Abstract
The innate immune response is a vital part of the body's antiviral defense system. The innate immune response is initiated by various receptor interactions, including danger associated molecular patterns (DAMPs). The S100A9 is a member of the DAMPs protein family and, is released by activated phagocytic cells such as neutrophils, monocytes, macrophages or endothelial cells, and S100A9 induces its effect through TLR4/MyD88 pathway.
Bovine viral diarrhea virus (BVDV) is one of the major devastating disease in the cattle industry worldwide. It shows its effect through immunosuppression and develops persistent infection in calves born from infected cows. The current study revealed that BVDV potentially induced immunosuppression by the interaction of BVDV Npro protein with cellular S100A9 protein. The Inhibition of S100A9 protein expression by small interfering RNA (siRNA) enhanced the virus replication in infected cells. Overexpression of bovine S100A9 enhanced the ncpBVDV2a 1373 mediated Type-I interferon production. A co-immunoprecipitation experiment demonstrated a strong interaction between ncp BVDV2a 1373 Npro protein and cellular S100A9 protein. This suggested that BVDV Npro reduced the S100A9 protein availability/activity in infected cells, resulting in reduced Type-I interferon production. A further study of S100A9-BVDV interaction will be need for better understanding of BVDV pathophysiology.
Keywords: S100A9 protein, BVDV Npro, Immunosuppression, Protein-protein interaction, Co-immunoprecipitation
Highlights
-
•
The bovine viral diarrhea virus (BVDV) nonstructural protein, Npro, is responsible for immunosuppression.
-
•
The mechanism of Npro immune immunosuppression is not well characterized.
-
•
S100A9, a cell protein that contains danger associated molecular patterns (DAMP), is important in innate immunity.
-
•
S100A9 protein and Npro protein associate while overexpression of S100A9 enhanced Type-I interferon production.
-
•
Inhibition of S100A9 by siRNA aided BVDV replication. Npro interacting with S100A9 may result in immunosuppression.
1. Introduction
The innate immune system is a key component of antiviral defense [1,2]. Danger associated molecular patterns (DAMPs), or alarmins [[3], [4], [5]] plays an important role in innate immunity. DAMPs are intracellular molecules that primarily regulate cell homeostasis, but also act as extracellularly when secreted by activated or damaged cells. The DAMPs interact with Pattern recognition receptors (PRRs) and initiate the production of cytokines and chemokines, which further amplify the inflammation [[6], [7], [8], [9]]. However, there is very little known about the role of DAMPs in inflammation or immunity during virus infection. A one protein family of DAMPs, known as S100 (i.e., S100A7, S100A15 [6], S100A12, S100A8, and S100A9) [[7], [8], [9]] have shown a direct effect on the innate immune system. The protein S100A9 initiates the immune response through the TLR4/MyD88 pathway [10]. S100A9 is expressed by a variety of cell types including neutrophils, dendritic cells, monocytes and endothelial cells [[10], [11], [12], [13]]. Activation of TLR4/MyD88 pathway by S100A9 triggers the downstream signaling cascade, leading to the activation of nuclear factor-кB (NF-кB), and subsequently activates type 1 interferon (IFN-α/β) [7,9].
BVDV belongs to the Pestivirus genus of the Flaviviridae family. It an immunosuppressive virus and one of the major threats to the cattle industry worldwide [[14], [15], [16], [17], [18]]. BVDV has a positive sense single stranded RNA genome with the size of 12.3 Kb [19]. The genome is translated into a single open reading frame (ORF) that encodes approximately 4000 codons comprising 10 to 11 posttranslational cleaved proteins [17,19]. The polyprotein is cleaved into 4 structural proteins [e.g. Erns, envelope proteins E1, E2, and capsid protein (C)] and 7 to 8 nonstructural proteins (e.g. Npro, p7, NS2-3 or NS2 and NS3, NS4A NS4B, NS5A and NS5B) by viral or/and host cell proteases [17,19,20]. The Flaviviridae family also includes Flavivirus and Hepacivirus genus. The Flavivirus genus has viruses with zoonotic importance, such as West Nile virus and dengue virus while Hepacivirus genus comprised solely of the hepatitis C virus (HCV) [21,22]. Of the three Flaviviridae genuses, the Pestivirus genome is unique because it encodes two proteins (e.g. Npro and Erns) that are not present in the other genus in Flaviviridae family [23]. Npro is the first protein coded in the ORF, which has protease activity, but is not necessary for viral replication [24,25]. Npro interferes with the innate immune mechanisms through inhibiting type one interferon (e.g. interferon alpha and beta: FN-α/β) in cell culture [23,26,27]. This inhibition is potentially associated with the immunotolerance and development of persistent infection (PI) in BVDV infection [25].
Studies with Npro deleted classical swine fever virus demonstrated that Npro is not required for viral replication in cell culture [25] while deletion facilitated the induction of type one IFN [26,27]. Vaccination with Npro deleted virus induced a strong antibody response with significantly increased protection against highly virulent CSFV [25,28]. Similarly, a BVDV virus with mutations in both the Npro protease coding sequence as well as the Erns RNase region, failed to induce PI in calves [29]. The Npro potentially reduces type I IFN by decreasing interferon regulatory factor-3 (IRF-3). The phosphorylation of IRF-3 resulted in its conformational change with increased binding ability to DNA. Phosphorylated IRF-3 binding to DNA up-regulated the IRF-3 transcriptional as well as interferon (IFN)-responsive genes to produce type I IFN [30,31]. While, BVDV Npro decreased the IRF-3 level via proteasomal degradation, leading to decreased expression of type I IFN [32,33].
The current study showed a strong association of BVDV Npro with cellular S100A9 protein. Inhibition of S100A9 protein expression by small interfering RNA (siRNA) enhanced the virus replication in infected cells. Overexpression of bovine S100A9 enhanced the ncpBVDV2a 1373 mediated Type-I interferon production. These results indicated that BVDV Npro reduced the S100A9 protein availability/activity in infected cells resulting in reduced Type-I interferon production.
