Abstract
A series of benzylidene hydrazides (1–20) was synthesized and tested, in vitro, for antibacterial, antifungal and antiviral activities. The microbial screening results indicated that compounds having chloro and nitro substituents were the most active ones. The antiviral evaluation depicted that compounds 9 and 19 were active against Vesicular stomatitis virus (VSV) in HeLa cell cultures. QSAR investigations indicated that the multi-target QSAR model was effective in describing the antimicrobial (antibacterial and antifungal) activity over the one-target QSAR models. Further the mt-QSAR model indicated that the topological parameters, second order molecular connectivity index (2χ) and third order Kier's alpha shape index (κα3) are effective in describing the antimicrobial activity of synthesized hydrazides.
Keywords: Hydrazides, QSAR, Antibacterial, Antifungal, Antiviral
Graphical abstract
Multi-target QSAR models developed for the antimicrobial activity of synthesized substituted hydrazides indicated the importance of topological parameters in demonstrating antimicrobial activity.
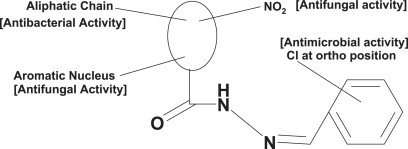
1. Introduction
Recently infectious diseases have dramatically increased and become a major threat to public health, despite tremendous progress in medicinal chemistry. The impact is more acute in developing countries due to non availability of desired medicines and emergence of widespread drug resistance [1]. Infections caused by fungal species are common in immunocompromised patients and carry significant treatment cost and mortality [2]. Antibacterial resistance is a growing problem which necessitates the discovery of newer antibiotics with activity against resistant strains [3].
Hydrazide derivatives have been claimed to possess antimicrobial [4], antimycobacterial [5], antitumour [6], anti-inflammatory [7], trypanocidal [8], antimalarial [9] and anti-HIV activities [10].
Quantitative structure activity relationship (QSAR), one of the most important area in medicinal chemistry, gives information that is useful for drug design. QSAR models are mathematical equations relating chemical structure to a wide variety of physical, chemical and biological properties. The derived relationship between molecular descriptors and activity is used to estimate the property of other molecules and/or to find the parameters affecting the biological activity [11].
Diseases caused by herpesviruses play an important role in viral infections of humans especially the incidence of infections by herpes simplex virus (HSV) is high throughout the world. Clinical manifestations of diseases due to HSV range from mucosal ulcerations with HSV-1 (Herpes labialis) to ulcerating vesicular lesions with HSV-2 (Herpes genitalis). More rarely, HSV infections can result in keratoconjunctivitis, eczema herpeticatum, herpes encephalitis, generalized disseminated diseases in neonates and immunosuppressed patients [12]. Vesicular stomatitis virus (VSV), a member of the Rhabdoviridae family, is an enveloped single-stranded RNA virus that causes an economically important disease in cattle, horses and swine [13].
The alphaviruses are pathogenic viruses with worldwide distribution. Infection can result in fever, rash, arthralgia or arthritis, lassitude, headache and myalgia. The prototypic alphavirus is Sindbis virus, which is transmitted to humans through mosquito bites [14]. Respiratory syncytial virus (RSV), a paramyxovirus, is an important cause of respiratory tract infection in infants, young children and adults [15].
The enterovirus, Coxsackie virus B3 (CVB3) is considered to be the most important etiological agent of virus-induced acute and chronic myocarditis. Since, more than 65 enterovirus serotypes exist, it was not possible to generate a vaccine until now and considerable interest exists to develop antiviral compounds. So far, these virus infections are treated symptomatically because there are no virus-specific prophylactics and drugs available for clinical use [16].
Inspired by the above facts and in continuation of our ongoing research program in the field of synthesis, antimicrobial activity and QSAR studies of medicinally important compounds [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], we hereby report the synthesis, antimicrobial activity, QSAR studies and antiviral evaluation of benzylidene hydrazides.
2. Chemistry
The synthesis of target compounds was carried out as depicted in Scheme 1, Scheme 2 . The different carboxylic acids were refluxed with ethanol in presence of sulphuric acid to yield their ethyl esters. The ethyl ester was refluxed with hydrazine hydrate in ethanol to yield the corresponding hydrazide which was then refluxed with benzaldehyde/o-chlorobenzaldehyde to yield the target acid hydrazones. The products were separated out by evaporating ethanol and recrystallized from ethanol to yield the pure compounds. The completion of reaction was monitored by single spot TLC on Merck silica gel G plates. All the compounds were obtained in appreciable yield and their physicochemical characteristics are presented in Table 1 .
Scheme 1.
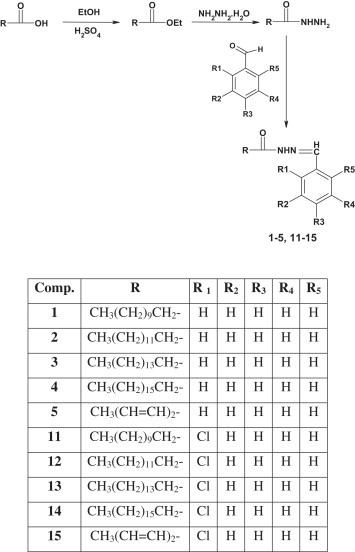
Synthetic route followed to obtain target compounds from aliphatic acids.
Scheme 2.
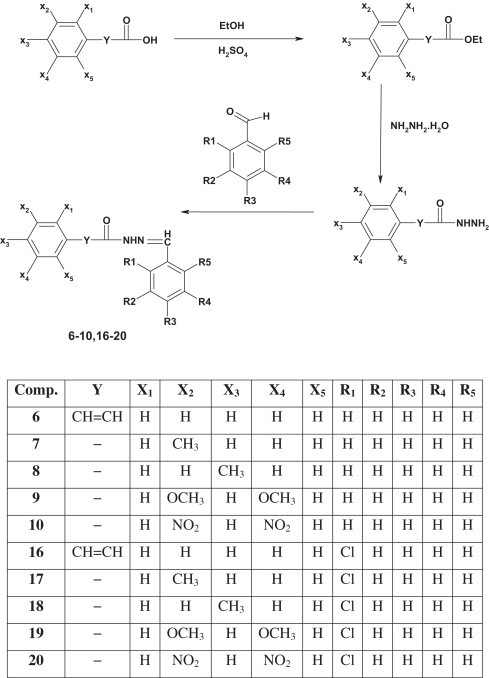
Synthetic route followed to obtain target compounds from aromatic acids.
Table 1.
Physicochemical characteristics of synthesized benzylidene hydrazides.
| Comp. | Mol. formula | M. wt. | Mp/Bp+ (°C) | Rf | % Yield |
|---|---|---|---|---|---|
| 1 | C19H30N2O | 302 | 60–64 | 0.62 | 67.8 |
| 2 | C21H34N2O | 330 | 65–69 | 0.43 | 58.3 |
| 3 | C23H38N2O | 358 | 93–97 | 0.65 | 62.3 |
| 4 | C25H42N2O | 386 | Semisolid | 0.57 | 59.3 |
| 5 | C13H14N2O | 214 | Semisolid | 0.72a | 45.1 |
| 6 | C16H14N2O | 250 | 92–96 | 0.76a | 40.0 |
| 7 | C15H14N2O | 238 | Semisolid | 0.62a | 79.0 |
| 8 | C15H14N2O | 238 | 220–224 | 0.82a | 54.3 |
| 9 | C16H16N2O3 | 284 | 228–232 | 0.83a | 74.0 |
| 10 | C 14H10N4O5 | 314 | 205–209 | 0.65a | 78.3 |
| 11 | C19H29N2OCl | 336 | 63–67 | 0.71 | 40.0 |
| 12 | C21H33N2OCl | 364 | 69–73 | 0.73 | 84.0 |
| 13 | C23H37N2OCl | 392 | 98–102 | 0.72 | 63.0 |
| 14 | C25H41N2OCl | 420 | 108–112 | 0.78 | 22.3 |
| 15 | C13H13N2OCl | 248 | 170–174+ | 0.71a | 61.1 |
| 16 | C16H13N2OCl | 284 | Semisolid | 0.50a | 80.2 |
| 17 | C15H13N2OCl | 272 | 155–159 | 0.75a | 40.1 |
| 18 | C15H13N2OCl | 272 | 241–245+ | 0.85a | 42.2 |
| 19 | C16H15N2O3Cl | 318 | 240–244 | 0.88a | 80.0 |
| 20 | C14H9N4O5Cl | 384 | 235–239 | 0.89a | 58.0 |
Mobile phase – Toluene:Chloroform (7:3).
Ethyl acetate:Hexane (6:4).
The structures of the synthesized compounds (1–20) were ascertained on the basis of their consistent IR and NMR spectral characteristics. The presence of C O functional group was marked by the appearance of stretching band around 1700 cm−1, which is the characteristic of an amide linkage. The appearance of stretching band around 3200 cm−1 indicated the presence of NH linkage of amide bond of hydrazone. The presence of peaks slightly above and below 3000 cm−1 indicated the presence of aromatic and aliphatic portion in synthesized compounds respectively. The appearance of aliphatic –C C– stretching band at 1658.5 cm−1 in compound 5 indicates the presence of unsaturated sorbic acid moiety in its structure. In compound 9, the ArOCH3 stretching at 1270.9 cm−1 reveals the presence of methoxy group of veratric acid. The aromatic nitro stretching around 1350 cm−1 (symmetric NO2 stretching) and 1540 cm−1 (asymmetric NO2 stretching) depicts the presence of nitro functional group in synthesized compounds 10 and 20. The appearance of singlet signal ranging from δ 8.10–10.47 ppm in compounds 1–20 confirms the presence of NH of hydrazide. The presence of aromatic hydrogens was confirmed by the multiplet signal in the range of δ 7.0–8.0 ppm. The appearance of triplet signal at δ 0.82–0.89 ppm in compounds 1 and 13 reveals the presence of terminal methyl group. Further, in compound 9, singlet signal at δ 3.92 ppm depicted the presence of methoxy group.
