins | [109] |
The most commonly known functions of each host cell protein are listed.
The immature form of the HCV core exists as a monomer and contains a hydrophobic domain at the carboxyl-terminus. Maturation of core requires processing by the host enzyme signal peptide peptidase to remove the hydrophobic carboxyl domain [14]. The mature form of the protein is often found in oligomeric complexes containing 24 or more copies of core. It is believed that the lack of multimeric structure in immature core makes it more amenable to interaction with host cell factors. Such interactions may be important for regulating cell cycle progression and intracellular signaling pathways that modulate apoptosis. Interestingly, the mature form of core also localizes to the nucleus [15], [16] and as such, both forms of the protein may have roles in virus–host interactions at the cellular level.
2.1.1. HCV core and apoptosis
Evidence supporting the idea that core is an important pathogenic determinant of HCV disease stems from the observation that expression of this viral protein in the livers of transgenic mice, induces liver steatosis and hepatocellular carcinoma [17], [18]. Most of the studies on HCV core have focused on its role in modulating apoptosis. Indeed, evidence suggests that apoptosis is an important factor in development of HCV-induced liver damage, particularly during chronic infection [19]. Whereas many reports indicate that capsid is a pro-apoptotic factor, a substantial number of studies suggest that this protein may also have anti-apoptotic effects.
2.1.2. Pro-apoptotic effects of HCV core protein
One of the first clues that apoptosis is relevant to HCV-induced liver disease was the observation that elevated levels of the pro-apoptotic protein Fas/CD95 and associated inflammation, correlated with expression of HCV antigens in the liver [20]. Specifically, among HCV patients, Fas/CD95 expression was higher in cells that had detectable HCV antigen. A subsequent study reported that expression of HCV core but not envelope glycoproteins or nonstructural proteins, sensitizes HepG2 cells to Fas/CD95 mediated apoptosis [21]. Seemingly at odds with the study by Hiramatsu et al. [20], Ruggleri et al. reported that core expression did not result in increased Fas/CD95 levels but rather induced upregulation of Fas ligand at the transcriptional level [22]. HCV core expression also sensitizes mammalian cells to apoptosis under serum-limiting conditions by a mechanism that involves upregulation of pro-apoptotic proteins including p53 and Bax [23]. Finally, core may also affect Fas-dependent apoptosis in hepatocytes by blocking survival signaling through the p38 mitogen activated kinase pathway [24].
Fas/CD95 is a death receptor that is part of the tumor necrosis factor alpha (TNF-α) receptor family. In addition to binding Fas/CD95, it is now clear that HCV core protein also directly interacts with the cytoplasmic domains of other TNF family death receptors (TNF-R1 and lymphotoxin-β receptor) [25]. Many similarities exist between Fas ligand and TNF-α-mediated apoptosis but it appears that core can affect each pathway differently (Fig. 3 ). Depending upon numerous factors including cell type and the physiological state of the cell, exposure to TNF-α can have pro- or anti-apoptotic effects. Immunohistochemical analyses first pointed to a role for TNF-α in liver inflammation of non-A, non-B hepatitis patients [26] and a subsequent study revealed that expression of HCV core potentiates TNF-α-dependent apoptosis likely through binding the cytoplasmic domain of the TNFR-1 [25]. An independent study also showed that core binds to the cytoplasmic domain and activates the related receptor, lymphotoxin-β receptor [27]. Similar to TNFR-1, signaling from the lymphotoxin-β receptor has been shown to induce cell death [28]. As well as activating death-inducing signaling through TNFR-1 and/or lymphotoxin-β receptors, HCV core reportedly inhibits TNF-α-dependent survival pathways that involve NF-κB signaling [29]. Together, these data support a role for HCV core protein in the induction and/or sensitization of cells to apoptosis. Through interaction with three distinct but related branches of extrinsic apoptotic signaling mechanisms, and inhibition of survival signals, core expression may be a central pathogenic determinant of liver disease. It is important to point out that apoptotic death of the host cell can have advantages for some viruses. Specifically, release of virions in apoptotic bodies that can be taken up by surrounding cells will limit inflammation thereby allowing the virus to spread relatively undetected by the immune system [2].
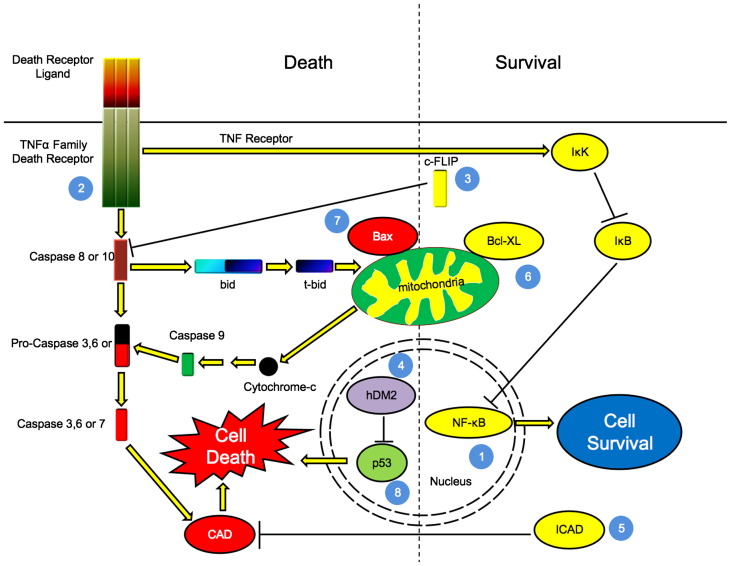
Fig. 3.
Flavivirus capsid proteins can affect apoptotic signalling pathways through multiple mechanisms. Numbered blue circles represent points at which capsid proteins act: 1: HCV core protein upregulates activity of transcription factor NF-κB, which leads to increased transcription of pro-survival genes. Core potentiated activation of TNFR may also lead to activation of IκB Kinase (IKK), which in turn phosphorylates and deactivates the NF-κB inhibitor IκB. This allows NF-κB to enhance transcripton of its target genes. 2: HCV core protein binds to the cytoplasmic domain of death receptors leading to potentiation of TNF-α mediated apoptosis, or alternatively, activation of NF-κB, a pathway that requires RIP and IKK. 3: Upregulation of c-FLIP by HCV core results in blocking apoptosis through inhibition of caspase-8 activation. 4: WNV capsid protein promotes p53 mediated apoptosis by blocking interaction of hDM2 with p53, thereby increasing the stability and consequent increase in transcription of pro-apoptotic target genes. 5: HCV core upregulates the levels of ICAD, thus inhibiting caspase mediated DNA cleavage. 6 and 7: The HCV core reportedly upregulates expression anti-apoptotic (Bcl-XL and pro-apoptotic (Bax) Bcl-2 family members that function at the level of mitochondria. 8: HCV core also increases the expression of the pro-apoptotic transcription p53, one of whose functions is to upregulate Bax. For more details, please see text. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Finally, there is evidence to suggest that core-induced Fas-dependent apoptosis in T lymphocytes may contribute to viral persistence. Chronic HCV infection is characterized by T-cell dysfunction and depletion [30] and interestingly, expression of core protein sensitizes cultured T-cells to Fas-mediated apoptosis [31]. Moreover, T-cells that are not directly infected with HCV may undergo apoptosis as the result of a bystander effect. Evidence for this notion stems from the observation that co-cultivation of a T-cell line with HepG2 cells expressing HCV core, resulted in increased sensitivity of the T-cells to Fas-mediated apoptosis [22].