2. Materials and methods
2.1. Cell cultures, virus and plasmids
BVDV-free Mardin Darby bovine kidney (MDBK) cells or human embryonic kidney 293T cells (American Tissue Culture Collection, Manassas, VA) [34] were grown and maintained in minimal essential medium (MEM, Gibco BRL, Grand Island, NY) (pH 7.0–7.4) supplemented with 10% BVDV free fetal bovine serum (FBS) (PPA, Pasching, Austria), penicillin (100 U/ml) and streptomycin solution (100 μg/ml) (Sigma-Aldrich, St. Louis MO). Bovine aortic endothelial cells (BAEC) (passage 3) were kindly provided by Dr. John Neill, National Animal Disease Center, United States Department of Agriculture, Agricultural Research Service, Ames, IA, USA and were grown in Bovine Endothelial Cell Growth Medium (Cell Applications, Inc., San Diego, CA) supplemented with 10% BVDV free fetal bovine serum (FBS) (PPA, Pasching, Austria), penicillin (100 U/ml) and streptomycin solution (100 μg/ml) (Sigma-Aldrich, St. Louis, MO).
The non-cytopathic (ncp) BVDV2a-1373 was propagated and titrated in MDBK cells. Titrated virus was aliquoted and stored at −80 °C until use. The p3XFLAG-CMV-10 (Sigma-Aldrich, St. Louis, MO) or pEGFP C1 (Clontech Laboratories Inc., Mountain View, CA) plasmid was transformed and amplified in DH5α strain of Escherichia coli (E coli) (Life Technologies, Grand Island NY). After amplification, plasmids were extracted through QIAGEN Plasmid Mini Kit (QIAGEN, Germantown, MD) and stored at −80 °C until use.
2.2. p3XFLAG-Npro Construction
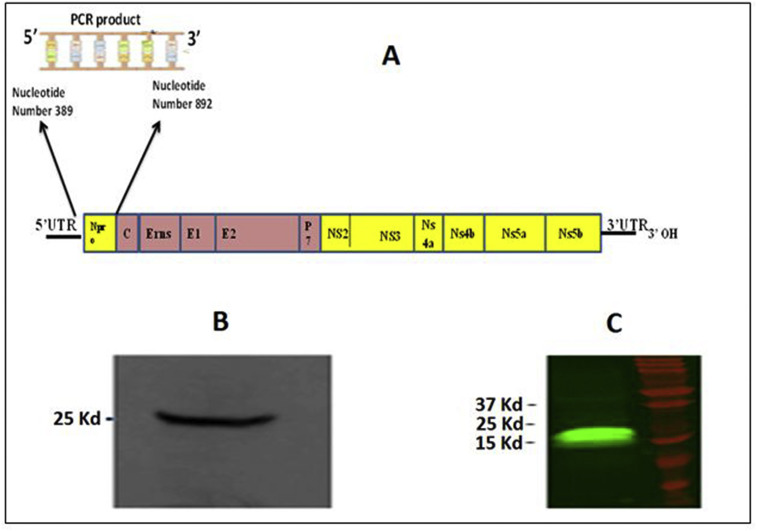
We chose to clone the Npro gene from ncp BVDV2a-1373 as BVDV suppresses type 1 IFN production in vitro [35] and the ncp BVDV2a-1373 is one of the high pathogenic BVDV strains [36]. To construct the Npro plasmid, total RNA extracted from ncp BVDV2a-1373 using QIAamp viral RNA mini-kit (QIAGEN, Germantown MD) was used as a template. The obtained RNA was used as a template to create Npro complementary DNA (cDNA). The BVDV Npro cDNA was made through reverse transcription–polymerase chain reaction (RT-PCR) using the OneStep RT-PCR kit (QIAGEN, Germantown MD). Npro-forward and -reverse primers such as 5′CGGAATTCTGCTGGAGTGAATACTGAAGAG ATAGT 3′ and 5′CGGGATCCATTAATCGTGGCGAAAATAATTACAA 3′ were designed respectively to amplify the segment encoding Npro protein from nucleotide 389 to 892, according to the sequence of ncp BVDV2a-1373 from the GenBank (Accession No., AF145967) (Fig. 1 A). Amplification of cDNA was performed using a temperature profile of 50 °C for 30 min, 95 °C for 15 min, followed by 30 cycles of (95 °C for 10 s, 52 °C for 30 s and 72 °C for 30 s) respectively. Upon completion of the 30 cycles, the resultant was prolonged for 7 min at 72 °C. The PCR product was analyzed on 1% agarose gel. The amplified DNA as well as p3XFLAG-CMV- 10 (6.4 Kb) plasmid were double digested by EcoRI, and BamH1 (Sigma, St. Louis, MO) for 2 h. The Npro cDNA was then ligated into the p3XFLAG-CMV- 10 vector using T4 ligase (Sigma, St. Louis, MO), to get p3XFLAG –Npro vector. The p3XFLAG –Npro was then transformed into E. coli- DH5α competent cells (Life Technologies, Grand Island, NY) for amplification. After amplification, the insertion of Npro to p3XFLAG-CMV in p3XFLAG –Npro vector was confirmed through restriction enzyme reaction digestion as well as by DNA sequencing. The restriction enzyme digestion of p3XFLAG –Npro vector with EcoRI, and BamH1 (Sigma, St. Louis, MO) gave 503 base pairs (bp) Npro and 6.4 Kb p3XFLAG-CMV vector portion.
Fig. 1.
p3XFLAG-Npro and p3XFLAG-S100A9 construction and knock down of S100A9 protein through siRNA. The BVDV Npro cDNA was made through RT-PCR targeting Npro protein from nucleotide 389 to 892 (A). The p3XFLAG-Npro or p3XFLAG-S100A9 vector were constructed and transfected in to 293T cells. After 24 h post transfection, cells were harvested and conformed for Npro-FLAG (B) or S100A9-FLAG (C) expression by western blot.