3. Results and discussion
3.1. Antibacterial and antifungal activity
Benzylidene hydrazides [1–20] were evaluated for their in vitro antibacterial activity against Gram-positive Staphylococcus aureus, Bacillus subtilis, Gram-negative Escherichia coli and antifungal activity against Candida albicans and Aspergillus niger by serial dilution method [27] using ciprofloxacin and fluconazole as reference standards for antibacterial and antifungal activity respectively and the results are presented in Table 2 .
Table 2.
Antibacterial and antifungal potential (μM/mL) of synthesized benzylidene hydrazides.
| Comp. | pMICsa | pMICbs | pMICec | pMICca | pMICan | pMICb | pMICf | pMICam |
|---|---|---|---|---|---|---|---|---|
| 1 | 1.38 | 1.30 | 1.38 | 1.21 | 1.21 | 1.35 | 1.21 | 1.30 |
| 2 | 1.42 | 1.42 | 1.36 | 1.24 | 1.24 | 1.40 | 1.24 | 1.34 |
| 3 | 1.46 | 1.46 | 1.50 | 1.76 | 0.86 | 1.47 | 1.31 | 1.41 |
| 4 | 1.49 | 1.49 | 1.79 | 2.09 | 1.20 | 1.59 | 1.65 | 1.61 |
| 5 | 1.23 | 1.23 | 1.23 | 0.93 | 1.54 | 1.23 | 1.24 | 1.23 |
| 6 | 1.30 | 1.30 | 1.30 | 1.90 | 1.60 | 1.30 | 1.75 | 1.48 |
| 7 | 1.28 | 1.28 | 1.28 | 1.88 | 1.58 | 1.28 | 1.73 | 1.46 |
| 8 | 1.28 | 1.28 | 1.28 | 1.34 | 1.88 | 1.28 | 1.61 | 1.41 |
| 9 | 1.36 | 1.36 | 1.36 | 1.96 | 1.66 | 1.36 | 1.81 | 1.54 |
| 10 | 1.40 | 1.40 | 1.40 | 1.70 | 1.70 | 1.40 | 1.70 | 1.52 |
| 11 | 1.43 | 1.43 | 1.80 | 1.73 | 1.43 | 1.55 | 1.58 | 1.56 |
| 12 | 1.47 | 1.47 | 1.47 | 1.12 | 1.12 | 1.47 | 1.12 | 1.33 |
| 13 | 1.50 | 1.49 | 1.50 | 2.10 | 1.27 | 1.50 | 1.69 | 1.57 |
| 14 | 1.53 | 1.53 | 1.53 | 1.83 | 1.29 | 1.53 | 1.56 | 1.54 |
| 15 | 1.30 | 1.30 | 1.30 | 1.60 | 1.50 | 1.30 | 1.55 | 1.40 |
| 16 | 1.36 | 1.36 | 1.36 | 1.76 | 1.76 | 1.36 | 1.76 | 1.52 |
| 17 | 1.34 | 1.34 | 1.34 | 1.34 | 1.51 | 1.34 | 1.43 | 1.37 |
| 18 | 1.34 | 1.34 | 1.34 | 1.61 | 1.61 | 1.34 | 1.61 | 1.45 |
| 19 | 1.41 | 1.41 | 1.41 | 2.01 | 1.56 | 1.41 | 1.79 | 1.56 |
| 20 | 1.45 | 1.45 | 1.45 | 2.05 | 1.60 | 1.45 | 1.83 | 1.60 |
| S.D.c | 0.08 | 0.08 | 0.15 | 0.35 | 0.25 | 0.10 | 0.22 | 0.11 |
| Std. | 2.61a | 2.61a | 2.61a | 2.64b | 2.64b | 2.61 | 2.64 | 2.62 |
Ciprofloxacin.
Fluconazole.
Standard deviation.
For antifungal activity against A. niger compounds 9, 10, 16 and 18 have shown marked antifungal potential having pMIC value greater than 1.60 as compared to other synthesized derivatives. In case of C. albicans compounds 4, 13, 19 and 20 were found to be most active with pMIC value 2.09, 2.10, 2.01 and 2.05 respectively.
In case of S. aureus compounds 13 and 14 emerged as effective antibacterial agent having pMIC value 1.50 and 1.53 respectively. In case of E. coli compounds 4 and 11 have shown good antibacterial capability at a pMIC value 1.79 and 1.80 respectively. For antibacterial activity against B. subtilis, compounds 4, 13 and 14 had shown good antibacterial potential with pMIC value of 1.49, 1.49 and 1.53 respectively. It can be seen from Table 2 that synthesized benzylidene hydrazides have higher antifungal potential as compared to their antibacterial activity.
The minimum bactericidal concentration/minimum fungicidal concentration (MBC/MFC) (Table 3 ) determination results revealed that the synthesized compounds were fungistatic against both fungi and bacteriostatic against S. aureus, but bactericidal against E. coli and B. subtilis. In general, the MBC and MFC of synthesized benzylidene derivatives were 3-fold higher than the MIC values which indicated that the synthesized compound were bacteriostatic and fungistatic in action except for B. subtilis and E. coli (a drug is considered to be bacteriostatic/fungistatic when its MBC and MFC values are 3-fold higher than its MIC values) [28]. In case of B. subtilis compounds 13 and 14 and for E. coli compounds 11–13 were found to be active bactericidal agents.
Table 3.
MBC/MFC values of synthesized benzylidene hydrazides.
| Comp. | MBC/MFC (μM/mL) |
||||
|---|---|---|---|---|---|
| S. aureus | B. subtilis | E. coli | C. albicans | A. niger | |
| 1 | >0.166 | >0.166 | 0.041 | >0.166 | >0.166 |
| 2 | >0.152 | >0.152 | >0.152 | >0.152 | >0.152 |
| 3 | >0.139 | 0.139 | >0.139 | >0.139 | >0.139 |
| 4 | 0.129 | <0.129 | 0.064 | <0.129 | 0.03 |
| 5 | >0.233 | >0.233 | 0.058 | >0.233 | >0.233 |
| 6 | >0.200 | >0.200 | 0.050 | >0.200 | >0.200 |
| 7 | >0.210 | 0.052 | >0.210 | >0.210 | >0.210 |
| 8 | >0.210 | 0.210 | 0.052 | >0.210 | >0.210 |
| 9 | 0.088 | 0.044 | 0.177 | 0.011 | 0.022 |
| 10 | >0.159 | 0.159 | >0.159 | 0.159 | 0.079 |
| 11 | >0.148 | 0.148 | 0.009 | >0.148 | >0.148 |
| 12 | >0.137 | >0.137 | 0.034 | >0.137 | >0.137 |
| 13 | >0.127 | 0.031 | 0.031 | >0.127 | >0.127 |
| 14 | >0.119 | 0.029 | >0.119 | >0.119 | 0.059 |
| 15 | >0.201 | 0.050 | 0.100 | >0.201 | >0.201 |
| 16 | 0.176 | 0.088 | >0.176 | >0.176 | >0.176 |
| 17 | >0.183 | 0.045 | 0.045 | >0.183 | >0.183 |
| 18 | 0.045 | 0.183 | 0.045 | >0.183 | 0.183 |
| 19 | >0.157 | 0.039 | >0.157 | 0.019 | >0.157 |
| 20 | >0.130 | 0.065 | >0.130 | >0.130 | >0.130 |
| Std. | 0.019 | 0.019 | 0.019 | 0.040 | 0.040 |
3.2. QSAR studies
3.2.1. Development of one-target QSAR model
In order to understand the experimental antibacterial and antifungal activity data of 20 substituted benzylidene derivatives on theoretical basis, we established a quantitative structure activity relationship (QSAR) between their in vitro activity and descriptors coding for lipophilic, electronic, steric and topological properties of the molecules under consideration using the linear free energy relationship model (LFER) described by Hansch and Fujita [11]. Biological activity data determined as MIC values were first transformed into pMIC values and used as dependent variable in QSAR study. The different molecular descriptors (independent variables) like log of octanol–water partition coefficient (log P), molar refractivity (MR), Kier's molecular connectivity (0 χ, 0 χv, 1 χ, 1 χv, 2 χ, 2 χv) and shape (κ 1, κα 1, κα 2, κα 3) topological indices, Randic topological index (R), Balaban topological index (J), Wiener topological index (W), Total energy (Te), energies of highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO), dipole moment (μ) and electronic energy (Ele.E), calculated for benzylidene hydrazides are presented in Table 4 [29], [30], [31], [32], [33], [34]. Units of the energies and dipole were electron volts (eV), and atomic units (a.u.), respectively [35].
Table 4.