2.1.3. Anti-apoptotic/pro-survival effects of HCV core protein
As discussed above, many studies have documented the pro-apoptotic effects of HCV core protein, however there is a considerable body of work indicating that this viral protein also functions to confer resistance to apoptosis. HCV infection frequently becomes persistent, thus it stands to reason that the virus utilizes strategies to prevent the premature death of at least a subset of infected cells. Virus-induced anti-apoptotic signaling through core-dependent mechanisms is thought to account for why the virus can avoid elimination by the immune system as well as the development of hepatocellular carcinoma.
An effective way to counteract apoptosis is to upregulate the expression of proteins that block programmed cell death. With respect to HCV core protein, it has been demonstrated that expression of this viral protein in HepG2 cells upregulates the anti-apoptotic Bcl-2 family member Bcl-XL at the transcriptional level [32], [33], [34], [35]. Regarding establishment of chronic HCV, it is interesting to note that over-expression of anti-apoptotic Bcl-2 family members can protect hepatocytes from Fas-mediated apoptosis [36]. Core expression can also activate the NF-κB pathway (Fig. 3). Indeed the anti-apoptotic effects of core are reportedly dependent upon activation of this pathway [37], [38]. The Bcl-XL gene has an NF-κB responsive element, but intriguingly, core-dependent upregulation of Bcl-XL does not depend on NF-κB.
A recent study found that HCV core inhibits TNF-α dependent apoptosis by sustaining the expression of FADD-like interleukin-1β-converting enzyme like inhibitory protein (c-FLIP) [39]. C-FLIP is an inhibitor of TNF-α mediated apoptosis and functions by inhibiting caspase-8 activity (Fig. 3). Core-dependent upregulation of c-FLIP does not involve MAP kinase or phosphoinositide-3 kinase signaling but may be a consequence of increased NF-κB activity [40], [41]. Reducing c-FLIP levels with small interfering RNAs can abrogate the anti-apoptotic effect of HCV core. This may indicate that inhibition of caspase activity is the most important mechanism by which core blocks apoptosis.
Finally, recent data have emerged which indicate that HCV core directly associates with the TNFR1–TRADD–TRAF2 signaling complex in order to potentiate TNF-α mediated NF-κB activation [37]. TRADD is a signal transducer that binds to the cytoplasmic domain of TNFR1 as well as TRAF2, a signaling protein that activates NF-κB [42]. Expression of a dominant negative TRAF2 construct inhibited the effect of HCV core on NF-κB signaling. The core protein may act to inhibit Fas and TNF-α mediated apoptosis by indirectly interfering with the function of the caspase-activated DNase (CAD), a nuclease that cleaves cellular DNA following activation by caspases [9], [39]. CAD is subject to regulation by an inhibitor protein (ICAD) whose activity level can influence the outcome of cell death programs (reviewed in [43]). Core protein increases the expression of ICAD leading to inhibition of TNF-α and Fas-mediated apoptotic death (Fig. 3).
2.1.4. So, is the HCV core pro- or anti-apoptotic?
The short answer is yes. Some of the apparent discrepancies regarding whether core is pro- or anti-apoptotic may be explained by the fact that the effects of core on signaling pathways are isoform dependent. Like many other RNA viruses, HCV is actually a quasi-species; a collection of closely related but not identical genetic variants that arise due to a combination of selection pressures and the low fidelity of the HCV RNA-dependent RNA polymerase [44]. Indeed, slight differences in core sequence can have dramatic differences in activation of signaling pathways. For example, core-dependent regulation of NF-κB signaling is genotype dependent. Core proteins from HCV 1a and 1b isolates differed by five fold in their abilities to affect transcription of a reporter gene under the control of a NF-κB-responsive promoter [45]. Further analyses revealed that as little as two amino acid residue changes within core were responsible for these differences. As NF-κB activation can inhibit the death of cells from apoptosis, it is possible that during disease progression, core variants that block cell death predominate, a situation that may contribute to development of hepatocellular carcinoma. Expression of core genes derived from variants of the same subtype of HCV can also have differential effects on signaling pathways. For instance, Polyak et al. demonstrated that expression of two core genes isolated from HCV genotype 1b infected patients, have dramatically different effects on type I interferon signal transduction [11].
Further data supporting the idea that quasi-species variation is important for viral persistence came from the study of core variants isolated from tumor and non-tumor tissue [46]. Variants from tumor tissue showed significantly greater abilities to inhibit the activation of the TGF-β pathway, an effect that is likely potentiated through direct interaction with SMAD3 (reviewed in [47]). Although sequence differences between core isolates can be found, there do not appear to be any hotspots in the core gene sequence per se. Instead it seems that environmental pressures can generate several types of mutations which, although having different sequences, can have similar biological properties [46].
2.1.5. Effect of HCV core on gene expression
The HCV core protein is thought to alter intracellular signaling by direct interaction with components of signaling pathways. In addition, it appears that the core protein can alter host cell gene expression programs by direct and indirect means. First, it has been reported that HCV core binds to and represses the p53 promoter, thus blocking p53 synthesis [48]. Recall that p53 is a transcription factor that can activate expression of pro-apoptotic Bcl-2 family members such as Bax. Subsequent studies have shown that core significantly affects the expression of more than 400 human genes [12], [13]. In hepatocytes, core upregulates anti-apoptotic and immune evasion genes such as PAK2, AP15, BH1 and Tax1BP1. In T-cells, genes involved in anergy and slowing the rate of cell cycle progression are activated by core. Many but not all of the changes in core-dependent gene expression are due to upregulation of the transcription factors NF-κB and NFAT. Another mechanism by which HCV core may affect global gene expression patterns is through the RNA interference pathway. Core protein reportedly binds to and inhibits the RNase III protein Dicer [49], an enzyme that processes small regulatory RNAs that fine tune the expression of up to 30% of human genes [50]. Together, these data suggest that core-dependent reprogramming of hepatocytes may contribute to viral persistence by suppressing apoptosis in hepatocytes and inducing T-cell anergy.
In summary, despite the inordinate amount of research on the effects of core expression on cellular signaling pathways, particularly those that influence apoptosis, a unifying mechanism to explain all of the core-specific effects is lacking. It is important to restate that many of these studies involved expression of core protein in the absence of other HCV proteins and as such, the relevance to the in vivo situation can rightfully be questioned. Moreover, the effect of core expression on signaling is surely dependent on core sequence, cell type, expression level and whether or not the expression is transient or stable. However, it is clear that core protein expression does affect multiple signaling pathways in host cells. Fortunately, with the recent development of infectious cDNA clones for HCV [51], [52], reverse genetic experiments will soon provide further insight into the role of this viral protein in pathogenesis.