2.3. pEGFP C1-Npro Vector construction
The pEGFP-C1 plasmid was kindly provided by Dr. Adam Hoppe, Department of Chemistry and Biochemistry, South Dakota State University, Brookings, SD. The pEGFP-C1 plasmid was utilized to construct pEGFP C1-Npro Vector by inserting Npro gene at EcoRI and Bam HI restriction site as described above in p3XFLAG-Npro Construction. The insertion of Npro gene was then confirmed by restriction digestion using EcoRI and Bam HI enzymes, which yield 503 bp Npro and 4.7 Kb pEGFP vector as well as by nucleotide sequencing of Npro.
2.4. p3XFLAG-S100A9 vector construction
The S100A9 gene was cloned into the p3XFLAG-CMV-10 plasmid for co-precipitation experiments. The total RNA was extracted from MDBK cells using TRIzol reagent (Invitrogen, Grand Island NY). The total RNA extracted from MDBK cells was used as template to generate to synthesize S100A9 cDNA using One step RT PCR kit (QIAGEN). The S100A9 forward:5′ GAAGATCTGATGACTTGCAAAATGTCGCAGCT3´; and S100A9 reverse: 5′CGGGATCCTTAGGGGGTGCCCTCCCC3 primers were designed to clone S100A9 gene from 108th nucleotide to 400th as described by the bovine S100A9 cDNA sequence published in GenBank (Accession Number: XM_005203730). The forward and reverse primers contained BglΙΙ and BamH1 restriction sites, respectively. Amplification of S100A9 cDNA was performed using a temperature profile of 50 °C for 30 min, 95 °C for 15 min, followed by 30 cycles of (95 °C for 10 s, 52 °C for 30 s and 72 °C for 30 s) respectively. Upon completion of the 30 cycles, the reaction was prolonged for 7 min at 72 °C. The obtained S100A9 cDNA was then inserted into p3XFLAG vector using BglΙΙ and Bam H1 restriction sites to get p3XFLAG- S100A9 plasmid. The insertion of S100A9 was then confirmed using BglΙΙ and Bam H1 restriction digestion as well as nucleotide sequencing. The restriction digestion yielded 292 bp S100A9 and 6.4 Kb p3XFLAG-CMV.
2.5. Transfections
The 293T cells have a higher transfection efficiency as compared to MDBK cells, therefore 293T cells were used to express the BVDV or bovine gene in those cells as per the practice used earlier [37,38]. 293T cells were transfected with p3XFLAG-S100A9, pEGFP C1-Npro or p3XFLAG-Npro plasmids carrying bovine S100A9 or BVDV Npro gene respectively. The 293T cells were transfected using X-tremeGENE HP DNA Transfection Reagent (Roche Applied Science, Indianapolis, IN), according to the manufacturer's protocol. Briefly, 293T cells were plated for 18 h before transfection at a concentration of 7.5 × 105 cells/well in six-well culture plates and were 85–95% confluent at the time of transfection. During transfection, the growth media was replaced with 500 μl of serum-free Opti-MEM (Life Technologies). The plasmid DNA was diluted in Opti-MEM to get a final concentration of 1 μg plasmid DNA/100 μl medium (0.01 μg/μl). Then 4 μl of the transfection reagent was added to the DNA- Opti-MEM mixture and mixed gently. The obtained transfection reagent-DNA- Opti-MEM mixture was added to 293T cells drop by drop. Then the cells were incubated at 37 °C for 6 h. After 6 h, an additional 2.4 ml of pre-warmed growth media [MEM, supplemented with 10% FBS, penicillin (100 U/ml) and streptomycin solution (100 μg/ml)] was added to each well and the plates were re-incubated at 37 °C with 5% CO2 for 24 h. The cells were then harvested for western blot analysis to confirm the protein expressions (Fig. 1 B and C) as per the method described below in section 2.6.
2.6. Western blot analysis
The protein expression of Npro-FLAG, Npro-GFP and, S100-FLAG in 293T transfected cells were confirmed by Western blot analysis. A total 5 × 106 transfected cells were cultured in 6-well plates for 48 h. The cells were washed once with PBS, then lysed with 0,5 ml ice-cold radioimmunoprecipitation assay (RIPA) buffer/well containing proteinase inhibitor (Sigma, St. Louis, MO) for 30 min. The cell lysate was transferred to 1.5 ml tube and centrifuged at 500 × g for 20 min. Supernatants were collected and total protein concentration was estimated using Lowry method [39]. A total 30 μg protein was loaded in each well of 12.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) for separation followed by blotting onto a nitrocellulose membrane (Invitrogen, Grand Island, NY). Membrane was blocked overnight at 4 °C with 3% nonfat milk in Tris Buffered Saline (TBS) (Sigma, St. Louis, MO). The membrane was then incubated overnight at 4 °C with either mouse anti-FLAG monoclonal antibody (Sigma-Aldrich) or rabbit anti-GFP polyclonal antibody (Abcam, Cambridge, MA) with 1:5000 dilution in 3% nonfat milk -TBS (pH of 8.0). The membrane was then washed three times with TBS and incubated with secondary goat anti-mouse antibody (IRDye® 800CW Goat, LI-COR, Lincoln NE) or goat anti-rabbit antibody (IRDye® 680RD, LI-COR) at a dilution of 1:5000 in 3% nonfat milk in TBS for 1 h at room temperature. Finally, the blot was analyzed using Odyssey fluorescent imaging instrument (LI-COR Biosciences, Lincoln, NE, USA) (Fig. 1B and C).
