Abstract
New on-demand multiplex molecular respiratory viral diagnostics offer superior performance although can be expensive and some platforms cannot process multiple specimens simultaneously. We performed a retrospective study reviewing results of patients tested for respiratory viruses following introduction of a two-stage testing algorithm incorporating an initial screen with Sofia® immunoassay then secondary Biofire Filmarray®, and compared to a period when only Filmarray® was used. Of 2976 testing episodes, 1814 underwent initial Sofia® then follow-up FilmArray®. A diagnosis of influenza was made by Sofia® in 282 patients, and by FilmArray® in an additional 163 (median time to result 1.12 hours versus 3.46 hours, P < 0.001). Significantly more patients received their diagnosis within 90 minutes in winter despite testing more samples (11.1% versus 3.4%, P < 0.001), and approximately $36,000 was saved. An algorithmic approach to respiratory viral diagnosis can combine the advantages of accuracy and speed and be cost saving.
Abbreviations: RVI, respiratory viral infection; ILI, influenza-like illness; COPD, chronic obstructive pulmonary disease; FilmArray, Biofire FilmArray® Respiratory Panel (Biofire Diagnostics Inc., Salt Lake City, UT); Sofia, Sofia Influenza A + B Fluorescent Immunoassay (Quidel, San Diego, CA); RSV, respiratory syncytial virus; HMPV, human metapneumovirus; NAAT, nucleic acid amplification test; DFA, direct fluorescent antibody; BAL, bronchoalveolar lavage
Keywords: Respiratory viral infection, Diagnosis, Influenza, PCR
1. Background
Respiratory viral infections such as influenza cause a large burden of morbidity and mortality worldwide, and are a common reason for patients to seek medical care. They are also an increasingly recognized cause of community-acquired pneumonia, particularly in patients who are older, have underlying comorbidities such as chronic obstructive pulmonary disease (COPD), or are immunocompromised (Gadsby et al., 2016, Ruuskanen et al., 2011). Clinical presentations are non-specific, so accurate and timely diagnostics are critical to optimizing antiviral use, limiting unnecessary antibiotics and reducing secondary spread of infection. Older diagnostic modalities such as serology, viral cultures, enzyme immunoassay and immunofluorescence tests are fraught with problems like poor sensitivity and slow turn-around time (Caliendo, 2011). Many institutions have relied on rapid antigen immunoassay tests, which are specific and can produce a result in a clinically useful timeframe, but lack sensitivity and are unable to detect other pathogens, limiting their utility in times of low influenza prevalence (Bruning et al., 2017). The revolution in the development, commercialization and dissemination of molecular multiplex assays for the rapid and accurate detection of respiratory viruses over the last decade has been a major step forward and has led to a greater appreciation of their ubiquity and contribution to many disease states (Gaydos, 2013, Gelfer et al., 2015, Varkey and Varkey, 2008). However, in the setting of a routine diagnostic microbiology laboratory, these assays can be labor intensive to implement and run, and are often expensive.
In many areas of medicine including modern diagnostic microbiology laboratories, algorithmic approaches to testing are being increasingly used to maximize advantages and minimize limitations and costs. An example of this is the diagnosis of Clostridium difficile infection, where many labs are now using an initial highly sensitive screening test followed by a specific secondary confirmatory molecular assay to improve overall sensitivity and specificity in a cost-effective manner (Makristathis et al., 2017). However, this approach has not been widely studied in the diagnosis of respiratory viral infections. In one study of patients with influenza, a combined testing approach such as this has been shown to decrease the time to result, cost, antibiotic use, ICU admission and an increase in timely administration of oseltamivir (Gonzalez-Del Vecchio et al., 2015).
With increasing pressure on our hospital systems to diagnose, treat and quickly discharge patients, rapid and accurate diagnostic tests are critical. With this in mind, multiplex PCR was introduced at our institution prior to the influenza season of 2014–15 as part of a 2-stage testing algorithm, replacing viral culture. The aim of this retrospective cohort study was to evaluate the performance of this two-stage testing approach in terms of diagnostic performance, timeliness and cost.