2.2. Japanese encephalitis virus serogroup
The Japanese encephalitis serogroup is divided into 12 antigenic serogroups which includes the human pathogens West Nile, St. Louis encephalitis, dengue, and yellow fever viruses [5], [53]. All viruses employ a zoonotic transmission cycle to move from pig or bird reservoirs to dead end hosts such as humans by using mosquito vectors. West Nile virus (WNV) is the etiologic agent of an acute febrile illness often characterized by the onset of meningo-encephalitis and in some cases, death. It is the most common cause of viral encephalitis in North America [54]. Japanese encephalitis virus (JEV) is endemic in south and southeast Asia and similar to WNV, can infect the central nervous system and has a high mortality rate [55]. Dengue virus is an extremely important pathogen that infects more than 100 million people each year [56]. It is the most common cause of arboviral disease in the world and there are no specific treatments for this disease. Although virus assembly occurs on ER membranes, a large fraction of the capsid pools in WNV, JEV and dengue infected cells accumulates in the nucleus/nucleolus (Fig. 2) and recent evidence is consistent with these viral proteins playing a role in pathogenesis via interactions with cellular signaling pathways [57], [58], [59], [60], [61].
2.2.1. West Nile and Japanese encephalitis virus capsid proteins
Shortly after the emergence of highly pathogenic WNV strains in North America in 1999, interest in the potential role of capsid as a pathogenic determinant intensified. Collectively, the data from the studies described below suggest that the WNV capsid is a pro-apoptotic factor. Specifically, expression of the capsid in the absence of other viral proteins induces apoptosis in a number of different in vitro and in vivo systems. For example, transient expression of capsid protein in multiple human cells lines results in loss of mitochondrial membrane potential and activation of caspase-9 and caspase-3 [62]. In vivo, expression of capsid in the brains of laboratory mice causes local inflammation and apoptotic cell death. The precise mechanism by which the capsid initiates apoptosis through the mitochondrial pathway is not well understood, however, targeting of capsid to the nucleus may be important for this process [59], [61], [63]. A recent report identified the nuclear protein HDM2 as a capsid-binding protein [63]. HDM2 is an E3 ubiquitin ligase that regulates levels of the transcription factor p53 [64], [65]. One way in which p53 can act in a pro-apoptotic manner is to upregulate levels of Bax, an apoptotis inducing member of the Bcl-2 family that acts at the level of mitochondria [66]. Inhibition of the E3 ligase activity of HDM2 by capsid results in accumulation of p53 and consequent transcriptional upregulation of Bax and subsequent loss of mitochondrial membrane potential (Fig. 3). Mapping studies revealed that capsid mutants lacking the carboxyl-terminal 18 amino acid residues do not bind HDM2 which coincidentally, is a region of the protein previously shown to be important for induction of apoptosis [62]. It has yet to be determined if this part of capsid is sufficient for apoptosis.
Nuclear localization of the JEV capsid protein also appears to be important for viral replication and pathogenesis [60]. A conserved glycine–proline motif in the central region of the capsid is essential for nuclear localization of capsid. Recombinant JEV strains with alanine substitution mutations in this motif exhibited severely reduced replication in mammalian and insect cells. Interestingly, whereas mutations in the GP motif had no effect on neurovirulence in mice, the neuroinvasiveness was attenuated. The authors concluded that nuclear localization of the capsid protein was important for pathogenesis of encephalitis, possibly by allowing the virus to breach the blood brain barrier. In this respect, it was recently demonstrated that TNF-α signaling is integral to the ability of WNV to infect cells of the central nervous system [67]. While it is clear that the GP motif is required for neuroinvasion, it remains to be determined however, if nuclear localization per se is required for neuropathogenesis. The GP motif does not resemble any known nuclear localization signals and it is possible that mutagenesis of the GP codons results in a conformational change that prevents the capsid from exposing the nuclear localization signal and/or potentially activating signaling pathways that result in secretion of neuroinflammatory cytokines such as TNF-α.
WNV infection results in transcriptional upregulation of TNF-α [68] but it has yet to be determined if capsid is the viral protein that mediates this process. However, it is known that expression of WNV capsid in the absence of other viral proteins in neurons and astrocytes leads neuronal apoptosis and release of several neuroinflammatory mediators including CXCL10 [69]. This study also demonstrated that capsid suppresses the expression of OASIS, a protein whose induction is associated with a protective response to ER stress [70].
WNV capsid expression upregulates phosphatase 2A (PP2A) activity [61]. Elevated PP2A activity has also been reported in HCV-infected cells [71], and is purportedly important for blocking interferon-α signaling. PP2A suppresses the activity of AP-1 [72], a transcription factor that upregulates type I interferon genes [73]. To our knowledge, the work of Hunt et al. [61] is the first to report that expression of a flavivirus capsid protein is sufficient to increase the activity of this phosphatase. Presumably, WNV capsid influences PP2A activity through interaction with I2 PP2A, also known as SET [74]. I2 PP2A is a major capsid-binding protein that like capsid, is concentrated in the nucleus [61]. The capsid-binding site overlaps with the region of I2 PP2A that is required for inhibition of PP2A activity. Whereas capsid expression negatively affects AP-1 activity likely through increased PP2A activity [61], I2 PP2A expression increases the transcriptional activity of the AP-1 complex [72]. We hypothesize that capsid-mediated downregulation of AP-1 serves to dampen or delay the innate immune response.
Finally, it is interesting to note that the capsid protein binding region of I2 PP2A includes amino acid residues 1–108, the region of I2 PP2A that is essential for interaction with the Cdk5-activating protein p35 (residues 29–94) [75], [76]. Binding of I2 PP2A to p35 is associated with activation of the Cdk5 complex which is required for signaling at the neuromuscular synapse [77]. Ultimately, it may be of interest to investigate whether capsid/I2 PP2A interactions contribute to neurological complications caused by WNV. However, these studies will require I2 PP2A null mice, which to our knowledge are not yet available.
2.2.2. Dengue virus capsid
A large pool of dengue virus capsid protein localizes to the nucleus [58] where it presumably functions in processes that are not directly related to virus assembly. Expression of dengue capsid protein sensitizes HepG2 cells to Fas-dependent apoptosis possibly by a mechanism that involves binding to Daxx, a cellular protein that shuttles between the nucleus and cytoplasm. Daxx is pro-apoptotic and functions in the Fas-dependent pathway by two different mechanisms. When localized to the nucleus it acts as a transcriptional repressor of Bcl-2 genes [78], [79]. In the cytoplasm, using different domains, Daxx stimulates apoptosis through interactions with Fas at the plasma membrane and as well as binding to and activating the kinase JNK [80]. It is believed that interactions between the capsid and Daxx occur in the nucleus and under this scenario, it is possible that capsid blocks translocation of Daxx to the cytoplasm without affecting its repressor activity in the nucleus. Given that liver damage as a result of virus-induced apoptosis may be a key component of dengue hemorrhagic fever and dengue shock syndrome [81], the capsid protein may be viewed as an important pathogenic determinant.