2.7. Mass spectrometry
2.7.1. Mass spectrometry analysis: cellular protein immunoprecipitation using anti-FLAG IgG1 mAb
Immunoprecipitation procedures were performed as previously described by Free et al. [40]. Briefly, 293T cells were plated in six, 6-well plates for 24 h before transfection. The plates were then divided into two groups with three plates each for three replicates. One group was transfected with p3XFLAG-Npro, while the other group was transfected with empty p3XFLAG, as described above. Twenty-four hr after transfection, cells were harvested and lysed using in 1 ml of ice-cold in-house solubilization/lysis buffer (50 mM [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid: HEPES], 1 mM EDTA, 10% glycerol, 1% Triton X-100, 150 mM NaCl, 50 mM NaF, and 40 mM Na4P2O7) with pH of 7.4 containing protease inhibitor cocktail (Sigma, St. Louis, MO), for 1 h at 4 °C. The cell lysates were then centrifuged at 20,000 × g at 4 °C for 5 min. The supernatant was collected and pre-cleared using protein G magnetic beads (Dynabeads® Protein G., Life Technologies) by incubation for 10 min at room temperature. The protein G magnetic beads were removed by centrifugation for 5 min at 1000–2500 × g and the supernatant was collected. The supernatants were then transferred into clean tubes with 100 μl of murine anti-FLAG IgG1 mAb-agarose slurry-beads (Sigma-Aldrich, Louis, MO). The mixture was incubated on a rocking platform overnight at 4 °C. The next day, the beads were washed three times with 1 ml ice-cold solubilization/lysis buffer. Finally, the proteins were eluted from the samples and the control beads by adding 5 packed gel volumes of 0.1 M glycine HCl buffer, pH 3.0. (Life Technologies, CA). The collected proteins were stored at −20 °C for further use.
2.7.2. Mass spectrometry analysis: protein precipitation after immunoprecipitation
A total of 340 μl of 100% trichloro acetic acid (TCA) (Sigma, St. Louis, MO) was added for every 1 ml of eluate for protein precipitation in on ice for 3 h. The eluate-TCA mixture was then centrifuged at 11,000 × g at 4 °C for 30 min to obtain protein pellet. The obtained protein pellets were dissolved in one ml of cold 98% acetone-0.05 N HCl solution and the tube was centrifuged again for 5 min at 16000 × g at 4 °C. The supernatant was removed carefully and the pellet was washed with one ml cold of acetone by centrifuging at 16000 × g for 5 min at 4 °C. The pellets were stored dry at −80 °C until mass spectroscopy analysis.
2.7.3. Mass spectrometry analysis and bioinformatics
The mass spectroscopic analysis was performed by Dr. Eduardo Callegari at the University of South Dakota Mass Spectrometry and Proteomics Core Facility, Vermillion, SD using nano ESI-Q-Tof micro mass spectrometer (Micromass, Manchester, UK) while data was analyzed with MassLynx software 4.1 (Micromass).
2.8. Co-immunoprecipitation
To confirm the interaction between the Npro and bovine S100A9, a co-immunoprecipitation experiment was performed. The 293T cells were seeded in six, 6-well plates with the concentration of 7.5 × 105 cells/well for 18 h before transfection. Plates were then divided into two groups with three plates each for three replicates. One group was co-transfected with (p3XFLAG empty plasmid and pEGFP-C1-Npro) together, while the other group was co-transfected with (p3XFLAG -S100A9 and pEGFP-C1-Npro) together. The plasmids were co-transfected as described above using 1 μg of each plasmid/well of transfection reaction. The plates were incubated for 24 h at 37 °C with 5% CO2 and the cells were lysed by 150 μl of lysis buffer (20 mM Tris HCl pH 8.0, 137 mM NaCl, 1% Triton X-100, and 2 mM EDTA) to perform the immunoprecipitation.
To confirm the interaction between Npro and S100A9, the protein was immunoprecipitated (isolated) using anti-FLAG beads in both the groups and analyzed for presence of FLAG as well as GFP (e.g. GFP-Nrop) through western blot. The western blot was performed using a mixture of both mouse anti-FLAG monoclonal antibody (Sigma, St. Louis, MO) or rabbit anti-GFP polyclonal antibody (Abcam, Cambridge MA) followed by goat anti-mouse antibody (IRDye® 800CW Goat, LI-COR, Lincoln NE) or goat anti-rabbit antibody (IRDye® 680RD, LI-COR) and finally blot was analyzed using Odyssey fluorescent imaging instrument (LI-COR Biosciences, Lincoln, NE, USA).
2.9. S100A9 knockdown and study of virus growth
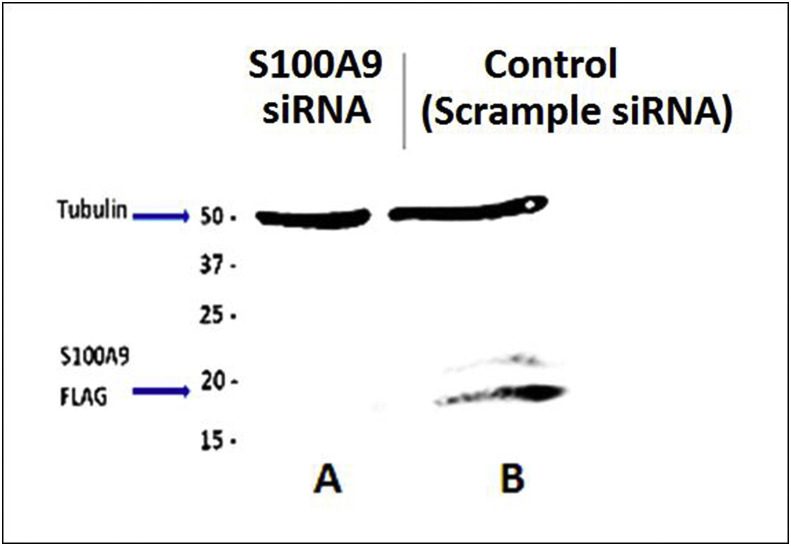
To determine if S100A9 has any effect on BVDV or IFNα/β production, the MDBK cells or bovine endothelial cells were knock out for S100A9 gene using anti-S100A9 Small interfering RNA (siRNA) (DNA Technology, Iowa City, IA). Briefly, cells were cultured for 3 days to obtain 80–90% confluency. The cells were harvested by trypsinization and then were washed with culture media. The cells were centrifuged and adjusted to final concentration as 1 × 106 cells/100 μl of Nucleofector® solution (LONZA, Basel, Switzerland) per sample. Thirty pmol of either S100A9 siRNA or control scrambled siRNA (siRNA of the reverse sequences of the bovine S100A9 siRNA) were added to the 100 μl cell suspension. The siRNA-cell suspension was then introduced into an Amaxa (LONZA) certified cuvette and the Nucleofector® Program X-001 was performed using the Nucleofactor® 2 device, (LONZA) according to manufacturer's instructions. Upon completion of the transfection, the cuvette was removed from the device and the samples were further incubated for 10 min at room temperature. Following incubation, cells were transferred to 6-well plates and media [MEM media containing 10% BVDV, penicillin (100 U/ml) and streptomycin solution (100 μg/ml)] volume was brought to 1.5 ml/well. The cells were cultured for 24 h and determined for S100A9 siRNA-mediated knockdown through western blot (Fig. 2 ).