2. Methods
2.1. Study design and data collection
The study population included all patients (children and adults) tested for respiratory viral infections at Tufts Medical Center, an academic medical center with approximately 290 adult and 128 pediatric beds in Boston, Massachusetts, from December 1, 2014 to December 28, 2015. Cases were identified by searching electronic medical records for positive and negative test results in patients tested for respiratory viral pathogens. Patients could be included more than once; each testing episode was considered independently. Samples were excluded if they were not processed through the usual algorithm or if final results were equivocal. Data was collected on patient age, testing location, time of nasopharyngeal swab collection, tests performed, final results and the time result was released. The Tufts Medical Center institutional review board approved the study and informed consent was not required given its minimal risk and retrospective nature.
2.2. Laboratory procedures
Nasopharyngeal flocked swabs were collected from patients with clinically suspected respiratory viral infections by trained nursing or medical personnel and transported to the laboratory in 1.5 mL Remel MicroTest™ M6™ Multi-Microbe Media (ThermoFisher Scientific, Lenexa, KS, USA) via an automated rapid transit system. In the winter period (mid December 2014 to April 2015), after an increase in the influenza prevalence, samples were first tested by Sofia® immunoassay (Quidel Corporation, San Diego, CA, USA) for influenza, and also RSV in children <5 years old or when requested (Sofia Influenza A+B FIA, Package Insert, 2015). Samples testing positive did not undergo further testing; negative samples were subsequently tested by FilmArray®. During the summer months (May-early December 2015), specimens were tested by Biofire FilmArray® (BioFire Diagnostics, Salt Lake City, a bioMérieux company, Marcy, l'Etoile, France) alone (FilmArray® Respiratory Panel Package Insert, 2014). Clinicians only had to order a ‘respiratory viral panel’ with specific testing performed determined by the laboratory in a standardized fashion. The manufacturer’s instructions were followed for all testing procedures; there were no protocol variations. No other respiratory viral testing was performed on these samples. Repeat testing was permitted if clinically indicated but was rarely performed within the same episode of care.
The Sofia® Fluorescent Immunoassay is a rapid, relatively inexpensive lateral flow assay for influenza A and B. It is reported to have a high specificity (94–98.3%) but low sensitivity (72.4–74%) compared to molecular assays (Bruning et al., 2017, Gomez et al., 2015, Hazelton et al., 2015, Noh et al., 2015). It requires less than 5 minutes hands-on setup time and has a 10–15 minute run-time. A separate test is available for RSV. The Biofire Filmarray® is a modern, simple, fast and accurate multiplex PCR-based test capable of detecting multiple respiratory viruses including influenza A and B, parainfluenza 1–4, human metapneumovirus, human rhinovirus/enterovirus, 4 coronaviruses, adenovirus, respiratory syncytial virus (RSV) as well as 3 bacteria (Bordetella pertussis, Chlamydophila pneumoniae, Mycoplasma pneumoniae). It requires a maximum of 5 minutes of hands-on preparation time and around 75 minutes of run-time. The sensitivity and specificity of this test reported to be 95 to 100% in most studies, dependent on the pathogen (FilmArray® Respiratory Panel Package Insert, 2014). Unlike other batched assays, only one patient sample can be tested per instrument at a given time.
During the study period, the on-site microbiology laboratory was staffed from 6 AM to midnight, with both Sofia® and FilmArray® testing performed during these hours. Two FilmArray® instruments were in use and they were not used for any other testing. From midnight to 6 AM, Sofia® testing alone was performed by the hematology laboratory. Although the Sofia® can be performed at the point of care, in our institution it was only performed in the laboratory given the availability of rapid specimen transport and significant burden of maintaining multiple instruments and training staff outside of the laboratory.