Values of selected descriptors used in the regression analysis.
| Comp. | log P | MR | 0χ | 0χv | 1χ | 1χv | 2χ | 2χv | 3χ | 3χv | κ1 | κα1 | κα2 | κα3 | R | J | W | Te | LUMO | HOMO | μ | Ele.E |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5.708 | 93.608 | 15.88 | 13.89 | 10.83 | 8.70 | 8.16 | 5.78 | 0.49 | 0.17 | 20.05 | 18.97 | 13.57 | 11.21 | 10.83 | 1.76 | 1568 | −3581.09 | −0.13 | −8.84 | 4.38 | −23877.40 |
| 2 | 6.500 | 102.810 | 17.30 | 15.30 | 11.83 | 9.70 | 8.86 | 6.49 | 0.49 | 0.17 | 22.04 | 20.96 | 15.45 | 12.98 | 11.83 | 1.73 | 2067 | −3893.06 | −0.21 | −9.24 | 2.39 | −27346.00 |
| 3 | 7.293 | 112.012 | 18.71 | 16.72 | 12.83 | 10.70 | 9.57 | 7.20 | 0.49 | 0.17 | 24.04 | 22.96 | 17.34 | 14.77 | 12.83 | 1.71 | 2662 | −4204.59 | −0.32 | −8.96 | 2.59 | −30530.90 |
| 4 | 8.086 | 121.214 | 20.13 | 18.13 | 13.83 | 11.70 | 10.28 | 7.90 | 0.49 | 0.17 | 26.04 | 24.96 | 19.25 | 16.59 | 13.83 | 1.69 | 3361 | −4516.26 | −0.32 | −8.96 | 2.59 | −33750.50 |
| 5 | 3.149 | 68.212 | 11.64 | 9.13 | 7.83 | 5.00 | 6.04 | 3.03 | 0.49 | 0.16 | 14.06 | 12.47 | 7.66 | 5.79 | 7.83 | 1.84 | 567 | −2589.03 | −0.57 | −8.75 | 4.71 | −14019.40 |
| 6 | 3.834 | 77.988 | 13.34 | 10.36 | 9.34 | 5.96 | 7.62 | 3.89 | 0.70 | 0.25 | 15.39 | 13.31 | 7.34 | 4.94 | 9.34 | 1.39 | 896 | −2972.83 | −0.54 | −8.80 | 4.74 | −18470.90 |
| 7 | 3.893 | 72.786 | 12.79 | 10.13 | 8.75 | 5.71 | 7.44 | 3.98 | 0.90 | 0.40 | 14.41 | 12.59 | 6.18 | 3.84 | 8.75 | 1.46 | 713 | −2845.59 | −0.35 | −8.83 | 4.58 | −16783.40 |
| 8 | 3.893 | 72.786 | 12.79 | 10.13 | 8.75 | 5.71 | 7.43 | 3.98 | 0.90 | 0.40 | 14.41 | 12.59 | 6.18 | 3.84 | 8.75 | 1.46 | 724 | −2845.60 | −0.39 | −8.82 | 5.00 | −16734.30 |
| 9 | 2.920 | 80.671 | 15.08 | 11.87 | 10.22 | 6.34 | 8.43 | 4.21 | 1.01 | 0.37 | 17.36 | 15.45 | 7.74 | 4.53 | 10.22 | 1.53 | 1066 | −3641.34 | −0.53 | −8.80 | 6.01 | −22455.00 |
| 10 | 3.333 | 82.394 | 16.82 | 11.58 | 10.97 | 6.29 | 9.89 | 4.36 | 1.61 | 0.45 | 19.33 | 16.63 | 7.50 | 4.62 | 10.97 | 1.59 | 1332 | −4351.04 | −2.16 | −9.32 | 5.20 | −25795.60 |
| 11 | 6.226 | 98.413 | 16.75 | 15.01 | 11.24 | 9.21 | 8.68 | 6.33 | 0.69 | 0.33 | 21.04 | 20.26 | 13.63 | 10.57 | 11.24 | 1.81 | 1740 | −3941.23 | −0.50 | −9.02 | 2.94 | −26585.40 |
| 12 | 7.018 | 107.615 | 18.17 | 16.42 | 12.24 | 10.21 | 9.38 | 7.04 | 0.69 | 0.33 | 23.04 | 22.25 | 15.46 | 12.24 | 12.24 | 1.78 | 2276 | −4252.93 | −0.32 | −9.21 | 2.99 | −29711.10 |
| 13 | 7.811 | 116.817 | 19.58 | 17.84 | 13.24 | 11.21 | 10.09 | 7.75 | 0.69 | 0.33 | 25.04 | 24.25 | 17.32 | 13.94 | 13.24 | 1.76 | 2912 | −4564.56 | −0.50 | −9.02 | 2.94 | −32956.10 |
| 14 | 8.604 | 126.019 | 21.00 | 19.25 | 14.24 | 12.21 | 10.80 | 8.45 | 0.69 | 0.33 | 27.03 | 26.25 | 19.19 | 15.68 | 14.24 | 1.73 | 3656 | −4876.23 | −0.50 | −9.02 | 2.94 | −36224.10 |
| 15 | 3.667 | 73.017 | 12.51 | 10.25 | 8.24 | 5.52 | 6.55 | 3.58 | 0.69 | 0.31 | 15.06 | 13.75 | 7.88 | 5.53 | 8.24 | 1.94 | 652 | −2948.97 | −0.59 | −8.78 | 5.12 | −15923.90 |
| 16 | 4.352 | 82.793 | 14.21 | 11.48 | 9.75 | 6.47 | 8.14 | 4.44 | 0.90 | 0.41 | 16.37 | 14.58 | 7.68 | 5.00 | 9.75 | 1.43 | 1014 | −3332.84 | −0.77 | −8.81 | 5.03 | −19365.60 |
| 17 | 4.411 | 77.591 | 13.67 | 11.25 | 9.16 | 6.22 | 7.96 | 4.53 | 1.09 | 0.56 | 15.39 | 13.85 | 6.55 | 3.95 | 9.16 | 1.50 | 814 | −3205.54 | −0.41 | −8.86 | 5.14 | −18787.40 |
| 18 | 4.411 | 77.591 | 13.67 | 11.25 | 9.16 | 6.22 | 7.95 | 4.53 | 1.09 | 0.56 | 15.39 | 13.85 | 6.55 | 3.95 | 9.16 | 1.51 | 826 | −3205.57 | −0.37 | −9.12 | 5.42 | −18727.20 |
| 19 | 3.438 | 85.476 | 15.95 | 12.99 | 10.63 | 6.86 | 8.95 | 4.76 | 1.21 | 0.53 | 18.34 | 16.72 | 8.11 | 4.68 | 10.63 | 1.56 | 1199 | −4001.38 | −0.37 | −9.10 | 6.54 | −24635.10 |
| 20 | 3.851 | 87.199 | 17.69 | 12.70 | 11.38 | 6.81 | 10.41 | 4.91 | 1.80 | 0.61 | 20.31 | 17.90 | 7.89 | 4.77 | 11.38 | 1.62 | 1487 | −4711.35 | −2.16 | −9.53 | 4.96 | −28085.80 |
In the present study, a data set of 20 benzylidene hydrazide derivatives was subjected to linear free energy regression analysis for model generation. The reference drugs were not included in model development as they belong to different structural series. Preliminary analysis was carried out in terms of correlation analysis. A correlation matrix constructed for antibacterial activity against S. aureus is presented in Table 5 . The correlations of different molecular descriptors with antibacterial and antifungal activity are presented in Table 6 . In general, high colinearity (r > 0.8) was observed between different parameters. The high interrelationship was observed between 0 χ and Ele.E (r = 0.999), and low interrelationship was observed between 1 χ and LUMO (r = 0.002). The correlation matrix indicated the predominance of topological parameters in describing the antibacterial and antifungal activity of synthesized compounds.
Table 5.
Correlation matrix for antibacterial activity of benzylidene hydrazides against S. aureus.
| pMICsa | log P | 0χ | 0χv | 1χ | 1χv | 2χ | 2χv | 3χ | 3χv | κα1 | κα2 | κα3 | Te | Ele.E | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pMICsa | 1.000 | ||||||||||||||
| log P | 0.784 | 1.000 | |||||||||||||
| 0χ | 0.923 | 0.822 | 1.000 | ||||||||||||
| 0χv | 0.899 | 0.940 | 0.950 | 1.000 | |||||||||||
| 1χ | 0.914 | 0.859 | 0.994 | 0.967 | 1.000 | ||||||||||
| 1χv | 0.861 | 0.971 | 0.917 | 0.991 | 0.944 | 1.000 | |||||||||
| 2χ | 0.864 | 0.636 | 0.941 | 0.808 | 0.916 | 0.749 | 1.000 | ||||||||
| 2χv | 0.881 | 0.968 | 0.929 | 0.993 | 0.951 | 0.994 | 0.787 | 1.000 | |||||||
| 3χ | −0.074 | −0.536 | −0.083 | −0.348 | −0.169 | −0.447 | 0.236 | −0.374 | 1.000 | ||||||
| 3χv | −0.096 | −0.447 | −0.179 | −0.331 | −0.248 | −0.427 | 0.109 | −0.334 | 0.873 | 1.000 | |||||
| κα1 | 0.901 | 0.920 | 0.965 | 0.991 | 0.975 | 0.982 | 0.822 | 0.978 | −0.323 | −0.365 | 1.000 | ||||
| κα2 | 0.801 | 0.957 | 0.865 | 0.958 | 0.897 | 0.981 | 0.652 | 0.957 | −0.562 | −0.575 | 0.963 | 1.000 | |||
| κα3 | 0.760 | 0.953 | 0.827 | 0.931 | 0.863 | 0.965 | 0.599 | 0.935 | −0.613 | −0.631 | 0.938 | 0.996 | 1.000 | ||
| Te | −0.890 | −0.646 | −0.958 | −0.837 | −0.924 | −0.773 | −0.973 | −0.799 | −0.183 | −0.041 | −0.865 | −0.702 | −0.649 | 1.000 | |
| Ele.E | −0.925 | −0.823 | −0.999 | −0.954 | −0.993 | −0.920 | −0.936 | −0.932 | 0.091 | 0.180 | −0.967 | −0.867 | −0.827 | 0.956 | 1.000 |
Table 6.