Fig. 2.
The knockout of S100A9 gene in MDBK cells or bovine endothelial cells. The S100A9 gene was knocked down using anti-S100A9 siRNA while scrambled siRNA (reverse sequences of the bovine S100A9) was used as negative control. The knock down of S100A9 gene was confirmed by western blot. The S100A9 knocked down cells did not show and protein band for S1009A on western blot analysis (A), while it was present in the Scramble control cell (B).
The S1009 knock out (transfected with S100A9 siRNA) or control MDBK cells (transfected with scrambled siRNA) were passaged two times and then infected with one MOI of ncp BVDV2a-1373. The supernatant from infected cells were collected at 12 h, 24 h, 36 h and 60 h post infection and stored at −80 °C until the viral titration. The virus titration was determined by serially dilutions of viral sample in MEM containing MDBK cells as described previously [41]. The end point for ncp BVDV2a-1373 was determined by staining the MDBK cells with anti-Erns BVDV mouse mAb (15C5, IDEXX Laboratories, Westbrook, ME, USA), followed by biotinylated rabbit anti-mouse IgG, (Zymed, Invitrogen Corporation, Frederick, MD, USA), Streptavidin-HRP, (Invitrogen Corporation, Camarillo, CA, USA) and AEC reagent (3 amino-9 ethyl-carbazole, Sigma, St. Louis, MO, USA). The highest dilution with staining was used as an end point to calculate the TCID50 according to method, as previously described [41]. The experiment to measure the BVDV production in S1009 knock out cells were repeated three times in three different sets of experiments for its repeatability and the data was analyzed using a paired t-test at 95% level of significance.
2.10. IFN expression
2.10.1. IFN-α/β assay
The IFNα/β production in BVDV-infected bovine endothelial cells was determined as per the method described by Rubinstein et al. with some modifications [42]. The p3XFLAG S100A9 transfected or S100A9 knock out bovine endothelial cells were infected with ncp BVDV2a-1373 at MOI of 5. The bovine endothelial cells treated with poly I:C at a concentration 1 μg/ml were used as a positive control for the assay while mock infected cells were used as negative control. After 24 h post infection, cell supernatant was collected and stored at 4 °C for IFN assay after BVDV inactivation. Each experiment was done in three replicates and reproduced three times. The data was analyzed using a paired t-test at 95% level of significance.
2.10.2. IFN reporter gene assay
The IFN - α/β activity was measured by using the NCL-1–ISRE–Luc–Hygro cells, (kindly provided by Dr. Clayton Kelling, University of Nebraska, Lincoln, NE). Briefly, 1.5 × 105 cells were seeded into a 12-well plate and incubated at 37 °C with 5% CO2 for 12 h. After 12 h incubation, the medium was replaced with 0.5 ml of the test supernatants and reincubated for another 8 h. The cells were harvested and lysed with 100 μl passive lysis buffer (Promega, WI), and the firefly luciferase activity in cell lysate was measured by luciferase reporter assay kit (Promega, WI) according to manufacturer's instructions. Briefly, 20 μl of cell lysate was added to each well of an opaque 96-well plate. A100μl of Luciferase Assay Reagent (Promega, WI) was added one well at a time, and each well was read immediately. The light produced for a period of 10 s was measured using a Microplate Reader (BioTek Synergy 2 Multi-detection, Winooski, VT). The experiment to measure IFN - α/β activity in S100A9 knock out or in S100A9 overexpressed cells was done in three replicates and reproduced three times. The data was analyzed using paired t-test at 95% level of significance.
3. Results
3.1. Identification of cellular protein interaction with BVDV2a 1373 Npro protein through mass spectrometry
Cellular protein interactions with ncp BVDV2a 1373 Npro were identified in p3XFLAG-Npro transfected 293T cells using co-immunoprecipitation technique. The proteins in cell lysate of p3XFLAG-Npro or p3XFLAG transfected 293T was pulled down (isolated) using anti-FLAG antibody and examined through mass spectrometry. A total 30 most abundant proteins were identified in p3XFLAG as well as in p3XFLAG-Npro transfected cells lysate which mainly included keratin protein for cytoskeletal, ATP synthase, chain B of Immunoglobulin and protein S100A9. In comparing these two groups, a total of sixteen proteins were identified which were present in p3XFLAG-Npro transfected cells that were absent in p3XFLAG transfected cells (Table 1 ). Among these proteins, S100A9 had the highest mass spectrometry score (i.e., 295) indicating a strong association between BVDV Npro and cellular S100A9 (Table 1). The further analysis of S100A9 protein using MassLynx software 4.1 (Micromass) revealed 7 peptide fragments (Table 2 ).
Table 1.