2.3. Statistical analysis
Basic descriptive statistics were calculated. Categorical data were reported as percentages, continuous data as means ± standard deviations if normally distributed and medians with ranges if non-normally distributed. Missing data for the variables of interest were negligible. For the purposes of analysis, final test results were categorized into 4 groups: negative, positive (influenza), positive (RSV), positive (other virus). Categorical variables were compared using chi-square tests. Odds ratios and associated P values were calculated using univariate logistic regression. Kaplan–Meier cumulative incidence curves were generated to represent time to result data and were compared using the log-rank test. P < 0.05 was considered statistically significant. Unfortunately sensitivity and specificity could not be accurately calculated as patients testing positive by Sofia® did not undergo confirmatory FilmArray® testing, though there was sufficient data available to calculate negative predictive value (NPV), using FilmArray® as the gold standard. All analyses were performed with the R statistical software platform version 3.4.1 (RStudio version 1.0.153).
Costs were estimated based on the cost per test in our laboratory (Sofia®, $15.75 and FilmArray®, $129).
3. Results
A total of 2987 testing episodes in 2625 unique patients were identified. Seven were excluded as samples were not processed through the 2-stage pathway appropriately and 4 because results were equivocal, leaving 2976 episodes from 2614 patients in our final analysis. Median age at the time of testing was 45 years (range 0–102 years), 1232 patients (47%) were male, and 520 patients (20%) were less than 18 years old. Specimens were most commonly received from inpatient units (n = 1497) followed by ambulatory clinics (n = 642), the emergency room (n = 602), bedded outpatients (n = 202), employee health (n = 29) and outpatient dialysis (n = 4). 1814 samples were processed during winter via the 2-stage algorithm, compared to 1162 during summer, which underwent FilmArray® only. More than one pathogen was detected in 77 cases throughout the study period. Overall, 1287 (43.2%) tests were positive. Results of the final diagnoses obtained stratified by testing season are shown in Table 1 .
Table 1.
Final test results by season/testing regimen.
| Summer (Biofire Filmarray® alone) n = 1162 | Winter (2-stage) n = 1814 | |
|---|---|---|
| Adenovirus | 21 (1.8%) | 17 (0.9%) |
| Coronavirus | 21 (1.8%) | 82 (4.5%) |
| Human metapneumovirus | 9 (0.8%) | 59 (3.3%) |
| Human rhino/enterovirus | 219 (18.8%) | 110 (6.1%) |
| Influenza A | 7 (0.6%) | 322 (17.8%) |
| Influenza A plus another virus | 0 | 10 (0.6%) |
| Influenza B | 8 (0.7%) | 110 (6.1%) |
| Influenza B plus another virus | 1 (0.1%) | 3 (0.2%) |
| Mycoplasma pneumoniae | 9 (0.8%) | 3 (0.2%) |
| Parainfluenza virus | 48 (4.1%) | 41 (2.3%) |
| Respiratory syncytial virus | 16 (1.4%) | 108 (6.0%) |
| 2 pathogens | 26 (2.2%) | 35 (1.9%) |
| 3 pathogens | 1 (0.1%) | 1 (0.1%) |
| No pathogens identified | 776 (66.8%) | 913 (50.3%) |
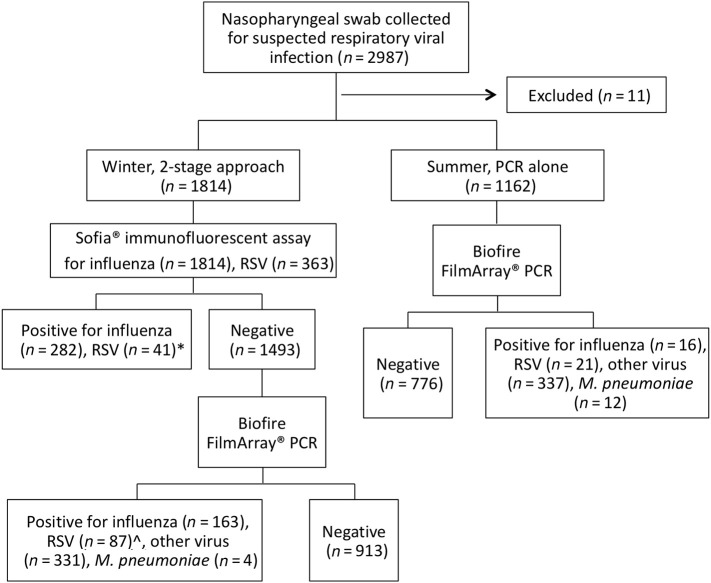