3. Togavirus capsid proteins
The family Togaviridae is comprised of two genera, Alphavirus and Rubivirus. The genus Alphavirus contains at least 28 species that are widely distributed throughout the world. Members of this genus are typically maintained in natural cycles involving transmission by an arthropod vector among susceptible vertebrate hosts [82]. Alphaviruses cause a wide range of diseases in mammals the most serious of which are neurological disorders and encephalitis following infection of the central nervous system. Evidence suggests that apoptosis in neurons is a primary mechanism of viral disease [83], [84], however to date, there is no evidence indicating that capsid proteins function in this process. A limited number of studies suggest alphavirus capsids function in signalling mechanisms that regulate translation of host cell proteins [85], [86], [87]. Independent of double strand RNA, purified alphavirus capsids stimulate the phosphorylation of the interferon-induced, double-stranded RNA-activated protein kinase, PKR. Subsequent PKR-dependent phosphorylation of the translation initiation factor (eIF-2), results in blocking host cell translation. These data indicate that alphavirus capsid proteins play a central role in shutoff of host cell protein synthesis. Unfortunately, follow up studies to address the mechanism of this phenomenon have not been published. Given that alphaviruses have served as excellent models for the study of virus replication and virus–host interactions, it is surprising that so little is known about capsids and signaling.
The genus Rubivirus contains only one member, Rubella virus (RV) and worldwide, this virus is responsible for more birth defects than any other infectious agent (reviewed in [88]). It has been proposed that the severe pathology associated with congenital rubella syndrome is at least partially the result of virus-induced apoptosis and/or the innate immune response [89], [90], [91], [92]. The role of capsid in modulating innate immunity has not been explored and its role in apoptosis is controversial. Data from Duncan et al. suggest that membrane-associated capsid protein is pro-apoptotic in certain cell types [93]. Contrary to these findings, Hofmann et al. reported that RV structural proteins including capsid do not induce apoptosis [94]. RV-induced apoptosis is highly dependent upon cell type [95] and as such, the apparent discrepancy between the findings of Duncan and Hoffmann may be due in large part to the fact that different types of expression systems (transient vs. stable) and cell lines were employed.
An interesting feature of the RV capsid is that it displays multiple intracellular localizations [96], [97]. A pool of capsid is targeted to the Golgi complex where it functions in virus assembly but in addition, a large fraction of the protein is associated with mitochondria (Fig. 2). Capsid expression induces perinuclear clustering of mitochondria [98] and there are a number of reasons to think that this may affect apoptotic signalling. First, the distribution of mitochondria is profoundly affected by over-expression of other proteins that modulate apoptosis including TNF receptors and Bax [99], [100]. Second, we and others have shown that capsid binds to the host cell protein p32 [101], [102], a mitochondrial protein that functions in apoptosis through its interactions with the BH3-only protein Hrk [103]. Data from earlier structural studies are consistent with a role for p32 in regulating intramitochondrial Ca+2 and the permeability transition pore complex [104]. Interestingly, Hrk interacts with the carboxyl-terminus of p32, which is the same region that binds capsid [98]. Studies are now underway to further understand how the RV capsid protein affects intracellular signalling from the level of mitochondria.
4. Coronavirus capsid proteins
Coronaviruses are the largest positive strand RNA viruses that have been characterized to date. The most well known human pathogen among this group is the severe acute respiratory syndrome coronavirus (SARS-CoV) [105]. In 2003, SARS outbreaks in Asia and North America resulted in the infection of thousands of people, many hundreds of which died. One of the key structural components of SARS-CoV virions is the N or nucleocapsid protein. Recent studies directed at understanding the pathogenic nature of SARS-CoV suggest that in addition to encapsidating the viral genome, the N protein functions as a regulator of virus–host interactions (reviewed in [106]). In particular, the N protein interferes with production of interferon by blocking the activities of IRF-3 and NF-κB [107]. Similar to NF-κB, IRF-3 is a transcription factor that activates the expression of interferon genes and as such, is an important component of the innate immune response. It is not clear how the SARS-CoV N protein inhibits NF-κB and IRF-3 activity but obviously interaction with host cell proteins is required for this process. At least four host cell-encoded N binding proteins have been identified.
Nuclear-localized N protein has no obvious function in virus assembly which occurs in the cytoplasm, and therefore this cohort of N may be engaged in nonstructural activities [108]. The nuclear localization of N protein is regulated by phosphorylation and binding to 14-3-3 proteins [109]. Reduction of 14-3-3 theta by RNAi results in increased nuclear translocation of N protein [109]. Given that N protein can stimulate transcription of the proinflammatory gene COX2 by binding to the NF-κB response element [110], regulating the amount of N protein in the nucleus by 14-3-3 proteins may be important for limiting virus-induced inflammation. Another way in which N protein may influence pathogenesis is through inhibition of the CDK4-cyclin D complex by binding to the cyclin D subunit [111]. N protein also binds to hnRNPA1 and cyclophilin A [112], [113], however, the importance of these interactions in virus biology has not been established. By understanding how coronavirus N proteins modulate virus–host interactions, it may be possible to use mutated N genes as the basis for recombinant vaccines that have few side effects.
5. Summary and perspectives
The idea of viral capsids as signaling molecules is a relatively new concept and this is reflected in the relative paucity of research in this area. However, if the HCV core protein is any indication, it is reasonable to expect that modulation of intracellular signaling pathways by other viral capsids (including flu viruses) is an important part of the disease process. In addition to understanding how specific viruses cause disease, the study of how capsids modulate intracellular signaling, may pave the way for rationale design of live attenuated vaccines that incorporate modified capsid genes. In addition, capsid proteins themselves may prove useful as therapeutic agents that can be used to upregulate or dampen specific signaling networks that malfunction as the result of disease.
Acknowledgements
The work was funded by a grant to T.C.H. from the Canadian Institutes of Health Research. T.C.H. is the recipient of a Medical Scientist Award from the Alberta Heritage Foundation for Medical Research. C.I. was supported in part by a graduate studentship award from the Faculty of Medicine & Dentistry.