Cellular protein interactions with ncp BVDV2 1373 Npro protein. A total 30 proteins were co-immunoprecipitated in the cells transfected with p3XFLAG (control) or p3XFLAG-Npro plasmid, using anti-FLAG antibody. The comparison between these two groups revealed a total of sixteen proteins which were present in p3XFLAG-Npro transfected cells that were absent in p3XFLAG transfected cells. The proteins are ranked according to mass spectrometry score e.g. highest to lowest in the table, indicating their interaction with viral (ncp BVDV2 1373) Npro protein.
| Protein | Cellular function | |
|---|---|---|
| 1. | Protein S 100 A9 | DAMPs – TLR4 agonist - chemotactic factor for phagocytes |
| 2. | Chaperonin | Protein folding |
| 3. | Chain A, Class P1 Glutathione S- Transferase | Cell detoxification |
| 4. | dUTPase | DNA synthesis |
| 5. | Dynein | Cell trafficking and Protein Transport |
| 6. | Myotrophin | NFkappa B signaling |
| 7. | Elongation factor 1-delta | Protein synthesis |
| 8. | Archain | Vesicle structure or trafficking |
| 9. | Programmed cell death 5 | Regulator in both apoptotic and paraptotic cell deaths |
| 10. | Chain A human Rap1a protein | Adhesion-related functions such as phagocytosis, cell-cell contacts and functional activation |
| 11. | Calmodulin | Regulator of NF-κB activation in lymphocytes |
| 12. | Spindin | Cell cycle and proliferation |
| 13. | KH domain –containing RNA binding –signal, signal transduction associated protein −1 | Required for TCR-induced tyrosine phosphorylation |
| 14. | NADH dehydrogenase | Component of complex I of the respiratory chain |
| 15. | Annexin A isoform | Cell motility – cell matrix – ion channel formation |
| 16. | Cofilin-1 | Actin polymerization – cytoskeleton |
Table 2.
The peptide fragments present in S100A9 protein. The cellular S100A9 protein was analyzed through nano ESI-Q-Tof micro mass spectrometer (Micromass, Manchester, UK) using MassLynx software 4.1 (Micromass). Software revealed seven peptide fragments in S100A9 protein.
| Protein S100-A9 [Homo sapiens] |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Observed | Mr (expt) | Mr (calc) | Delta | Miss | Score | Expect | Rank | Unique | Peptide |
| 503.2925 | 1004.5704 | 1004.5655 | 0.0050 | 1 | 46 | 6.1 | 5 | U | R.KDLQNFLK.K |
| 815.8993 | 1629.7840 | 1629.7895 | −0.0054 | 0 | 70 | 0.021 | 1 | K.QLSFEEFIMLMAR.L + Ox1 | |
| 871.9211 | 1741.8276 | 1741.8192 | 0.0084 | 0 | 100 | 1.8e-06 | 1 | U | K.VIEHIMEDLDTNADK.Q |
| 602.9818 | 1805.9236 | 1805.9312 | −0.0076 | 0 | (69) | 0.024 | 1 | R.NIETIINTFHQISVK.L | |
| 903.9719 | 1805.9292 | 1805.9312 | −0.0019 | 0 | 76 | 0.0045 | 1 | R.NIETIINTFHQISVK.L | |
| 602.9838 | 1805.9299 | 1805.9312 | −0.0013 | 0 | (47) | 3.5 | 1 | R.NIETIINTFHQISVK.L | |
| 903.9783 | 1805.9420 | 1805.9312 | 0.0109 | 0 | (76) | 0.0048 | 1 | R.NIETIINTFHQISVK.L | |
3.2. Co-immunoprecipitation confirms the interaction of BVDV2a 1373 Npro and S100A9 cellular protein
To further confirm the interaction between the BVDV2a 1373 Npro and cellular S100A9, the 293T cells were co-transfected with pEGFP-C1-Npro and p3XFLAG -S100A9 plasmid or pEGFP-C1-Npro and p3XFLAG (empty FLAG plasmid without S100A9). The transfected cells were lysed and proteins were isolated using the anti-FLAG beads. Proteins isolated by anti-FLAG beads were analyzed for presence of GFP (e.g GFP-Npro) as well as FLAG using western blot. The mock transfected 293T cells were used as negative control while cells transfected with pEGFP-C1-Npro plasmid were used as GFP-Npro positive control.
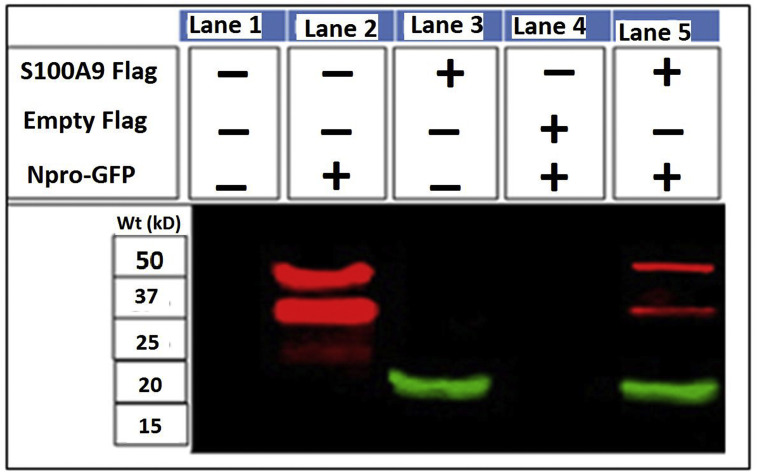
Cells co-transfected with p3XFLAG (without S100A9) with pEGFP C1-Npro did not show the presence of Npro while the cells co-transfected with p3XFLAG S100A9 along with pEGFP C1-Npro presence of both S100A9 as well as Npro proteins (Fig. 3 ). These resulted further confirmed a strong association of cellular S100A9 and BVDV Npro proteins.
Fig. 3.
Interaction of BVDV2a 1373 Npro and S100A9 cellular protein. The interaction of BVDV Npro with S100A9 protein in cells were confirmed by co immunoprecipitation assays. The 293T cells were co-transfected with pEGFP-C1-Npro and p3XFLAG -S100A9 plasmid or pEGFP-C1-Npro and p3XFLAG. At 24 h post transfection, cells were lysed and the proteins were isolated (by co-immunoprecipitation) using the anti-FLAG beads. Isolated proteins were analyzed for presence of GFP (e.g GFP-Npro) as well as FLAG using western blot using anti-GPF or anti-FLAG antibodies. The mock transfected 293T cells were used as negative control while cells transfected with pEGFP-C1-Npro plasmid were used as GFP-Npro positive control. Lane (1) The 293T cells without co-transfection did not show the presence of GFP-Npro or FLAG- S100A9, Lane (2) cells transfected with pEGFP-C1-Npro alone and showed the presence of GFP-Npro, Lane (3) cells transfected with p3XFLAG -S100A9 and showed the presence of S100A9, Lane (4) cells were co transfected with empty p3XFLAG along with pEGFP C1-Npro and did not show the presence of Npro (the FLAG size was 8 aa and was seen at very lower side of the gel). Lane (5) Cell lysates from cells co-transfected with p3XFLAG S100A9 along with pEGFP C1-Npro which showed the presence of S100A9 as well as Npro.