Of the 1814 patients tested by the 2-stage algorithm, 321 (17.7%) were diagnosed by Sofia®, with 282 cases of influenza (205 influenza A and 75 influenza B) and 41 of RSV (including 2 patients testing positive for both influenza A and RSV). Results are shown in Fig. 1 . The 1493 patients with a negative Sofia influenza antigen went on to FilmArray®, and of these 580 (38.8%) were positive: 163 for influenza (125 influenza A and 38 influenza B), 87 for RSV (including 5 patients with influenza and RSV), 331 for another virus and 4 for Mycoplasma pneumoniae. Overall, 37% (163/445) of patients diagnosed with influenza tested negative by Sofia®. Sofia® testing for RSV was performed in 363 patients (all children <5 years old and on request in others), and 28% of these (16/57) were Sofia® negative but FilmArray® RSV positive. An additional 71 cases of RSV were diagnosed amongst patients who did not have Sofia®-RSV antigen testing performed, 5 of whom were also positive for influenza. Negative predictive values (averaging prevalence across the whole period of testing) were 89.4% (1369/1532) for influenza and 95% (306/322) for RSV.
Fig. 1.
Diagram illustrating breakdown of testing performed and results obtained over the study period. PCR, polymerase chain reaction. RSV, respiratory syncytial virus. *Two patients tested positive for both Influenza A and RSV by Sofia®. ^16 cases positive for RSV by FilmArray® were Sofia® negative, the remaining 71 did not undergo RSV testing by Sofia®. Five were positive for both influenza and RSV by FilmArray®.
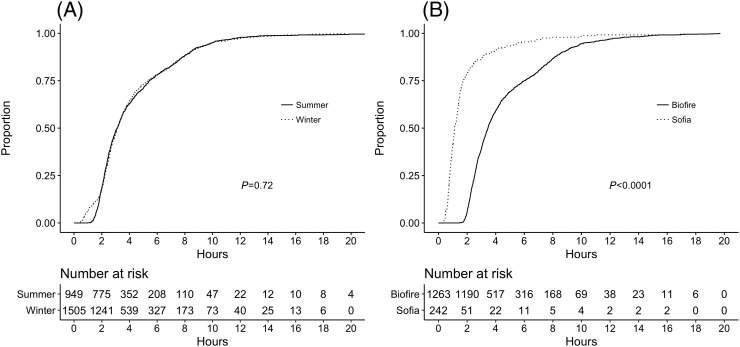
Specimen collection times were available for 2454 samples, 1505 of which were tested during winter and 949 during summer. Kaplan–Meier curves comparing these two time periods are shown in Fig. 2(a). While the two curves look similar overall, there were a significantly higher proportion of final test results available in less than 90 minutes in the winter compared to the summer, due to the use of the faster antigen test during that season (11.1% versus 3.4%, odds ratio 3.58, 95% confidence interval 2.46–5.36, P < 0.001). This corresponded to 144 cases of influenza and 21 cases of RSV in which a diagnosis was made in less than 90 minutes. A comparison of the time to result by test type in only the 1505 patients tested using the 2-stage algorithm is shown in Fig. 2(b). Median time to result was 1.12 hours (interquartile range, IQR 0.78–1.73 hours) if Sofia® was positive, compared to 3.46 hours (IQR 2.53–6.00 hours) if patients went on to have a FilmArray® (P < 0.001).
Fig. 2.
Time to final result represented on a Kaplan–Meier cumulative incidence curve (A) comparing summer months using Biofire FilmArray® only to the winter 2-stage approach, and (B) patients during the winter who reached their final diagnosis by Sofia® compared to Biofire FilmArray®. P-values reflect log-rank test results.
Based on costs of these individual tests in our laboratory, if FilmArray® alone had been performed on all samples, the total cost over the influenza season would have been an additional $36,353. Instead, FilmArray® was avoided in all those patients positive for a respiratory virus by the less expensive Sofia® test.