References
- 1.Kitler M.E., Gavinio P., Lavanchy D. Vaccine. 2002;20(Suppl 2):S5–S14. doi: 10.1016/s0264-410x(02)00121-4. [DOI] [PubMed] [Google Scholar]
- 2.Roulston A., Marcellus R.C., Branton P.E. Annu. Rev. Microbiol. 1999;53:577–628. doi: 10.1146/annurev.micro.53.1.577. [DOI] [PubMed] [Google Scholar]
- 3.Iinuma M., Nagai Y., Maeno K., Yoshida T., Matsumoto T. J. Gen. Virol. 1971;12(3):239–247. doi: 10.1099/0022-1317-12-3-239. [DOI] [PubMed] [Google Scholar]
- 4.Kaukinen P., Vaheri A., Plyusnin A. Arch. Virol. 2005;150(9):1693–1713. doi: 10.1007/s00705-005-0555-4. [DOI] [PubMed] [Google Scholar]
- 5.Lindenbach B.D., Thiel H.-J., Rice C.M. In: Fields Virology. Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., Straus S.E., editors. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1101–1152. [Google Scholar]
- 6.Polyak S.J. Clin. Liver Dis. 2003;7(1):67–88. doi: 10.1016/s1089-3261(02)00075-2. [DOI] [PubMed] [Google Scholar]
- 7.Fisher J.M., Sossin W., Newcomb R., Scheller R. Cell. 1988;54:813–822. doi: 10.1016/s0092-8674(88)91131-2. [DOI] [PubMed] [Google Scholar]
- 8.Benali-Furet N.L., Chami M., Houel L., De Giorgi F., Vernejoul F., Lagorce D., Buscail L., Bartenschlager R., Ichas F., Rizzuto R., Paterlini-Brechot P. Oncogene. 2005;24(31):4921–4933. doi: 10.1038/sj.onc.1208673. [DOI] [PubMed] [Google Scholar]
- 9.Sacco R., Tsutsumi T., Suzuki R., Otsuka M., Aizaki H., Sakamoto S., Matsuda M., Seki N., Matsuura Y., Miyamura T., Suzuki T. Virology. 2003;317(1):24–35. doi: 10.1016/j.virol.2003.08.028. [DOI] [PubMed] [Google Scholar]
- 10.Miller K., McArdle S., Gale M.J., Jr., Geller D.A., Tenoever B., Hiscott J., Gretch D.R., Polyak S.J. J. Interferon Cytokine Res. 2004;24(7):391–402. doi: 10.1089/1079990041535647. [DOI] [PubMed] [Google Scholar]
- 11.Polyak S.J., Klein K.C., Shoji I., Miyamura T., Lingappa J.R. In: Hepatitis C Viruses: Genomes and Molecular Biology. Tan S.-L., editor. Horizon Scientific Press; Norwich, UK: 2006. pp. 89–119. [Google Scholar]
- 12.Dominguez-Villar M., Munoz-Suano A., Anaya-Baz B., Aguilar S., Novalbos J.P., Giron J.A., Rodriguez-Iglesias M., Garcia-Cozar F. J. Leukoc. Biol. 2007;82(5):1301–1310. doi: 10.1189/jlb.0507335. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen H., Sankaran S., Dandekar S. Virology. 2006;354(1):58–68. doi: 10.1016/j.virol.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 14.McLauchlan J., Lemberg M.K., Hope G., Martoglio B. EMBO J. 2002;21(15):3980–3988. doi: 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo S.Y., Masiarz F., Hwang S.B., Lai M.M., Ou J.H. Virology. 1995;213(2):455–461. doi: 10.1006/viro.1995.0018. [DOI] [PubMed] [Google Scholar]
- 16.Yasui K., Wakita T., Tsukiyama-Kohara K., Funahashi S.I., Ichikawa M., Kajita T., Moradpour D., Wands J.R., Kohara M. J. Virol. 1998;72(7):6048–6055. doi: 10.1128/jvi.72.7.6048-6055.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moriya K., Fujie H., Shintani Y., Yotsuyanagi H., Tsutsumi T., Ishibashi K., Matsuura Y., Kimura S., Miyamura T., Koike K. Nat. Med. 1998;4(9):1065–1067. doi: 10.1038/2053. [DOI] [PubMed] [Google Scholar]
- 18.Moriya K., Yotsuyanagi H., Shintani Y., Fujie H., Ishibashi K., Matsuura Y., Miyamura T., Koike K. J. Gen. Virol. 1997;78(Pt 7):1527–1531. doi: 10.1099/0022-1317-78-7-1527. [DOI] [PubMed] [Google Scholar]
- 19.Calabrese F., Pontisso P., Pettenazzo E., Benvegnu L., Vario A., Chemello L., Alberti A., Valente M. Hepatology. 2000;31(5):1153–1159. doi: 10.1053/he.2000.7123. [DOI] [PubMed] [Google Scholar]
- 20.Hiramatsu N., Hayashi N., Katayama K., Mochizuki K., Kawanishi Y., Kasahara A., Fusamoto H., Kamada T. Hepatology. 1994;19(6):1354–1359. [PubMed] [Google Scholar]
- 21.Ruggieri A., Harada T., Matsuura Y., Miyamura T. Virology. 1997;229(1):68–76. doi: 10.1006/viro.1996.8420. [DOI] [PubMed] [Google Scholar]
- 22.Ruggieri A., Murdolo M., Rapicetta M. Virus Res. 2003;97(2):103–110. doi: 10.1016/j.virusres.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 23.Honda M., Kaneko S., Shimazaki T., Matsushita E., Kobayashi K., Ping L.H., Zhang H.C., Lemon S.M. Hepatology (Baltim. Md.) 2000;31(6):1351–1359. doi: 10.1053/jhep.2000.7985. [DOI] [PubMed] [Google Scholar]
- 24.Yang S.H., Lee C.G., Lee C.W., Choi E.J., Yoon S.K., Ahn K.S., Sung Y.C. Mol. Cells. 2002;13(3):452–462. [PubMed] [Google Scholar]
- 25.Zhu N., Khoshnan A., Schneider R., Matsumoto M., Dennert G., Ware C., Lai M.M. J. Virol. 1998;72(5):3691–3697. doi: 10.1128/jvi.72.5.3691-3697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshioka K., Kakumu S., Arao M., Tsutsumi Y., Inoue M., Wakita T., Ishikawa T., Mizokami M. J. Clin. Pathol. 1990;43(4):298–302. doi: 10.1136/jcp.43.4.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C.M., You L.R., Hwang L.H., Lee Y.H. J. Virol. 1997;71(12):9417–9426. doi: 10.1128/jvi.71.12.9417-9426.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Browning J.L., Miatkowski K., Sizing I., Griffiths D., Zafari M., Benjamin C.D., Meier W., Mackay F. J. Exp. Med. 1996;183(3):867–878. doi: 10.1084/jem.183.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shrivastava A., Manna S.K., Ray R., Aggarwal B.B. J. Virol. 1998;72(12):9722–9728. doi: 10.1128/jvi.72.12.9722-9728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thimme R., Bukh J., Spangenberg H.C., Wieland S., Pemberton J., Steiger C., Govindarajan S., Purcell R.H., Chisari F.V. Proc. Natl. Acad. Sci. U. S. A. 2002;99(24):15661–15668. doi: 10.1073/pnas.202608299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hahn C.S., Cho Y.G., Kang B.S., Lester I.M., Hahn Y.S. Virology. 2000;276(1):127–137. doi: 10.1006/viro.2000.0541. [DOI] [PubMed] [Google Scholar]
- 32.Otsuka M., Kato N., Taniguchi H., Yoshida H., Goto T., Shiratori Y., Omata M. Virology. 2002;296(1):84–93. doi: 10.1006/viro.2002.1371. [DOI] [PubMed] [Google Scholar]