3.3. Inhibition of bovine S100A9 by small interfering RNA (siRNA)-enhanced virus replication
The activity of S100A9 on BVDV replication was measured in S100A9 knock out MDBK cells. The S100 A9 knock out or control MDBK cells were infected with ncpBVDV2a 1373 with MOI of 1. The supernatant was collected at 12 h, 24 h, 36 h and 60 h post infection and analyzed for BVDV viral titer. The results showed an increase in viral production by 1.5 log10 at 24 h (p < 0.05) and 0.5 log10 at 36 h in S100A9 knock out cells, as compared to scrambled siRNA-transfected cells (Fig. 4 ). BVDV infected S100A9 gene knock out cells had virus production of 2.5 ± 0.75 log10/ml, 5.0 ± 0.75 log10/ml, 6 ± 0.75 log10/ml, 6.25 ± 0.75 log10/ml, 5.5 ± 0.75 log10/ml at 12 h, 24 h, 36 h, 48 h and 60 h post infection respectively. In comparison, the scrambled siRNA-transfected cells had virus production of 3.25 ± 0.75 log10/ml, 3.5 ± 0.75 log10/ml, 5.5 ± 0.75 log10/ml, 6.5 ± 0.75 log10/ml and 5.5 ± 0.75 log10/ml 12 h, 24 h, 36 h and 60 h post infection respectively.
Fig. 4.
Inhibition of bovine S100A9 by small interfering RNA (siRNA)-enhanced virus replication. The S100 A9 knock down or control MDBK cells were infected with ncpBVDV2a 1373 with MOI of 1. The supernatant was collected at 12 h, 24 h, 36 h or 60 h post infection and analyzed for BVDV viral titer. There was a significant increase in viral production by 1.5 log10 at 24 h (p < 0.05) in S100A9 knocked out cells while at 36 h in S100A9 knock out cells also increased (0.5 log10) virus production compared to scrambled siRNA-transfected cells. Error bars are SEM.
3.4. Overexpression of bovine S100A9 counteracted the interferon Type-I inhibition induced by ncp BVDV2a 1373
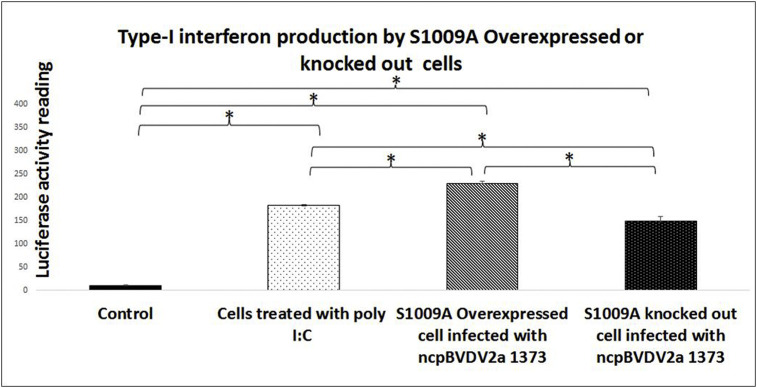
To determine whether the interaction of BVDV Npro with S100A9 affected the ability of Npro to inhibit type 1 (α/β) IFN, an interferon reporter assay was performed in S100A9 over expressed or S100A9 knock out cells [23]. The S100A9 over expressed or S100A9 knock out cells were infected with ncp BVDV2a-1373 for 24 h. After 24 h infection, the supernatant was collected and examined for type 1 interferon expression through firefly luciferase activity in NCL-1–ISRE–Luc–Hygro cells. There was significantly higher type 1 interferon expression in S100A9 overexpressed cells as compared to S100A9 knockout or poly I:C treated cells (p < 0.05) (Fig. 5 ) or BVDV2a 1373 infected normal cells without any. S100A9 overexpressed cells infected with ncpBVDV2a 1373 showed the luciferase activity reading as 230 ± 5.0 as compared to 148.0 ± 10.0 or 182.0 ± 2.0 in S100A9 knockout infected with ncpBVDV2a 1373 or cell treated with poly I:C respectively (Fig. 5).
Fig. 5.
Overexpression of bovine S100A9 counteract the interferon Type-I interferon (IFN α/β) inhibition induced by ncp BVDV2a 1373. The S100A9 over expressed or S100A9 knock out cells were infected with ncp BVDV2a-1373 with MOI of 5 for 24 h while control cells were mock treated or treated with poly I:C. After 24 h infection/treatment, supernatant was collected and examined for type 1 interferon expression through firefly luciferase activity in NCL-1–ISRE–Luc–Hygro cells. The control cell treated with poly I:C, S100A9 over expressed or S100A9 knock out cells infected with ncp BVDV2a-1373 showed significantly higher IFN α/β as compare to mock-infected/treated control cell {*}, p < 0.05. While S100A9 overexpressed cells infected with ncp BVDV2a-1373 had significantly higher IFN α/β as compared to S100A9 knock out cells infected with ncp BVDV2a-1373 or cells treated with poly I:C {**}, p < 0.05.
4. Discussion
In this study, immunoprecipitation coupled with mass spectrometry demonstrated the interaction of viral ncp BVDV2a 1373 Npro protein with the cellular S100A9 protein. The interaction was confirmed first by a co-immunoprecipitation- mass spectrometry experiment targeting BVDV Npro protein, which showed the co-presence of cellular S1009A. It was reconfirmed by western blot analysis demonstrated cellular S1009A protein interaction with BVDV Npro protein. Additionally, SiRNA-knockdown experiments showed a significant increase in BVDV titers (p < 0.05) by more than one log in S100A9 knock down cells at 24 h post infection while overexpression of S100A9 enhanced type 1 interferon production, indicating a functional relationship between BVDV pathogenesis and cellular S100A9 protein.