4. Discussion
Prior to the onset of the 2014–15 influenza season, a decision was made to introduce a new respiratory viral testing algorithm in our diagnostic microbiology laboratory. The goal of this was to combine the benefits of an inexpensive, rapid, specific test with the accuracy of a more sensitive one, reducing the overall false negative rate. We have retrospectively evaluated the performance of this protocol change and have described significant benefits in terms of overall performance, timeliness and cost. While a variety of 2-stage testing approaches, including those using viral culture and older batched multiplex molecular assays have been described, our study is the first to evaluate this approach in the modern era using a new on-demand molecular diagnostic test. In our cohort, if we had only performed Sofia® antigen testing on all samples, we would have missed 158 cases of influenza, 82 of RSV (although some of these may have been diagnosed with more usage of the RSV antigen), 5 influenza-RSV coinfections, 331 other viruses, and 4 cases of M. pneumoniae. Conversely, if we had used Filmarray® alone for all samples, 280 cases of influenza, 39 RSV cases and 2 influenza/RSV coinfections would have had their diagnoses delayed by at least several hours, at significant additional cost. Despite processing almost twice as many specimens, during the winter the median time to result was similar to the summer and a significantly higher number of final results were available before 2 hours.
The diagnostic algorithmic testing strategy we have described has many advantages. Firstly, it decreases the overall turnaround time in the busy winter months. While a 1–2 hour time saving may not seem like much, having a result available within this timeframe means that the test is much more likely to be used for real-time clinical decision-making in the context of a busy emergency department or outpatient setting. It facilitates the rapid diagnosis of influenza and RSV, which would hopefully translate into earlier administration of antiviral therapy, which has significant therapeutic benefit (Aoki et al., 2003), and could decrease use of unnecessary antibiotics. In addition, it could mean earlier initiation of appropriate infection control precautions and reduced secondary transmission. This effect will be amplified when applied across many patients. Secondly, it not only maximizes diagnostic accuracy for influenza, but also detects many other respiratory viral pathogens that can be clinically indistinguishable from each other. This provides both patients and clinicians with the clarity of a definitive microbiologic diagnosis and reassurance that a favorable prognosis is likely without the need for antibiotics. Furthermore, due to differences between transmission characteristics between respiratory pathogens, CDC guidelines for isolation precautions differ for each diagnosis (Siegel et al., 2007). Identifying the specific pathogen may increase the likelihood that the appropriate precautions are implemented, reducing nosocomial transmission. Finally, it facilitates increased testing volumes within existing laboratory technician time constraints, and does not require input from the ordering clinician, who may not be familiar with the nuances of the different respiratory viral tests available. Using the Sofia® as a laboratory-based test rather than a true point-of-care test does increase turn-around time somewhat (minimal in locations with on-site laboratories and rapid specimen transport systems), but obviates the need to train non-laboratory based staff and maintain instruments outside the diagnostic laboratory.
Like any diagnostic testing approach, there are some potential drawbacks of the 2-stage approach that should be considered. False positive results with the Sofia® antigen test clearly occur, and this could lead to misdiagnosis, unnecessary administration of oseltamivir, and potentially missing an alternative diagnosis. Patients testing positive by Sofia could be coinfected with a second virus (which is unlikely to have a significant impact on management decisions or outcome) or a bacterial pathogen (which if missed, could lead to failure to prescribe antibiotics). Rates of bacterial pathogens in our study were low, and many patients did receive empiric antibiotics despite a diagnosis of a virus to cover for the possibility of secondary bacterial infection. To minimize the risks of these errors occurring, we recommend only performing 2-stage testing during the influenza season when prevalence is high (to minimize the false positive rate), and making individualized decisions regarding use of antibiotics incorporating all available clinical data. Backup molecular testing should be available on clinician request for difficult cases.