Correlation of molecular descriptors with antimicrobial (antibacterial and antifungal) activity.
| Mol. descriptor | pMICsa | pMICbs | pMICec | pMICca | pMICan | pMICb | pMICf | pMICam |
|---|---|---|---|---|---|---|---|---|
| log P | 0.796 | 0.757 | 0.681 | 0.097 | −0.796 | 0.784 | −0.369 | 0.129 |
| MR | 0.922 | 0.889 | 0.739 | 0.266 | −0.769 | 0.887 | −0.221 | 0.308 |
| 0χ | 0.981 | 0.955 | 0.740 | 0.381 | −0.655 | 0.923 | −0.066 | 0.455 |
| 0χv | 0.935 | 0.900 | 0.750 | 0.260 | −0.769 | 0.899 | −0.225 | 0.311 |
| 1χ | 0.967 | 0.939 | 0.740 | 0.372 | −0.683 | 0.914 | −0.089 | 0.432 |
| 1χv | 0.889 | 0.846 | 0.733 | 0.192 | −0.804 | 0.861 | −0.299 | 0.230 |
| 2χ | 0.939 | 0.933 | 0.657 | 0.511 | −0.430 | 0.864 | 0.162 | 0.609 |
| 2χv | 0.913 | 0.874 | 0.742 | 0.222 | −0.773 | 0.881 | −0.258 | 0.274 |
| 3χ | −0.016 | 0.036 | −0.163 | 0.338 | 0.580 | −0.074 | 0.591 | 0.442 |
| 3χv | −0.055 | 0.010 | −0.172 | 0.293 | 0.583 | −0.096 | 0.558 | 0.399 |
| κ1 | 0.953 | 0.918 | 0.749 | 0.288 | −0.745 | 0.908 | −0.189 | 0.346 |
| κ2 | 0.823 | 0.774 | 0.698 | 0.120 | −0.844 | 0.804 | −0.378 | 0.135 |
| κ3 | 0.765 | 0.711 | 0.668 | 0.058 | −0.860 | 0.754 | −0.435 | 0.061 |
| κα1 | 0.939 | 0.900 | 0.752 | 0.244 | −0.778 | 0.901 | −0.243 | 0.298 |
| κα2 | 0.818 | 0.768 | 0.699 | 0.103 | −0.850 | 0.801 | −0.394 | 0.120 |
| κα3 | 0.770 | 0.716 | 0.674 | 0.055 | −0.863 | 0.760 | −0.439 | 0.060 |
| R | 0.967 | 0.939 | 0.740 | 0.372 | −0.683 | 0.914 | −0.089 | 0.432 |
| J | 0.320 | 0.273 | 0.348 | −0.300 | −0.589 | 0.342 | −0.564 | −0.276 |
| W | 0.906 | 0.881 | 0.720 | 0.296 | −0.732 | 0.871 | −0.176 | 0.334 |
| Te | −0.962 | −0.948 | −0.689 | −0.449 | 0.498 | −0.890 | −0.075 | −0.552 |
| Ele.E | −0.980 | −0.956 | −0.745 | −0.383 | 0.659 | −0.925 | 0.066 | −0.456 |
| LUMO | −0.090 | −0.131 | 0.019 | −0.270 | −0.386 | −0.055 | −0.429 | −0.381 |
| HOMO | −0.568 | −0.592 | −0.289 | −0.168 | 0.135 | −0.476 | −0.058 | −0.313 |
| μ | −0.608 | −0.593 | −0.602 | 0.070 | 0.774 | −0.643 | 0.488 | 0.045 |
The antibacterial activity of synthesized benzylidene hydrazides against S. aureus is explained by the topological parameter, zero order molecular connectivity index (Eq. (1)).
QSAR model for antibacterial activity against S. aureus
| (1) |
Here and thereafter, n – number of data points, r – correlation coefficient, q 2 – cross-validated r 2 obtained by leave one out method, s – standard error of the estimate and F – Fischer statistics.
As the coefficient of 0 χ in Eq. (1) is positive, therefore the antibacterial activity against S. aureus will increase with increase in value of 0 χ. This is clearly evident from Table 4 that compounds 13 and 14 having high 0 χ values, 19.58 and 21.00 (Table 4), have increased antibacterial activity values 1.50 and 1.53 (Table 2) respectively. Similarly, compounds 5, 7 and 8 having minimum 0 χ values 11.64, 12.79 and 12.79 (Table 4), have minimum antibacterial activity against S. aureus (Table 2). Topological indices are numerical quantifier of molecular topology and are sensitive to bonding pattern, symmetry, content of heteroatom as well as degree of complexity of atomic neighborhoods [36]. The zero order molecular connectivity index, 0 χ, represents the molecules with unbranched structure.
The QSAR model expressed by Eq. (1) was cross-validated by its high q 2 values (q 2 = 0.953) obtained by leave one out (LOO) method. The value of q 2 greater than 0.5 is the basic requirement for qualifying a QSAR model to be valid one [37]. The comparison of observed and predicted antibacterial activities is presented in Table 7 . It can be seen from the results that the observed and predicted antibacterial activities lie close to each other as evidenced by their low residual values (Table 7).
Table 7.
Comparison of observed and predicted antibacterial and antifungal activity obtained by ot-QSAR model.
| Comp. | pMICsa (Eq. (1)) |
pMICbs (Eq. (2)) |
pMICec (Eq. (3)) |
pMICca (Eq. (4)) |
pMICan (Eq. (5)) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Obs. | Pre. | Res. | Obs. | Pre. | Res. | Obs. | Pre. | Res. | Obs. | Pre. | Res. | Obs. | Pre. | Res. | |
| 1 | 1.38 | 1.39 | −0.01 | 1.30 | 1.38 | −0.08 | 1.38 | 1.45 | −0.07 | 1.21 | 1.59 | −0.38 | 1.21 | 1.32 | −0.11 |
| 2 | 1.42 | 1.43 | −0.01 | 1.42 | 1.42 | 0.00 | 1.36 | 1.50 | −0.14 | 1.24 | 1.69 | −0.45 | 1.24 | 1.23 | 0.01 |
| 3 | 1.46 | 1.47 | −0.01 | 1.46 | 1.46 | 0.00 | 1.50 | 1.55 | −0.05 | 1.76 | 1.79 | −0.03 | 0.86 | 1.15 | −0.29 |
| 4 | 1.49 | 1.51 | −0.02 | 1.49 | 1.50 | −0.01 | 1.79 | 1.60 | 0.19 | 2.09 | 1.88 | 0.21 | 1.20 | 1.07 | 0.13 |
| 5 | 1.23 | 1.26 | −0.03 | 1.23 | 1.26 | −0.03 | 1.23 | 1.29 | −0.06 | 0.93 | 1.30 | −0.37 | 1.54 | 1.57 | −0.03 |
| 6 | 1.30 | 1.31 | −0.01 | 1.30 | 1.31 | −0.01 | 1.30 | 1.31 | −0.01 | 1.90 | 1.52 | 0.38 | 1.60 | 1.60 | 0.00 |
| 7 | 1.28 | 1.30 | −0.02 | 1.28 | 1.29 | −0.01 | 1.28 | 1.29 | −0.01 | 1.88 | 1.50 | 0.38 | 1.58 | 1.66 | −0.08 |
| 8 | 1.28 | 1.30 | −0.02 | 1.28 | 1.29 | −0.01 | 1.28 | 1.29 | −0.01 | 1.34 | 1.49 | −0.15 | 1.88 | 1.66 | 0.22 |
| 9 | 1.36 | 1.36 | 0.00 | 1.36 | 1.36 | 0.00 | 1.36 | 1.36 | 0.00 | 1.96 | 1.63 | 0.33 | 1.66 | 1.62 | 0.04 |
| 10 | 1.40 | 1.41 | −0.01 | 1.40 | 1.40 | 0.00 | 1.40 | 1.39 | 0.01 | 1.70 | 1.83 | −0.13 | 1.70 | 1.62 | 0.08 |
| 11 | 1.43 | 1.41 | 0.02 | 1.43 | 1.41 | 0.02 | 1.80 | 1.48 | 0.32 | 1.73 | 1.66 | 0.07 | 1.43 | 1.35 | 0.08 |
| 12 | 1.47 | 1.45 | 0.02 | 1.47 | 1.45 | 0.02 | 1.47 | 1.53 | −0.06 | 1.12 | 1.76 | −0.64 | 1.12 | 1.27 | −0.15 |
| 13 | 1.50 | 1.50 | 0.00 | 1.49 | 1.49 | 0.00 | 1.50 | 1.58 | −0.08 | 2.10 | 1.86 | 0.24 | 1.27 | 1.19 | 0.08 |
| 14 | 1.53 | 1.54 | −0.01 | 1.53 | 1.53 | 0.00 | 1.53 | 1.63 | −0.10 | 1.83 | 1.95 | −0.12 | 1.29 | 1.11 | 0.18 |
| 15 | 1.30 | 1.29 | 0.01 | 1.30 | 1.28 | 0.02 | 1.30 | 1.32 | −0.02 | 1.60 | 1.37 | 0.23 | 1.50 | 1.58 | −0.08 |
| 16 | 1.36 | 1.34 | 0.02 | 1.36 | 1.32 | 0.04 | 1.36 | 1.34 | 0.02 | 1.76 | 1.59 | 0.17 | 1.76 | 1.60 | 0.16 |
| 17 | 1.34 | 1.32 | 0.02 | 1.34 | 1.32 | 0.02 | 1.34 | 1.32 | 0.02 | 1.34 | 1.57 | −0.23 | 1.51 | 1.65 | −0.14 |
| 18 | 1.34 | 1.32 | 0.02 | 1.34 | 1.32 | 0.02 | 1.34 | 1.32 | 0.02 | 1.61 | 1.56 | 0.05 | 1.61 | 1.65 | −0.04 |
| 19 | 1.41 | 1.39 | 0.02 | 1.41 | 1.39 | 0.02 | 1.41 | 1.39 | 0.02 | 2.01 | 1.70 | 0.31 | 1.56 | 1.62 | −0.06 |
| 20 | 1.45 | 1.44 | 0.01 | 1.45 | 1.43 | 0.02 | 1.45 | 1.42 | 0.03 | 2.05 | 1.90 | 0.15 | 1.60 | 1.61 | −0.01 |
Eqs. (2), (3), (4), (5) were developed to predict the antibacterial and antifungal activity of benzylidene hydrazides against B. subtilis, E. coli, C. albicans and A. niger.