- 33.Marusawa H., Hijikata M., Chiba T., Shimotohno K. J. Virol. 1999;73(6):4713–4720. doi: 10.1128/jvi.73.6.4713-4720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.You L.R., Chen C.M., Lee Y.H.W. J. Virol. 1999;73(2):1672–1681. doi: 10.1128/jvi.73.2.1672-1681.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida H., Kato N., Shiratori Y., Otsuka M., Maeda S., Kato J., Omata M. J. Biol. Chem. 2001;276(19):16399–16405. doi: 10.1074/jbc.M006671200. [DOI] [PubMed] [Google Scholar]
- 36.Lacronique V., Mignon A., Fabre M., Viollet B., Rouquet N., Molina T., Porteu A., Henrion A., Bouscary D., Varlet P., Joulin V., Kahn A. Nat. Med. 1996;2(1):80–86. doi: 10.1038/nm0196-80. [DOI] [PubMed] [Google Scholar]
- 37.Chung Y.M., Park K.J., Choi S.Y., Hwang S.B., Lee S.Y. Biochem. Biophys. Res. Commun. 2001;284(1):15–19. doi: 10.1006/bbrc.2001.4936. [DOI] [PubMed] [Google Scholar]
- 38.Tai D.I., Tsai S.L., Chen Y.M., Chuang Y.L., Peng C.Y., Sheen I.S., Yeh C.T., Chang K.S., Huang S.N., Kuo G.C., Liaw Y.F. Hepatology (Baltim. Md.) 2000;31(3):656–664. doi: 10.1002/hep.510310316. [DOI] [PubMed] [Google Scholar]
- 39.Saito K., Meyer K., Warner R., Basu A., Ray R.B., Ray R. J. Virol. 2006;80(9):4372–4379. doi: 10.1128/JVI.80.9.4372-4379.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Micheau O., Lens S., Gaide O., Alevizopoulos K., Tschopp J. Mol. Cell. Biol. 2001;21(16):5299–5305. doi: 10.1128/MCB.21.16.5299-5305.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kreuz S., Siegmund D., Scheurich P., Wajant H. Mol. Cell. Biol. 2001;21(12):3964–3973. doi: 10.1128/MCB.21.12.3964-3973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsu H., Shu H.B., Pan M.G., Goeddel D.V. Cell. 1996;84(2):299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 43.Nagata S. Annu. Rev. Immunol. 2005;23:853–875. doi: 10.1146/annurev.immunol.23.021704.115811. [DOI] [PubMed] [Google Scholar]
- 44.Pawlotsky J.M. Clin. Liver Dis. 2003;7(1):45–66. doi: 10.1016/s1089-3261(02)00065-x. [DOI] [PubMed] [Google Scholar]
- 45.Ray R.B., Steele R., Basu A., Meyer K., Majumder M., Ghosh A.K., Ray R. Virus Res. 2002;87(1):21–29. doi: 10.1016/s0168-1702(02)00046-1. [DOI] [PubMed] [Google Scholar]
- 46.Pavio N., Battaglia S., Boucreux D., Arnulf B., Sobesky R., Hermine O., Brechot C. Oncogene. 2005;24(40):6119–6132. doi: 10.1038/sj.onc.1208749. [DOI] [PubMed] [Google Scholar]
- 47.Shi Y., Massague J. Cell. 2003;113(6):685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 48.Ray R.B., Steele R., Meyer K., Ray R. J. Biol. Chem. 1997;272(17):10983–10986. doi: 10.1074/jbc.272.17.10983. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y., Kato N., Jazag A., Dharel N., Otsuka M., Taniguchi H., Kawabe T., Omata M. Gastroenterology. 2006;130(3):883–892. doi: 10.1053/j.gastro.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 50.Lewis B.P., Burge C.B., Bartel D.P. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 51.Lindenbach B.D., Evans M.J., Syder A.J., Wolk B., Tellinghuisen T.L., Liu C.C., Maruyama T., Hynes R.O., Burton D.R., McKeating J.A., Rice C.M. Science. 2005;309(5734):623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 52.Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z., Murthy K., Habermann A., Krausslich H.G., Mizokami M., Bartenschlager R., Liang T.J. Nat. Med. 2005;11(7):791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heinz F.X., Purcell M.S., Gould E.A., Howard C.R., Houghton M. In: Virus Taxonomy. Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L., Carstens E.B., Estes M.K., editors. Academic; San Diego: 2000. pp. 860–878. [Google Scholar]
- 54.Brinton M.A. Annu. Rev. Microbiol. 2002;56:371–402. doi: 10.1146/annurev.micro.56.012302.160654. [DOI] [PubMed] [Google Scholar]
- 55.Gubler D.J., Kuno G., Markoff L. In: Fields Virology. Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., Straus S.E., editors. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1153–1252. [Google Scholar]
- 56.Wilder-Smith A., Schwartz E. N. Engl. J. Med. 2005;353(9):924–932. doi: 10.1056/NEJMra041927. [DOI] [PubMed] [Google Scholar]
- 57.Bulich R., Aaskov J.G. J. Gen. Virol. 1992;73(Pt 11):2999–3003. doi: 10.1099/0022-1317-73-11-2999. [DOI] [PubMed] [Google Scholar]
- 58.Tadano M., Makino Y., Fukunaga T., Okuno Y., Fukai K. J. Gen. Virol. 1989;70(Pt 6):1409–1415. doi: 10.1099/0022-1317-70-6-1409. [DOI] [PubMed] [Google Scholar]
- 59.Westaway E.G., Khromykh A.A., Kenney M.T., Mackenzie J.M., Jones M.K. Virology. 1997;234(1):31–41. doi: 10.1006/viro.1997.8629. [DOI] [PubMed] [Google Scholar]
- 60.Mori Y., Okabayashi T., Yamashita T., Zhao Z., Wakita T., Yasui K., Hasebe F., Tadano M., Konishi E., Moriishi K., Matsuura Y. J. Virol. 2005;79(6):3448–3458. doi: 10.1128/JVI.79.6.3448-3458.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hunt T.A., Urbanowski M.D., Kakani K., Law L.M., Brinton M.A., Hobman T.C. Cell. Microbiol. 2007;9(11):2756–2766. doi: 10.1111/j.1462-5822.2007.01046.x. [DOI] [PubMed] [Google Scholar]
- 62.Yang J.S., Ramanathan M.P., Muthumani K., Choo A.Y., Jin S.H., Yu Q.C., Hwang D.S., Choo D.K., Lee M.D., Dang K., Tang W., Kim J.J., Weiner D.B. Emerg. Infect. Dis. 2002;8(12):1379–1384. doi: 10.3201/eid0812.020224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang M.R., Lee S.R., Oh W., Lee E.W., Yeh J.Y., Nah J.J., Joo Y.S., Shin J., Lee H.W., Pyo S., Song J. Cell. Microbiol. 2007;10(1):165–176. doi: 10.1111/j.1462-5822.2007.01027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kubbutat M.H., Jones S.N., Vousden K.H. Nature. 1997;387(6630):299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 65.Haupt Y., Maya R., Kazaz A., Oren M. Nature. 1997;387(6630):296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 66.Miyashita T., Reed J.C. Cell. 1995;80(2):293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 67.Wang T., Town T., Alexopoulou L., Anderson J.F., Fikrig E., Flavell R.A. Nat. Med. 2004;10(12):1366–1373. doi: 10.1038/nm1140. [DOI] [PubMed] [Google Scholar]