The strength of our study is that we evaluated this diagnostic strategy in a large cohort of patients over a busy influenza season, in a real-world context. However there are some limitations that should be considered when interpreting our findings. Measuring clinical outcomes and downstream effects were beyond the scope of this study. However, other published studies have demonstrated the clinical impact of various molecular diagnostic tests and have shown earlier antiviral use, decreased ED and hospital length of stay, reduced secondary transmission and decreased antibiotic use (Aoki et al., 2003, Chu et al., 2015, Gelfer et al., 2015, Mayer et al., 2017). We would anticipate a similar impact in our patient population. At the same time however, the requirement for a private room or cohorting of patients found to have respiratory viruses requiring isolation, which previously would have gone undiagnosed, will undoubtedly result in costly throughput issues, a consideration which must not be underestimated. Because patients testing positive by Sofia® did not undergo further testing, we could not determine the true sensitivity and specificity of this test compared to FilmArray®. Previous studies have shown Sofia® specificity to be high, with a recent meta-analysis reporting a pooled specificity of 95.3% but sensitivity of only 75.3% for influenza A & B, and pooled specificity of 97.8% and sensitivity of 80% for RSV (Bruning et al., 2017, Gomez et al., 2015, Hazelton et al., 2015, Noh et al., 2015). Similarly, we assumed that all patients testing negative by PCR were true negatives; it is possible that some of these patients could have had false negative tests, i.e. that sensitivity of the FilmArray® is not truly 100%. While our findings are likely to be generalizable to other hospital-based clinical microbiology laboratories, the performance will be affected by a number of site-specific variables including testing volume, number of instruments (which affects ability to perform testing in parallel and therefore queuing), staffing, and the choice of specific rapid/molecular tests, with many options now commercially available. Finally, a formal cost-effectiveness analysis incorporating secondary costs such as laboratory technician time, hospital and drug costs was beyond the scope of this study, but could be considered to more fully evaluate this testing strategy relative to others. Financing within the US healthcare system is particularly complex and given intra-hospital budget ‘silos’, actual billing reimbursement charges as opposed to costs, reagent/instrument rental deals and purchasing arrangements, many other factors need to be considered to fully understand the economic implications of this approach.
New multiplex PCR assays such as the Biofire FilmArray® are a revolutionary leap forward in the microbiology laboratory based diagnosis of respiratory viral infections. This on-demand test has significant advantages over older multiplex PCR platforms that often require batching and specialized personnel to establish and run. However like any test there are shortcomings, including cost and inability to handle simultaneous testing of multiple samples. Our findings demonstrate a simple way to maximize performance of this test using a combined testing approach with a rapid immunoassay. As the array of licensed rapid molecular diagnostic tests for respiratory viral infections increases, including those available for use at the point of care, hospitals will need to carefully consider the best testing approaches based on their local epidemiology and available resources. A combination testing approach as we have described is one way to optimize diagnostic performance and minimize the limitations, including costs, of individual tests.
Funding
This work was supported by the Tufts Medical Center Division Geographic Medicine and Infectious Disease Francis P. Tally MD Fellowship.
Conflicts of interest
All authors report no potential conflicts of interest.
Acknowledgements
The authors would like to acknowledge the assistance of Susan Bollinger, Shelley Bame-Aldred and Kaitlin Smith for their assistance with data collection, and the Tufts Medical Center Microbiology Laboratory who performed all of the testing and were happy to share their experience.