QSAR model for antibacterial activity against B. subtilis
| (2) |
QSAR model for antibacterial activity against E. coli
| (3) |
QSAR model for antifungal activity against C. albicans
| (4) |
QSAR model for antifungal activity against A. niger
| (5) |
In case of B. subtilis, the developed QSAR model indicated the predominance of electronic energy in describing the antibacterial activity. The coefficient of electronic energy (Ele.E) is negative, which shows that the antibacterial activity will increase with the decrease in electronic energy of the synthesized compound, which can be clearly seen from the results of antibacterial activity against B. subtilis (Table 2) and values of electronic energy presented in Table 4.
For antibacterial activity against E. coli, the developed QSAR model (Eq. (3)) describes the importance of Kier's alpha shape topological index (κα 1). In this case, the positive correlation was observed between κα 3 and antibacterial activity against E. coli. The model described by Eq. (4) depicted the importance of second order molecular connectivity index, 2 χ, in describing the antifungal activity against C. albicans.
The model described by Eq. (5) demonstrated the importance of Kier's alpha shape topological index (κα 3) in describing the antifungal activity against A. niger. The negative correlation of molecular descriptor with antifungal activity reveals that decrease in value of κα 3 (Table 4) will lead to increase in antifungal activity against A. niger. The low residual values presented in Table 7 are in agreement with the models expressed by Eqs. (2), (3), (4), (5).
As in case of Eq. (1), the high q 2 values supported the validity of developed QSAR models described by Eqs. (2), (3), (5). It is important to note that Eqs. (2), (3), (4), (5) were derived using the entire data set as there were no outliers in the data set. Even though the sample size and the ‘Rule of Thumb’ allowed us to go for development of tetra-parametric model in multiple linear regression analysis, the high colinearity [21] among the parameters restricted us to go for mono-parametric model only. The ‘Rule of Thumb’ gives information about the number of parameters to be selected for regression analysis in QSAR based on the number of compounds [38]. According to this rule for QSAR model development one should select one parameter for a five-compound data set.
Generally for QSAR studies, the biological activities of compounds should span 2–3 orders of magnitude. But in the present study the range of antibacterial and antifungal activities of the synthesized compounds is within one order of magnitude. But it is important to note that the predictability of the QSAR models developed in the present study is high evidenced by their low residual values. This is in accordance with results suggested by the Bajaj et al. [39], who stated that the reliability of the QSAR model lies in its predictive ability even though the activity data are in the narrow range. Further, recent literature reveals that the QSAR have been applied to describe the relationship between narrow range of biological activity and physicochemical properties of the molecules [24], [40], [41], [42]. When biological activity data lies in the narrow range, the presence of minimum standard deviation of the biological activity justifies its use in QSAR studies [24], [25]. The minimum standard deviation (Table 2) observed in the antimicrobial activity data justifies its use in QSAR studies. Some of the other statistically important ot-QSAR models are given in Table 8 .
Table 8.
Regression analysis and quality of correlation for modeling antibacterial and antifungal activity of synthesized benzylidene hydrazides.
| S. no. | QSAR model (pMIC=) | n | r | q2 | s | F |
|---|---|---|---|---|---|---|
| S. aureus | ||||||
| 1. | −0.000012Ele.E + 1.087 | 20 | 0.980 | 0.951 | 0.017 | 446.56 |
| 2. | 0.0431χ + 0.927 | 20 | 0.967 | 0.919 | 0.022 | 258.85 |
| 3. | 0.043R + 0.927 | 20 | 0.967 | 0.919 | 0.022 | 258.85 |
| 4. | −0.000011Te + 0.962 | 20 | 0.962 | 0.909 | 0.023 | 225.84 |
| B. subtilis | ||||||
| 5. | 0.02950χ + 0.918 | 20 | 0.955 | 0.906 | 0.026 | 185.42 |
| 6. | −0.00011Te + 0.956 | 20 | 0.948 | 0.886 | 0.028 | 159.12 |
| 7. | 0.042R + 0.927 | 20 | 0.939 | 0.858 | 0.030 | 133.33 |
| 8. | 0.0421χ + 0.927 | 20 | 0.939 | 0.858 | 0.030 | 133.33 |
| E. coli | ||||||
| 9. | 0.0370χv + 0.928 | 20 | 0.750 | 0.463 | 0.103 | 23.13 |
| 10. | 0.027κ1 + 0.901 | 20 | 0.749 | 0.474 | 0.104 | 22.94 |
| 11. | −0.000017Ele.E + 1.004 | 20 | 0.745 | 0.456 | 0.104 | 22.40 |
| 12. | 0.0702χv + 1.04 | 20 | 0.742 | 0.456 | 0.105 | 22.03 |
| C. albicans | ||||||
| 13. | −0.00022Te + 0.829 | 20 | 0.494 | −0.033 | 0.320 | 4.535 |
| A. niger | ||||||
| 14. | 0.0370χv + 0.928 | 20 | 0.750 | 0.463 | 0.103 | 23.13 |
| 15. | 0.027κ1 + 0.901 | 20 | 0.749 | 0.474 | 0.104 | 22.94 |
| 16. | −0.000017Ele.E + 1.004 | 20 | 0.745 | 0.456 | 0.104 | 22.40 |
| 17. | 0.0702χv + 1.047 | 20 | 0.742 | 0.456 | 0.105 | 22.03 |
3.2.2. Development of multi-target QSAR model
According to the above ot-QSAR models one should use five different equations with different errors to predict the activity of a new compound against the five microbial species. The ot-QSAR models, which are almost in all the literature, become unpractical or at less complicated to use when we have to predict to each compound results for more than one target. In these cases we have to develop one ot-QSAR for each target. However, very recently the interest has been increased in development of multi-target QSAR (mt-QSAR) models. In opposition to ot-QSAR, the mt-QSAR model is a single equation that considers the nature of molecular descriptors which are common and essential for describing the antibacterial and antifungal activity [43], [44], [45], [46], [47].
In the present study we have attempted to develop three different types of mt-QSAR models viz. mt-QSAR model for describing antibacterial activity of synthesized compounds against S. aureus, B. subtilis and E. coli, mt-QSAR model for describing antifungal activity of synthesized compounds against C. albicans and A. niger as well a common mt-QSAR model for describing the antimicrobial (overall antibacterial and antifungal) activity of substituted benzylidene hydrazides against all the above mentioned microorganisms.
In order to develop mt-QSAR models, initially we have calculated the average antibacterial activity [pMICb = (pMICsa + pMICbs + pMICec)/3], antifungal activity [pMICf = (pMICca + pMICan)/2] and antimicrobial activity values [pMICam = (pMICsa + pMICbs + pMICec + pMICca + pMICan)/5] of substituted hydrazide derivatives which are presented in Table 2. These average activity values were also correlated with the molecular descriptors of synthesized compounds (Table 6).
The mt-QSAR model for antibacterial activity displayed the importance of both electronic energy (r = 0.925) and zero order molecular connectivity index, χ 0 (r = 0.923), in describing the antibacterial activity of substituted benzylidene hydrazides, as the value of correlation coefficients in developed models (Eqs. (6), (7)) is nearly same.
mt-QSAR model for antibacterial activity
| (6) |
| (7) |
The mt-QSAR model for antifungal activity revealed the importance of third order molecular connectivity index (3 χ) in describing antifungal activity.
mt-QSAR model for antifungal activity
| (8) |
The mt-QSAR model of antimicrobial activity (Eq. (9)) depicted the importance of second order molecular connectivity index (2 χ) in describing the antimicrobial activity of synthesized substituted benzylidene hydrazide derivatives.
mt-QSAR model for antimicrobial activity
| (9) |
| (10) |
Further, in case of mt-QSAR model for antimicrobial activity higher regression coefficient was obtained when we go for multiple linear regression analysis. In multiple linear regression (MLR) analysis, the coefficient of regression (r) increases to 0.719 from 0.609 (in linear regression), thus leading to an increase in predictability of mt-QSAR models, which can be observed by the low residual values (Table 9 ). It was observed from mt-QSAR models (Eqs. (6), (7), (8), (9), (10)) that the antibacterial, antifungal and overall antimicrobial activity of synthesized substituted benzylidene hydrazides is governed by second order molecular connectivity index (2 χ) and third order Kier's alpha shape index (κα 3).
Table 9.
Comparison of observed and predicted antimicrobial activity obtained by mt-QSAR models.