- 68.Cheng Y., King N.J., Kesson A.M. Virology. 2004;329(2):361–370. doi: 10.1016/j.virol.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 69.van Marle G., Antony J., Ostermann H., Dunham C., Hunt T., Halliday W., Maingat F., Urbanowski M.D., Hobman T., Peeling J., Power C. J. Virol. 2007;81(20):10933–10949. doi: 10.1128/JVI.02422-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kondo S., Murakami T., Tatsumi K., Ogata M., Kanemoto S., Otori K., Iseki K., Wanaka A., Imaizumi K. Nat. Cell. Biol. 2005;7(2):186–194. doi: 10.1038/ncb1213. [DOI] [PubMed] [Google Scholar]
- 71.Duong F.H., Filipowicz M., Tripodi M., La Monica N., Heim M.H. Gastroenterology. 2004;126(1):263–277. doi: 10.1053/j.gastro.2003.10.076. [DOI] [PubMed] [Google Scholar]
- 72.Al-Murrani S.W., Woodgett J.R., Damuni Z. Biochem. J. 1999;341(Pt 2):293–298. [PMC free article] [PubMed] [Google Scholar]
- 73.Ludwig S., Ehrhardt C., Neumeier E.R., Kracht M., Rapp U.R., Pleschka S. J. Biol. Chem. 2001;276(14):10990–10998. doi: 10.1074/jbc.M009902200. [DOI] [PubMed] [Google Scholar]
- 74.Li M., Makkinje A., Damuni Z. J. Biol. Chem. 1996;271(19):11059–11062. doi: 10.1074/jbc.271.19.11059. [DOI] [PubMed] [Google Scholar]
- 75.Qu D., Li Q., Lim H.Y., Cheung N.S., Li R., Wang J.H., Qi R.Z. J. Biol. Chem. 2002;277(9):7324–7332. doi: 10.1074/jbc.M107270200. [DOI] [PubMed] [Google Scholar]
- 76.Saito S., Miyaji-Yamaguchi M., Shimoyama T., Nagata K. Biochem. Biophys. Res. Commun. 1999;259(2):471–475. doi: 10.1006/bbrc.1999.0790. [DOI] [PubMed] [Google Scholar]
- 77.Fu A.K., Fu W.Y., Cheung J., Tsim K.W., Ip F.C., Wang J.H., Ip N.Y. Nat. Neurosci. 2001;4(4):374–381. doi: 10.1038/86019. [DOI] [PubMed] [Google Scholar]
- 78.Li H., Leo C., Zhu J., Wu X., O'Neil J., Park E.J., Chen J.D. Mol. Cell. Biol. 2000;20(5):1784–1796. doi: 10.1128/mcb.20.5.1784-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li R., Pei H., Watson D.K., Papas T.S. Oncogene. 2000;19(6):745–753. doi: 10.1038/sj.onc.1203385. [DOI] [PubMed] [Google Scholar]
- 80.Yang X., Khosravi-Far R., Chang H.Y., Baltimore D. Cell. 1997;89(7):1067–1076. doi: 10.1016/s0092-8674(00)80294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marianneau P., Cardona A., Edelman L., Deubel V., Despres P. J. Virol. 1997;71(4):3244–3249. doi: 10.1128/jvi.71.4.3244-3249.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Griffin D.E. In: Fields Virology. Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., Straus S.E., editors. Lippincott Willilams & Wilkins; Philadelphia: 2007. pp. 1023–1068. [Google Scholar]
- 83.Levine B., Huang Q., Isaacs J.T., Reed J.C., Griffin D.E., Hardwick J.M. Nature. 1993;361(6414):739–742. doi: 10.1038/361739a0. [DOI] [PubMed] [Google Scholar]
- 84.Ubol S., Park S., Budihardjo I., Desnoyers S., Montrose M.H., Poirier G.G., Kaufmann S.H., Griffin D.E. J. Virol. 1996;70(4):2215–2220. doi: 10.1128/jvi.70.4.2215-2220.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Favre D., Studer E., Michel M.R. Biosci. Rep. 1996;16(6):485–511. doi: 10.1007/BF01198464. [DOI] [PubMed] [Google Scholar]
- 86.van Steeg H., Kasperaitis M., Voorma H.O., Benne R. Eur. J. Biochem. 1984;138(3):473–478. doi: 10.1111/j.1432-1033.1984.tb07940.x. [DOI] [PubMed] [Google Scholar]
- 87.Elgizoli M., Dai Y., Kempf C., Koblet H., Michel M.R. J. Virol. 1989;63(7):2921–2928. doi: 10.1128/jvi.63.7.2921-2928.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hobman T., Chantler J. In: Fields Virology. Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., Straus S.E., editors. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1069–1100. [Google Scholar]
- 89.Pugachev K.V., Frey T.K. Virology. 1998;250(2):359–370. doi: 10.1006/viro.1998.9395. [DOI] [PubMed] [Google Scholar]
- 90.Hofmann J., Pletz M.W., Liebert U.G. J. Gen. Virol. 1999;80(Pt 7):1657–1664. doi: 10.1099/0022-1317-80-7-1657. [DOI] [PubMed] [Google Scholar]
- 91.Megyeri K., Berencsi K., Halazonetis T.D., Prendergast G.C., Gri G., Plotkin S.A., Rovera G., Gonczol E. Virology. 1999;259(1):74–84. doi: 10.1006/viro.1999.9757. [DOI] [PubMed] [Google Scholar]
- 92.Adamo M.P., Zapata M., Frey T.K. Virology. 2008;370(1):1–11. doi: 10.1016/j.virol.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Duncan R., Esmaili A., Law L.M., Bertholet S., Hough C., Hobman T.C., Nakhasi H.L. Virology. 2000;275(1):20–29. doi: 10.1006/viro.2000.0467. [DOI] [PubMed] [Google Scholar]
- 94.Hofmann J., Pletz M.W., Liebert U.G. J. Gen. Virol. 1999;80(Pt 7):1657–1664. doi: 10.1099/0022-1317-80-7-1657. [DOI] [PubMed] [Google Scholar]
- 95.Duncan R., Muller J., Lee N., Esmaili A., Nakhasi H.L. Virology. 1999;255(1):117–128. doi: 10.1006/viro.1998.9562. [DOI] [PubMed] [Google Scholar]
- 96.Hobman T.C., Lundstrom M.L., Mauracher C.A., Woodward L., Gillam S., Farquhar M.G. Virology. 1994;202(2):574–585. doi: 10.1006/viro.1994.1379. [DOI] [PubMed] [Google Scholar]
- 97.Lee J.Y., Marshall J.A., Bowden D.S. Virology. 1999;265(1):110–119. doi: 10.1006/viro.1999.0016. [DOI] [PubMed] [Google Scholar]
- 98.Beatch M.D., Everitt J.C., Law L.J., Hobman T.C. J. Virol. 2005;79(16):10807–10820. doi: 10.1128/JVI.79.16.10807-10820.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.De Vos K., Goossens V., Boone E., Vercammen D., Vancompernolle K., Vandenabeele P., Haegeman G., Fiers W., Grooten J. J. Biol. Chem. 1998;273(16):9673–9680. doi: 10.1074/jbc.273.16.9673. [DOI] [PubMed] [Google Scholar]
- 100.Desagher S., Martinou J.C. Trends Cell Biol. 2000;10(9):369–377. doi: 10.1016/s0962-8924(00)01803-1. [DOI] [PubMed] [Google Scholar]
- 101.Beatch M.D., Hobman T.C. J. Virol. 2000;74(12):5569–5576. doi: 10.1128/jvi.74.12.5569-5576.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mohan K.V., Ghebrehiwet B., Atreya C.D. Virus Res. 2002;85(2):151–161. doi: 10.1016/s0168-1702(02)00030-8. [DOI] [PubMed] [Google Scholar]
- 103.Sunayama J., Ando Y., Itoh N., Tomiyama A., Sakurada K., Sugiyama A., Kang D., Tashiro F., Gotoh Y., Kuchino Y., Kitanaka C. Cell Death Differ. 2004;11(7):771–781. doi: 10.1038/sj.cdd.4401418. [DOI] [PubMed] [Google Scholar]
- 104.Jiang J., Zhang Y., Krainer A.R., Xu R.M. Proc. Natl. Acad. Sci. U. S. A. 1999;96(7):3572–3577. doi: 10.1073/pnas.96.7.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 106.M. Surjit, S.K. Lal, Infect. Genet. Evol. (in press) (Electronic publication ahead of print).