References
- Aoki F.Y., Macleod M.D., Paggiaro P., Carewicz O., El Sawy A., Wat C. Early administration of oral oseltamivir increases the benefits of influenza treatment. J Antimicrob Chemother. 2003;51:123–129. doi: 10.1093/jac/dkg007. [DOI] [PubMed] [Google Scholar]
- Bruning A.H.L., Leeflang M.M.G., Vos J., Spijker R., de Jong M.D., Wolthers K.C. Rapid Tests for Influenza, Respiratory Syncytial Virus, and Other Respiratory Viruses: A Systematic Review and Meta-analysis. Clin Infect Dis. 2017;65:1026–1032. doi: 10.1093/cid/cix461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliendo A.M. Multiplex PCR and emerging technologies for the detection of respiratory pathogens. Clin Infect Dis. 2011;52(Suppl. 4):S326–330. doi: 10.1093/cid/cir047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H.Y., Englund J.A., Huang D., Scott E., Chan J.D., Jain R. Impact of rapid influenza PCR testing on hospitalization and antiviral use: A retrospective cohort study. J Med Virol. 2015;87:2021–2026. doi: 10.1002/jmv.24279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FilmArray® Respiratory Panel Package Insert. Biofire Diagnostics SLC; UT: 2014. [Google Scholar]
- Gadsby N.J., Russell C.D., McHugh M.P., Mark H., Conway Morris A., Laurenson I.F. Comprehensive Molecular Testing for Respiratory Pathogens in Community-Acquired Pneumonia. Clin Infect Dis. 2016;62:817–823. doi: 10.1093/cid/civ1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaydos C.A. What is the role of newer molecular tests in the management of CAP? Infect Dis Clin North Am. 2013;27:49–69. doi: 10.1016/j.idc.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfer G., Leggett J., Myers J., Wang L., Gilbert D.N. The clinical impact of the detection of potential etiologic pathogens of community-acquired pneumonia. Diagn Microbiol Infect Dis. 2015;83:400–406. doi: 10.1016/j.diagmicrobio.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez S., Prieto C., Vera C., RO J., Folgueira L. Evaluation of a new rapid diagnostic test for the detection of influenza and RSV. Enferm Infecc Microbiol Clin. 2015;34:298–302. doi: 10.1016/j.eimc.2015.05.007. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Del Vecchio M., Catalan P., de Egea V., Rodriguez-Borlado A., Martos C., Padilla B. An algorithm to diagnose influenza infection: evaluating the clinical importance and impact on hospital costs of screening with rapid antigen detection tests. Eur J Clin Microbiol Infect Dis. 2015;34:1081–1085. doi: 10.1007/s10096-015-2328-7. [DOI] [PubMed] [Google Scholar]
- Hazelton B., Nedeljkovic G., Ratnamohan V.M., Dwyer D.E., Kok J. Evaluation of the Sofia Influenza A + B fluorescent immunoassay for the rapid diagnosis of influenza A and B. J Med Virol. 2015;87:35–38. doi: 10.1002/jmv.23976. [DOI] [PubMed] [Google Scholar]
- Makristathis A., Zeller I., Mitteregger D., Kundi M., Hirschl A.M. Comprehensive evaluation of chemiluminescent immunoassays for the laboratory diagnosis of Clostridium difficile infection. Eur J Clin Microbiol Infect Dis. 2017;36:1253–1259. doi: 10.1007/s10096-017-2916-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer L.M., Kahlert C., Rassouli F., Vernazza P., Albrich W.C. Impact of viral multiplex real-time PCR on management of respiratory tract infection: a retrospective cohort study. Pneumonia (Nathan) 2017;9:4. doi: 10.1186/s41479-017-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh J.Y., Choi W.S., Lee J., Kim H.L., Song J.Y., Cheong H.J. Clinical performance of the Sofia Influenza A+B FIA in adult patients with influenza-like illness. Diagn Microbiol Infect Dis. 2015;83:130–132. doi: 10.1016/j.diagmicrobio.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuskanen O., Lahti E., Jennings L.C., Murdoch D.R. Viral pneumonia. Lancet. 2011;377:1264–1275. doi: 10.1016/S0140-6736(10)61459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel J.D., Rhinehart E., Jackson M., Chiarello L., the Healthcare Infection Control Practices Advisory Committee Guideline for Isolation Precautions: Preventing Transmission of Infectious Agents in Healthcare Settings. 2007. https://www.cdc.gov/infectioncontrol/guidelines/isolation/ [Retrieved Sept 06, 2017, from] [DOI] [PMC free article] [PubMed]
- Sofia Influenza A+B FIA, Package Insert. CA; Quidel SD: 2015. [Google Scholar]
- Varkey J.B., Varkey B. Viral infections in patients with chronic obstructive pulmonary disease. Curr Opin Pulm Med. 2008;14:89–94. doi: 10.1097/MCP.0b013e3282f4a99f. [DOI] [PubMed] [Google Scholar]