| Comp. | pMICb (Eq. (6)) |
pMICf (Eq. (8)) |
pMICam (Eq. (9)) |
pMICam MLR (Eq. (10)) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Obs. | Pre. | Res. | Obs. | Pre. | Res. | Obs. | Pre. | Res. | Obs. | Pre. | Res. | |
| 1 | 1.35 | 1.39 | −0.04 | 1.21 | 1.43 | −0.22 | 1.30 | 1.44 | −0.14 | 1.30 | 1.39 | −0.09 |
| 2 | 1.40 | 1.44 | −0.04 | 1.24 | 1.43 | −0.19 | 1.34 | 1.47 | −0.13 | 1.34 | 1.42 | −0.08 |
| 3 | 1.47 | 1.49 | −0.02 | 1.31 | 1.43 | −0.12 | 1.41 | 1.51 | −0.10 | 1.41 | 1.46 | −0.05 |
| 4 | 1.59 | 1.53 | 0.06 | 1.65 | 1.43 | 0.22 | 1.61 | 1.54 | 0.07 | 1.61 | 1.49 | 0.12 |
| 5 | 1.23 | 1.25 | −0.02 | 1.24 | 1.43 | −0.19 | 1.23 | 1.33 | −0.10 | 1.23 | 1.29 | −0.06 |
| 6 | 1.30 | 1.32 | −0.02 | 1.75 | 1.50 | 0.25 | 1.48 | 1.41 | 0.07 | 1.48 | 1.42 | 0.06 |
| 7 | 1.28 | 1.29 | −0.01 | 1.73 | 1.57 | 0.16 | 1.46 | 1.40 | 0.06 | 1.46 | 1.42 | 0.04 |
| 8 | 1.28 | 1.29 | −0.01 | 1.61 | 1.57 | 0.04 | 1.41 | 1.40 | 0.01 | 1.41 | 1.42 | −0.01 |
| 9 | 1.36 | 1.37 | −0.01 | 1.81 | 1.62 | 0.19 | 1.54 | 1.45 | 0.09 | 1.54 | 1.48 | 0.06 |
| 10 | 1.40 | 1.42 | −0.02 | 1.70 | 1.83 | −0.13 | 1.52 | 1.52 | 0.00 | 1.52 | 1.59 | −0.07 |
| 11 | 1.55 | 1.43 | 0.12 | 1.58 | 1.50 | 0.08 | 1.56 | 1.46 | 0.10 | 1.56 | 1.44 | 0.12 |
| 12 | 1.47 | 1.48 | −0.01 | 1.12 | 1.50 | −0.38 | 1.33 | 1.50 | −0.17 | 1.33 | 1.47 | −0.14 |
| 13 | 1.50 | 1.52 | −0.02 | 1.69 | 1.50 | 0.19 | 1.57 | 1.53 | 0.04 | 1.57 | 1.50 | 0.07 |
| 14 | 1.53 | 1.57 | −0.04 | 1.56 | 1.50 | 0.06 | 1.54 | 1.57 | −0.03 | 1.54 | 1.54 | 0.00 |
| 15 | 1.30 | 1.28 | 0.02 | 1.55 | 1.50 | 0.05 | 1.40 | 1.36 | 0.04 | 1.40 | 1.34 | 0.06 |
| 16 | 1.36 | 1.33 | 0.03 | 1.76 | 1.57 | 0.19 | 1.52 | 1.44 | 0.08 | 1.52 | 1.46 | 0.06 |
| 17 | 1.34 | 1.32 | 0.02 | 1.43 | 1.64 | −0.21 | 1.37 | 1.43 | −0.06 | 1.37 | 1.46 | −0.09 |
| 18 | 1.34 | 1.32 | 0.02 | 1.61 | 1.64 | −0.03 | 1.45 | 1.43 | 0.02 | 1.45 | 1.46 | −0.01 |
| 19 | 1.41 | 1.40 | 0.01 | 1.79 | 1.69 | 0.10 | 1.56 | 1.48 | 0.08 | 1.56 | 1.52 | 0.04 |
| 20 | 1.45 | 1.45 | 0.00 | 1.83 | 1.90 | −0.07 | 1.60 | 1.55 | 0.05 | 1.60 | 1.63 | −0.03 |
3.3. Structure activity relationship
From the results of antimicrobial activity, the following structure activity relationship can be drawn:
-
1.
The compounds have shown marked antifungal potential as compared to antibacterial activity, which shows that different structural requirements are essential for antibacterial and antifungal activity. These results are similar to Sortino et al. [48].
-
2.
The synthesized compounds derived from o-chlorobenzaldehyde have shown higher activity as compared to those derived from benzaldehyde. This observation reveals the fact that presence of chloro substituent at ortho position increases the antibacterial and antifungal potential of the compounds. This fact is supported by the observations of Guven et al. [49].
-
3.
It can also be seen from the results of antibacterial and antifungal activity that the activity increases with increase in chain length of acid portion of synthesized compounds (1–4, 11–14). This is similar to one of our previous reports [23].
-
4.
Compounds 6 and 16 derived from cinnamic acid have shown better activity as compared to other synthesized compounds. This may be due to the presence of extended conjugation in cinnamic acid portion and aromatic nucleus of aldehydes, which may be involved in binding of synthesized compound with the target site. This fact is supported by one of our earlier studies [24].
-
5.
The presence of electron withdrawing group (NO2) in compounds 10 and 20 makes them highly active antibacterial and antifungal agents. The role of electron withdrawing group in increasing the antibacterial and antifungal activity is similar to the results of P. Sharma et al. [50]. The SAR results are summarized in Fig. 1 .
Fig. 1.
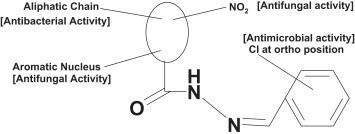
Structural requirements for the antimicrobial activity benzylidene/2-chlorobenzylidene hydrazides.
3.4. Antiviral activity
The antiviral screening of benzylidene/2-chlorobenzylidene hydrazides (1–20) was performed using an MTS-based CPE reduction assay [51] against Feline Corona virus (FIPV), Feline Herpes virus in CRFK cell culture; Herpes simplex virus-1 (KOS) [HSV-1 KOS], Herpes simplex virus-2 (G) [HSV-2G], Vaccinia virus [VV], Vesicular stomatitis virus [VSV], Herpes simplex virus-1 TK− KOS ACVr [HSV-1 TK− KOS ACVr] in HEL cell cultures; Vesicular stomatitis virus (VSV), Coxsackie virus B4 [CV-B4], Respiratory syncytial virus [RSV] in HeLa cell cultures; Influenza A virus H1N1 subtype, Influenza A virus H3N2 subtype and influenza B virus in MDCK cell cultures; Para-influenza-3 virus [PI-3V], Reovirus-1 [RV-1], Sindbis virus [SV], Coxsackie virus B4 [CV-B4], Punta Toro virus [PTV] in Vero cell cultures.
None of the synthesized benzylidene hydrazide derivatives was found to be active against the tested viral strains. The compounds 9 and 19 being exceptional have shown antiviral potential against Vesicular stomatitis virus (VSV) in HeLa cells (Table 10 ). No specific antiviral effects (MCC ≥ 5-fold higher than EC50) were noted for any of the compounds in HEL, CRFK, MDCK and VERO cell cultures. Compounds 5, 16 and 20 also have shown some antiviral potential but they have got high cytotoxic concentration values.
Table 10.
Cytotoxicity and antiviral activity of synthesized benzylidene hydrazides in HeLa cell cultures.
| Comp. | Cytotoxicity (μg/mL) |
Antiviral EC50c (μg/mL) |
||||||
|---|---|---|---|---|---|---|---|---|
| CC50a | Minimum cytotoxic conc.b | Vesicular stomatitis virus |
Coxsackie virus B4 |
Respiratory syncytial virus |
||||
| Visual CPE score | MTS | Visual CPE score | MTS | Visual CPE score | MTS | |||
| 9 | 61.2 | 100 | 9 | 6.5 | >20 | >20 | >20 | >20 |
| 19 | 64.0 | 100 | 12 | 13.2 | >20 | >20 | >20 | >20 |
| DS-5000 | >100 | >100 | >100 | >100 | >100 | >100 | 4 | 1.7 |
| (S)-DHPA (μM) | >250 | >250 | >250 | >250 | >250 | >250 | >250 | >250 |
| Ribavirin (μM) | >250 | >250 | 25 | 17.2 | 112 | 73.1 | 10 | 6.6 |
50% Cytotoxic concentration, as determined by measuring the cell viability with the colorimetric formazan-based MTS assay.
Minimum compound concentration that causes a microscopically detectable alteration of normal cell morphology.
50% Effective concentration, or concentration producing 50% inhibition of virus-induced cytopathic effect, as determined by visual scoring of the CPE, or by measuring the cell viability with the colorimetric formazan-based MTS assay.
4. Conclusion
A series of benzylidene hydrazides (1–20) were tested for their in vitro antimicrobial activity against S. aureus, B. subtilis, E. coli, C. albicans and A. niger. The results of antimicrobial activity indicated that compounds having chloro and nitro substituents were the most active ones. The results of MBC/MFC revealed that some of the compounds viz. 13, 14 and 11–13 were bactericidal against B. subtilis and E. coli respectively. The antiviral evaluation depicted that compounds 9 and 19 were active against Vesicular stomatitis virus (VSV) in HeLa cell cultures. To understand the relationship between physicochemical parameters and antibacterial and antifungal activity of substituted benzylidene hydrazide derivatives, QSAR investigation was performed by development of one-target and multi-target models. The multi-target model was found to be effective in describing the antimicrobial activity of substituted hydrazides in comparison to the one-target models. Further it indicated the importance of the topological parameters, second order molecular connectivity index (2 χ) and third order Kier's alpha shape index (κα 3) in describing the antimicrobial activity of substituted hydrazides.
5. Experimental
Melting points were determined in open capillary tubes on a sonar melting point apparatus and are uncorrected. Reaction progress was monitored by thin layer chromatography on silica gel sheets (Merck silica gel G) and purity of compounds was ascertained by single spot on TLC sheet. 1H nuclear magnetic resonance (1H NMR) spectra were recorded on Bruker Avance II 400 NMR spectrometer using appropriate deuterated solvents and are expressed in parts per million (δ, ppm) downfield from tetramethylsilane (internal standard). Infrared (IR) spectra were recorded on Perkin Elmer FTIR spectrometer using KBr pellets.
5.1. General procedure for synthesis of ester
The mixture of corresponding acid (0.08 mol) and ethanol (0.74 mol) was heated under reflux in the presence of sulphuric acid till the completion of reaction. Then, the reaction mixture was added to 200 mL ice cold water and the residual acid was removed by treatment with sodium bicarbonate. The ester formed was extracted with ether (50 mL). The ether layer was separated which on evaporation yielded the crude ester that was recrystallized from alcohol.
5.2. General procedure for synthesis of acid hydrazides
Hydrazine hydrate (99%) (0.015 mol) was added in ethanolic solution of ester (0.01 mol) and refluxed for 5 h. The reaction mixture was then cooled and the precipitates were filtered off, washed with water, dried and recrystallized from ethanol.
5.3. General procedure for synthesis hydrazones
The solution of acid hydrazide (0.01 mol) and appropriate aldehyde (0.01 mol) in ethanol was refluxed for 4–5 h. The precipitates obtained were filtered off, washed with water and recrystallized from ethanol.