- 107.Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M., Baric R.A., Palese P. J. Virol. 2007;81(2):548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.You J., Dove B.K., Enjuanes L., DeDiego M.L., Alvarez E., Howell G., Heinen P., Zambon M., Hiscox J.A. J. Gen. Virol. 2005;86(Pt 12):3303–3310. doi: 10.1099/vir.0.81076-0. [DOI] [PubMed] [Google Scholar]
- 109.Surjit M., Kumar R., Mishra R.N., Reddy M.K., Chow V.T., Lal S.K. J. Virol. 2005;79(17):11476–11486. doi: 10.1128/JVI.79.17.11476-11486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yan X., Hao Q., Mu Y., Timani K.A., Ye L., Zhu Y., Wu J. Int. J. Biochem. Cell Biol. 2006;38(8):1417–1428. doi: 10.1016/j.biocel.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 111.Surjit M., Liu B., Chow V.T., Lal S.K. J. Biol. Chem. 2006;281(16):10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Luo C., Luo H., Zheng S., Gui C., Yue L., Yu C., Sun T., He P., Chen J., Shen J., Luo X., Li Y., Liu H., Bai D., Shen J., Yang Y., Li F., Zuo J., Hilgenfeld R., Pei G., Chen K., Shen X., Jiang H. Biochem. Biophys. Res. Commun. 2004;321(3):557–565. doi: 10.1016/j.bbrc.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Luo H., Ye F., Sun T., Yue L., Peng S., Chen J., Li G., Du Y., Xie Y., Yang Y., Shen J., Wang Y., Shen X., Jiang H. Biophys. Chem. 2004;112(1):15–25. doi: 10.1016/j.bpc.2004.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Oh W., Yang M.R., Lee E.W., Park K.M., Pyo S., Yang J.S., Lee H.W., Song J. J. Biol. Chem. 2006;281(40):30166–30174. doi: 10.1074/jbc.M602651200. [DOI] [PubMed] [Google Scholar]
- 115.Sabile A., Perlemuter G., Bono F., Kohara K., Demaugre F., Kohara M., Matsuura Y., Miyamura T., Brechot C., Barba G. Hepatology. 1999;30(4):1064–1076. doi: 10.1002/hep.510300429. [DOI] [PubMed] [Google Scholar]
- 116.Shi S.T., Polyak S.J., Tu H., Taylor D.R., Gretch D.R., Lai M.M. Virology. 2002;292(2):198–210. doi: 10.1006/viro.2001.1225. [DOI] [PubMed] [Google Scholar]
- 117.Kittlesen D.J., Chianese-Bullock K.A., Yao Z.Q., Braciale T.J., Hahn Y.S. J. Clin. Invest. 2000;106(10):1239–1249. doi: 10.1172/JCI10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ohkawa K., Ishida H., Nakanishi F., Hosui A., Ueda K., Takehara T., Hori M., Hayashi N. J. Biol. Chem. 2004;279(12):11719–11726. doi: 10.1074/jbc.M308560200. [DOI] [PubMed] [Google Scholar]
- 119.Mamiya N., Worman H.J. J. Biol. Chem. 1999;274(22):15751–15756. doi: 10.1074/jbc.274.22.15751. [DOI] [PubMed] [Google Scholar]
- 120.Owsianka A.M., Patel A.H. Virology. 1999;257(2):330–340. doi: 10.1006/viro.1999.9659. [DOI] [PubMed] [Google Scholar]
- 121.Hsieh T.Y., Matsumoto M., Chou H.C., Schneider R., Hwang S.B., Lee A.S., Lai M.M. J. Biol. Chem. 1998;273(28):17651–17659. doi: 10.1074/jbc.273.28.17651. [DOI] [PubMed] [Google Scholar]
- 122.Hosui A., Ohkawa K., Ishida H., Sato A., Nakanishi F., Ueda K., Takehara T., Kasahara A., Sasaki Y., Hori M., Hayashi N. J. Biol. Chem. 2003;278(31):28562–28571. doi: 10.1074/jbc.M210485200. [DOI] [PubMed] [Google Scholar]
- 123.Chen C.-M., You L.-R., Hwang L., Lee Y.-H. J. Virol. 1997;71:9417–9426. doi: 10.1128/jvi.71.12.9417-9426.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Otsuka M., Kato N., Lan K., Yoshida H., Kato J., Goto T., Shiratori Y., Omata M. J. Biol. Chem. 2000;275(44):34122–34130. doi: 10.1074/jbc.M000578200. [DOI] [PubMed] [Google Scholar]
- 125.Alisi A., Giambartolomei S., Cupelli F., Merlo P., Fontemaggi G., Spaziani A., Balsano C. Oncogene. 2003;22(17):2573–2580. doi: 10.1038/sj.onc.1206333. [DOI] [PubMed] [Google Scholar]
- 126.Moriishi K., Okabayashi T., Nakai K., Moriya K., Koike K., Murata S., Chiba T., Tanaka K., Suzuki R., Suzuki T., Miyamura T., Matsuura Y. J. Virol. 2003;77(19):10237–10249. doi: 10.1128/JVI.77.19.10237-10249.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tsutsumi T., Suzuki T., Shimoike T., Suzuki R., Moriya K., Shintani Y., Fujie H., Matsuura Y., Koike K., Miyamura T. Hepatology. 2002;35(4):937–946. doi: 10.1053/jhep.2002.32470. [DOI] [PubMed] [Google Scholar]
- 128.Cheng P.L., Chang M.H., Chao C.H., Lee Y.H. Oncogene. 2004;23(47):7821–7838. doi: 10.1038/sj.onc.1208066. [DOI] [PubMed] [Google Scholar]
- 129.Watashi K., Hijikata M., Tagawa A., Doi T., Marusawa H., Shimotohno K. Mol. Cell. Biol. 2003;23(21):7498–7509. doi: 10.1128/MCB.23.21.7498-7509.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yoshida T., Hanada T., Tokuhisa T., Kosai K., Sata M., Kohara M., Yoshimura A. J. Exp. Med. 2002;196(5):641–653. doi: 10.1084/jem.20012127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhu N., Ware C.F., Lai M.M. Virology. 2001;283(2):178–187. doi: 10.1006/viro.2001.0896. [DOI] [PubMed] [Google Scholar]
- 132.Aoki H., Hayashi J., Moriyama M., Arakawa Y., Hino O. J. Virol. 2000;74(4):1736–1741. doi: 10.1128/jvi.74.4.1736-1741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li X.D., Makela T.P., Guo D., Soliymani R., Koistinen V., Vapalahti O., Vaheri A., Lankinen H. J. Gen. Virol. 2002;83(Pt 4):759–766. doi: 10.1099/0022-1317-83-4-759. [DOI] [PubMed] [Google Scholar]
- 134.Fan Z., Zhuo Y., Tan X., Zhou Z., Yuan J., Qiang B., Yan J., Peng X., Gao G.F. J. Med. Virol. 2006;78(11):1365–1373. doi: 10.1002/jmv.20707. [DOI] [PMC free article] [PubMed] [Google Scholar]