5.3.1. Analytical data of compound 1
Mp (°C) 60–64; Yield – 67.80%; 1H NMR (400 MHz, CDCl3) δ ppm: 0.82–0.86 (t, 3H, CH3), 1.19–2.63 (m, 20H, CH2), 7.01–7.67 (m, 5H, ArH), 8.10 (s, 1H, NH). IR (KBr pellets) ν cm−1: 3069.79 (CH str., aromatic), 2923.03 (CH str., aliphatic), 1616.80 (C O str.), 1434.25 (C C str., aromatic).
5.3.2. Analytical data of compound 5
Mp (°C) Semisolid; Yield – 45.10%; 1H NMR (400 MHz, CDCl3) δ ppm: 1.32–1.38 (t, 3H, CH3), 5.72 (m, 1H, CH of CHCH3), 6.20 (t, 1H, terminal CH of CH CHCH3), 7.43–7.45 (m, 5H, ArH), 8.67 (s, 1H, NH). IR (KBr pellets) ν cm−1: 3211.03 (NH str., amide), 3028.7 (CH str., aromatic), 2974.8 (CH str., aromatic), 1725.9 (C O str.), 1658.5 (C C str., aliphatic).
5.3.3. Analytical data of compound 9
Mp (°C) 228–232; Yield – 74.00%; 1H NMR (400 MHz, CDCl3) δ ppm: 3.92 (s, 6H, OCH3), 6.86 (s, 1H, CH of ArOCH3), 7.27 (s, 2H, CH of C2 and C6 of ArOCH3), 7.46–7.69 (m, 5H, ArH), 8.67 (s, 1H, NH). IR (KBr pellets) ν cm−1: 3211.03 (NH str., amide), 3061.3 (CH str., aromatic), 2976.7 (CH str., aromatic), 1709.7 (C O str.), 1625.5 (C C str., aliphatic).
5.3.4. Analytical data of compound 13
Mp (°C) 98–102; Yield – 63.00%; 1H NMR (400 MHz, CDCl3) δ ppm: 0.87–0.89 (t, 3H, CH3), 1.21–1.28 (m, 28H, CH2), 7.24–7.43 (m, 3H, CH of C4, C5 and C6 of ArH), 7.65–7.67 (d, 1H, CH of C3 of ArH), 10.45–10.47 (s, 1H, NH). IR (KBr pellets) ν cm−1: 3299.72 (NH str., amide), 3039.73 (CH str., Aromatic), 2949.53 (CH str., aliphatic.), 1702.40 (C O str.), 793.73 (C–Cl str.).
5.3.5. Analytical data of compound 14
Mp (°C) 108–112; Yield – 22.30%; 1H NMR (400 MHz, CDCl3) δ ppm: 1.36–1.40 (s, 3H, CH3), 4.33–4.37 (s, 1H, N CH), 7.25–7.44 (m, 4H, ArH of Ar–CH3), 7.85–8.20 (m, 4H, ArH of Ar–Cl), 9.08 (s, 1H, NH). IR (KBr pellets) ν cm−1: 3299.72 (NH str., amide), 3038.91 (CH str., Aromatic), 2949.74 (CH str., aliphatic), 1718.40 (C O str.), 794.92 (C–Cl str.).
5.3.6. Analytical data of compound 16
Mp (°C) Semisolid; Yield – 80.20%; 1H NMR (400 MHz, CDCl3) δ ppm: 5.60–5.62 (d, 2H, CH of CH CH–C O), 6.35 (s, 1H, N CH), 7.21–7.30 (m, 5H, ArH of cinnamic acid), 7.32–7.46 (m, 4H, Ar–Cl), 9.28 (s, 1H, NH). IR (KBr pellets) ν cm−1 : 3229.10 (NH str., amide), 3062.6 (CH str., aromatic), 2976.8 (CH str., aliphatic), 1669.4 (C O str.), 755.8 (C–Cl str.).
5.4. Evaluation of antimicrobial activity
5.4.1. Determination of MIC
The antimicrobial activity was performed against Gram-positive bacteria: S. aureus, B. subtilis, Gram-negative bacterium: E. coli and fungal strains: C. albicans and A. niger. The standard and test samples were dissolved in DMSO to give a concentration of 100 μg/mL. The minimum inhibitory concentration (MIC) was determined by tube dilution method [27]. Dilutions of test and standard compounds were prepared in double strength nutrient broth – I.P. (bacteria) or Sabouraud dextrose broth I.P. [52] (fungi). The samples were incubated at 37 °C for 24 h (bacteria), at 25 °C for 7 days (A. niger) and at 37 °C for 48 h (C. albicans), respectively, and the results were recorded in terms of MIC (the lowest concentration of test substance which inhibited the growth of microorganisms).
5.4.2. Determination of MBC/MFC
The minimum bactericidal concentration (MBC) and fungicidal concentration (MFC) were determined by subculturing on fresh medium 100 μL of culture from each tube that remained clear in the MIC determination. MBC and MFC values represent the lowest concentration of compound that produces a 99.9% end point reduction [53].
5.5. QSAR studies
The details of molecular descriptors are available in literature and therefore they are not discussed over here [29], [30], [31], [32], [33], [34]. The structures of benzylidene hydrazide derivatives are first pre-optimized with the Molecular Mechanics Force Field (MM+) procedure included in Hyperchem 6.03 [54] and the resulting geometries are further refined by means of the semiempirical method PM3 (parametric Method-3). We chose a gradient norm limit of 0.01 kcal/Å for the geometry optimization. The lowest energy structure was used for each molecule to calculate physicochemical properties using TSAR 3.3 software for Windows [55]. Further, the regression analysis was performed using the SPSS software package [56]. The predictive powers of the equation were validated by determination of cross-validated r 2 (q 2) using leave one out (LOO) cross-validation method [57].
5.6. Evaluation of antiviral activity
5.6.1. Antiviral assay
The antiviral screening of benzylidene/2-chlorobenzylidene hydrazides (1–20) was performed against Feline Corona virus (FIPV), Feline Herpes virus in CRFK cell culture; Herpes simplex virus-1 (KOS) [HSV-1 KOS], Herpes simplex virus-2 (G) [HSV-2G], Vaccinia virus [VV], Vesicular stomatitis virus [VSV], Herpes simplex virus-1 TK− KOS ACVr [HSV-1 TK− KOS ACVr] in HEL cell cultures; Vesicular stomatitis virus (VSV), Coxsackie virus B4 [CV-B4], Respiratory syncytial virus [RSV] in HeLa cell cultures; Para-influenza-3 virus [PI-3V], Reovirus-1 [RV-1], Sindbis virus [SV], Coxsackie virus B4 [CV-B4], Punta Toro virus [PTV] in Vero cell cultures; Influenza A virus H1N1 subtype, Influenza A virus H3N2 subtype and influenza B virus in MDCK cell cultures and the results were expressed as the 50% effective concentration (EC50). Cells, grown to confluency in 96-well plates, were infected with 100 CCID50 of virus, one CCID50 being the 50% cell culture infective dose. After an adsorption period of 2 h at 37 °C, virus was removed and serial dilutions of the compounds were added. The cultures were further incubated at 37 °C for 3 days, until complete CPE was observed in the infected and untreated virus control.
5.6.2. Cytotoxic assay
The cytotoxicity of the compounds was evaluated in parallel with their antiviral activity in uninfected cells, and is expressed as minimum cytotoxic concentration to cause a microscopically detectable alteration of normal cell morphology (HEL cells, HeLa cells, CRFK cells, MDCK cells and Vero cells).
Acknowledgements
We would like to thank Mrs. Leentje Persoons, Rega Institute for Medical Research, Belgium for her excellent technical assistance in the evaluation of antiviral activity.
References
- 1.Yu K.G., Li D.H., Zhou C.H., Diao J.L. Chin. Chem. Lett. 2009;20:411–414. [Google Scholar]
- 2.Ezabadi I.R., Camoutsis C., Zoumpoulakis P., Geronikaki A., Sokovic M., Glamocilija J., Ciric A. Bioorg. Med. Chem. 2008;16:1150–1161. doi: 10.1016/j.bmc.2007.10.082. [DOI] [PubMed] [Google Scholar]
- 3.Murphy S.T., Case H.L., Ellsworth E., Hagen S., Huband M., Joannides T., Limberakis C., Marotti K.R., Ottolini A.M., Rauckhorst M., Starr J., Stier M., Taylor C., Zhu T., Blaser A., Denny W.A., Lu G.L., Smaillc J.B., Rivault F. Bioorg. Med. Chem. Lett. 2007;17:2150–2155. doi: 10.1016/j.bmcl.2007.01.090. [DOI] [PubMed] [Google Scholar]
- 4.Vittorio F., Ronsisvalle G., Marrazzo A., Blandini G. Farmaco. 1995;50:265–272. [PubMed] [Google Scholar]
- 5.Nayyar A., Monga V., Malde A., Coutinho E., Jain R. Bioorg. Med. Chem. 2007;15:626–640. doi: 10.1016/j.bmc.2006.10.064. [DOI] [PubMed] [Google Scholar]
- 6.Sztanke K., Pasterhak K., Rzymowska J., Sztanke M., Kandefer-Szerszen M. Eur. J. Med. Chem. 2007;43(2):404–419. doi: 10.1016/j.ejmech.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 7.Radhwan M.A.A., Ragab E.A., Sabry N.M., El-Shenawy S.M. Bioorg. Med. Chem. 2007;15:3832–3841. doi: 10.1016/j.bmc.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 8.Leite A.C.L., Lima R.S.D., Moreira D.R., Cardoso M.V., Brito A.C.G.D., Santos L.M.F.D., Hernandes M.Z., Kipustok A.C., Lima R.S.D., Soares M.B.P. Bioorg. Med. Chem. 2006;14:3749–3757. doi: 10.1016/j.bmc.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 9.Gemma S., Kukreja G., Fattorusso C., Persico M., Romano M.P., Altarelli M., Savini L., Campiani G., Fattorusso E., Basilico N., Taramelli D., Yardley V., Butini S. Bioorg. Med. Chem. Lett. 2006;16:5384–5388. doi: 10.1016/j.bmcl.2006.07.060. [DOI] [PubMed] [Google Scholar]
- 10.Al-Mawsawi L.Q., Dayam R., Taheri L., Witvrouw M., Debyser Z., Neamati N. Bioorg. Med. Chem. Lett. 2007;17:6472–6475. doi: 10.1016/j.bmcl.2007.09.102. [DOI] [PubMed] [Google Scholar]
- 11.Hansch C., Fujita T. J. Am. Chem. Soc. 1964;86:1616–1626. [Google Scholar]
- 12.Meerbach A., Klocking R., Meier C., Lomp A., Helbig B., Wutzler P. Antiviral Res. 2000;45:69–77. doi: 10.1016/s0166-3542(99)00076-5. [DOI] [PubMed] [Google Scholar]
- 13.Romanutti C., Castilla V., Cato C.E., Wachsman M.B. Int. J. Antimicrob. Agents. 2007;29:2667–2679. doi: 10.1016/j.ijantimicag.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Kim H.Y., Patkar C., Warrier R., Kuhn R., Cushman M. Bioorg. Med. Chem. Lett. 2005;15:3207–3211. doi: 10.1016/j.bmcl.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 15.Andries K., Moermans M., Grevers T., Willebroads R., Sommen C., Lacrampe J., Janssens F., Wyde P.R. Antiviral Res. 2003;60:209–219. doi: 10.1016/j.antiviral.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Makarov V.A., Riabova O.B., Granik V.G., Dahse H.M., Stelzner A., Wutzler P., Schmidtke M. Bioorg. Med. Chem. Lett. 2005;15:37–39. doi: 10.1016/j.bmcl.2004.10.043. [DOI] [PubMed] [Google Scholar]
- 17.Kumar P., Narasimhan B., Sharma D. ARKIVOC. 2008;xiii:159–178. [Google Scholar]
- 18.Kumar P., Narasimhan B., Sharma D., Judge V., Narang R. Eur. J. Med. Chem. 2009;44:1853–1863. doi: 10.1016/j.ejmech.2008.10.034. [DOI] [PubMed] [Google Scholar]
- 19.Narasimhan B., Kothawade U.R., Pharande D.S., Mourya V.K., Dhake A.S. Indian J. Chem. 2003;42(B):2828–2834. [Google Scholar]
- 20.Narasimhan B., Belsare D., Pharande P., Mourya V.K., Dhake A.S. Eur. J. Med. Chem. 2004;39(10):827–834. doi: 10.1016/j.ejmech.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 21.Narasimhan B., Mourya V.K., Dhake A.S. Bioorg. Med. Chem. Lett. 2006;16:3023–3029. doi: 10.1016/j.bmcl.2006.02.056. [DOI] [PubMed] [Google Scholar]
- 22.Narasimhan B., Mourya V.K., Dhake A.S. Pharm. Chem. J. 2007;3:120–125. [Google Scholar]
- 23.Narasimhan B., Narang R., Judge V., Ohlan S., Ohlan R. ARKIVOC. 2007;xv:112–126. [Google Scholar]
- 24.Narasimhan B., Judge V., Narang R., Ohlan S., Ohlan R. Bioorg. Med. Chem. Lett. 2007;17:5836–5845. doi: 10.1016/j.bmcl.2007.08.037. [DOI] [PubMed] [Google Scholar]
- 25.Kumar A., Narasimhan B., Kumar D. Bioorg. Med. Chem. 2007;15:4113–4124. doi: 10.1016/j.bmc.2007.03.074. [DOI] [PubMed] [Google Scholar]
- 26.Narasimhan B., Dhake A.S., Mourya V.K. ARKIVOC. 2007;i:189–204. [Google Scholar]
- 27.Cappucino J.G., Sherman N. Addison Wesley Longman Inc; California: 1999. Microbiology – A Laboratory Manual. 263. [Google Scholar]
- 28.Emami S., Falhati M., Banifafemi A., Shafiee A. Bioorg. Med. Chem. 2004;12:5881–5889. doi: 10.1016/j.bmc.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 29.Hansch C., Leo A., Unger S.H., Kim K.H., Nikaitani D., Lien E.J. J. Med. Chem. 1973;16(11):1207–1216. doi: 10.1021/jm00269a003. [DOI] [PubMed] [Google Scholar]
- 30.Kier L.B., Hall L.H. Academic Press; New York: 1976. Molecular Connectivity in Chemistry and Drug Research. [Google Scholar]
- 31.Randic M. J. Am. Chem. Soc. 1975;97:6609–6615. [Google Scholar]
- 32.Balaban A.T. Chem. Phys. Lett. 1982;89:399–404. [Google Scholar]
- 33.Wiener H. J. Am. Chem. Soc. 1947;69:17–20. doi: 10.1021/ja01193a005. [DOI] [PubMed] [Google Scholar]
- 34.Randic M. Croat. Chem. Acta. 1993;66:289–312. [Google Scholar]
- 35.Dai J., Sun C., Han S., Wang L. Bull. Environ. Contam. Toxicol. 1999;62:530–538. doi: 10.1007/s001289900908. [DOI] [PubMed] [Google Scholar]
- 36.Lather V., Madan A.K. Bioorg. Med. Chem. 2005;13:1599–1604. doi: 10.1016/j.bmc.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 37.Golbraikh A., Tropsha A. J. Mol. Graph. Model. 2002;20:269–276. doi: 10.1016/s1093-3263(01)00123-1. [DOI] [PubMed] [Google Scholar]
- 38.Narasimhan B., Kumari M., Jain N., Dhake A.S., Sundaravelan C. Bioorg. Med. Chem. Lett. 2006;16:4951–4958. doi: 10.1016/j.bmcl.2006.06.046. [DOI] [PubMed] [Google Scholar]
- 39.Bajaj S., Sambi S.S., Madan A.K. Croat. Chem. Acta. 2005;78(2):165–174. [Google Scholar]
- 40.Sharma P., Kumar A., Sharma M. Eur. J. Med. Chem. 2006;41:833–840. doi: 10.1016/j.ejmech.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 41.Hatya S.A., Aki-sener E., Tekiner-Gulbas B., Yildiz I., Temiz-Arpaci O., Yalcin I., Altanlar N. Eur. J. Med. Chem. 2006;41:1398–1404. doi: 10.1016/j.ejmech.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 42.Kumar A., Sharma P., Gurram V.K., Rane N. Bioorg. Med. Chem. Lett. 2006;16:2484–2491. doi: 10.1016/j.bmcl.2006.01.080. [DOI] [PubMed] [Google Scholar]
- 43.Prado-Prado F.J., Gonzalez-Diaz H., Vega O.M.D.L., Ubeira F.M., Chou K.C. Bioorg. Med. Chem. 2008;16(11):5871–5880. doi: 10.1016/j.bmc.2008.04.068. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalez-Diaz H., Prado-Prado F.J. J. Comput. Chem. 2008;29(4):656–667. doi: 10.1002/jcc.20826. [DOI] [PubMed] [Google Scholar]
- 45.Cruz-Monteagudo M., Gonzalez-Diaz H., Aguero-Chapin G., Santana L., Borges F., Dominguez E.R., Podda G., Uriarte E. J. Comput. Chem. 2007;28(11):1909–1923. doi: 10.1002/jcc.20730. [DOI] [PubMed] [Google Scholar]
- 46.Gonzalez-Diaz H., Vilar S., Santana L., Uriarte E. Curr. Top. Med. Chem. 2007;7(10):1015–1029. doi: 10.2174/156802607780906771. [DOI] [PubMed] [Google Scholar]
- 47.Gonzalez-Diaz H., Gonzalez-Diaz Y., Santana L., Ubeira F.M., Uriarte E. Proteomics. 2008;8(4):750–778. doi: 10.1002/pmic.200700638. [DOI] [PubMed] [Google Scholar]
- 48.Sortino M., Delgado P., Jaurez S., Quiroga J., Abonia R., Insuasey B., Rodero M.N., Garibotto F.M., Enriz R.D., Zaccino S.A. Bioorg. Med. Chem. Lett. 2007;15:484–494. doi: 10.1016/j.bmc.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 49.Guven O.O., Erdogan T., Goker H., Yildiz S. Bioorg. Med. Chem. 2007;17:2233–2236. doi: 10.1016/j.bmcl.2007.01.061. [DOI] [PubMed] [Google Scholar]
- 50.Sharma P., Rane N., Gurram V.K. Bioorg. Med. Chem. Lett. 2004;14:4185–4190. doi: 10.1016/j.bmcl.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 51.De Palma A.M., Heggermont W., Leyssen P., Purstinger G., Wimmer E., De Clercq E., Rao A., Monforte A.M., Chimirri A., Neyts J. Biochem. Biophys. Res. Commun. 2007;353:628–632. doi: 10.1016/j.bbrc.2006.12.063. [DOI] [PubMed] [Google Scholar]
- 52.vol. I. Controller of Publications, Ministry of Health Department, Govt. of India; New Delhi: 2007. (Pharmacopoeia of India). p. 37. [Google Scholar]
- 53.Rodriguez-Arguelles M.C., Lopez-Silva E.C., Sanmartin J., Pelagatti P., Zani F. J. Inorg. Biochem. 2005;99:2231–2239. doi: 10.1016/j.jinorgbio.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 54.Hypercube, Inc.; Florida: 1993. Hyperchem 6.0. [Google Scholar]
- 55.Oxford Molecular Limited; 2000. TSAR 3D Version 3.3. [Google Scholar]
- 56.SPSS Inc.; Bangalore, India: 1999. SPSS for Windows, Version 10.05. [Google Scholar]
- 57.Schaper K.J. Quant. Struct. Act. Relat. 1999;18:354–360. [Google Scholar]