Currently there is very little is known about the role of S100A9 in viral infections. A previous study showed that S100A9 was upregulated in response to poly I:C, a synthetic analog of viral double-stranded RNA (dsRNA) with increase type 1 IFN [43]. The human papillomavirus infection also increased systemic levels of S100A8 and S100A9 [44]. Likewise, S100A9 increased 60-fold in patients with coronavirus induced severe acute respiratory syndrome [45].
S100A9 exhibits its effect through two well characterized PRRs: TLR4 and receptor advanced glycation end products (RAGE) [10,46]. Both PRRs use independent downstream pathways [13]. When TLRs are stimulated by alarmins (i.e., PAMPs and DAMPs), they recruit one or more adapter proteins, such as MyD88 (a downstream signaling protein). Activated MyD88 further activates signaling molecules, including phosphatidylinositol-3–kinase (PI3K), mitogen-activated protein kinases (MAPK) (i.e., c-Jun NH2-terminal kinase [JNK], p38, and extracellular signal-regulated kinase (ERKs), v-akt murine thymoma viral oncogene homolog (Akt), interferon regulatory factor (IRF), β-catenin [47], as well as nuclear factor-кB (NF-кB) [48]. Increased NF-кB expression increased the pro-inflammatory cytokines including chemokine ligand 2(CCL2), interleukin-1 (IL-1), interleukin-6 (IL-6), and tumor necrosis factor-α (TNF-α), interleukin-8 (IL-80, monocyte-chemo attractant protein–1, chemokine ligand 1 (CXCL1 or GRO-α), type 1 IFN [[49], [50], [51]]; vascular endothelial growth factor (VEGF), granulocyte-macrophage colony-stimulating factor (GMCSF) and matrix metalloproteinases (MMP2 and MMP9) [52] with upregulated adhesion molecules and antiapoptotic proteins such as B-cell CLL/lymphoma-2, Bcl-XL [53].
Since S100A9 is integral to so many important immune pathways, it is a “logical” target for viral protein interference with cell defense mechanisms [54]. In this study, virus titer increased by one log in the absence of S100A9; whereas, overexpression of S100A9 in cells infected with ncp BVDV2a-1373 overcame the interferon inhibition. This is consistent with Npro inhibition of IFN-1 through a dual mechanism that includes degradation of IRF-3 in the proteasome [33].
The results of this study indicated that S100A9 interacted with ncp BVDV2a-1373 Npro. The interaction of BVDV2a-1373 Npro with cellular S100A9, potentially reduced the availability of S100A9 and therefore reduced the type 1 interferon production. Hilton et al., 2006 [55] and Chen et al., 2007 [56] found that NPro blocks IRF-3 from binding to DNA. Additionally, NPro has the property of targeting IRF-3 for polyubiquitination and subsequent destruction by cellular multicatalytic proteasomes, indicating that Npro induces immune suppression through multiple pathways that need to be further explored.
It has been suggested that BVDV infection, in persistently infected (PI) animals, may lead to immunosuppression through neutrophil dysfunction [57]. Modified live BVDV cp strains caused a decrease in circulating lymphocytes and neutrophils and also suppressed neutrophil functions [58,59]. These effects could be explained by the interaction of S100A9 with the virus Npro, as S100A9 has a major role in the migration of phagocytes [60,61] and neutrophil oxidative burst [62]. Additionally, an important sequela of BVDV infection is the increase in secondary bacterial infection [63]. Interaction of Npro with S100A9 protein could have potent antimicrobial and chemotactic effects that could increase the incidence of secondary infections [60].
S100A8/S100A9 functions as an endogenous TLR4 ligand that is essential to the amplification of lipopolysaccharide (LPS) effects on phagocytes upstream of TNF-α [64]. One study showed a decline in S100A8/S100A9 levels in patients that survived sepsis, whereas non-survivors had increased serum levels of S100A8/S100A9 [65]. This higher mortality with BVDV mucosal disease where sudden death often occurs could also be due to an increase level of S100A9 that amplifies the inflammatory response, thus increasing the chance of endotoxic shock. The interaction of BVDV Npro with S100A9 found in this study creates several questions that need to be answered regarding BVDV pathogenesis and role of DAMPs in viral infection.
5. Conclusion
Bovine viral diarrhea virus (BVDV) is an immunosuppressive virus. There are a number of studies which demonstrated that the nonstructural protein Npro, is responsible for its immunosuppressive effect. However, very little is known of the mechanism of how Npro protein induces immunosuppression. The current study demonstrated that Npro interacts with the cellular S100A9 protein. S100A9 is a member of the danger associated molecular patterns (DAMPs) and play an important role in innate immune response. Interaction of BVDV Npro with cellular S100A9 protein, potentially reduced its availability/activity in virus infected cells leading to reduced type 1 interferon production and higher virus production.
Sources of funding
This work was supported by the Department of Veterinary and Biomedical Sciences/Animal Disease Research and Diagnostic Laboratory, SDSU, Brookings, SD, USA; SDSU Experimental Station and the Center for Biological Control and Analysis by Applied Photonics Department of Chemistry and Biochemistry, SDSU, Brookings SD, USA.
Author contributions
CCLC and MFD designed the study. CCLC, JSR, and LJB coordinated the study. MFD, MKSR and LJB carried out the experiments. MFD analyzed the data. CCLC and MFD wrote while CCLC, MDF and MKSR edited the manuscript. All authors approved the manuscript.
Competing interests
The authors have no conflict of interest to declare.
Acknowledgements
The authors thank Dr. Adam Hoppe, Department of Chemistry and Biochemistry, SDSU for providing the plasmids. Authors are also thankful to Dr. Eduardo Callegari, University of South Dakota, Mass Spectrometry and Proteomics Core Facility, Vermillion, SD for his help in mass spectrometry analysis. Authors also show their gratitude to Dr. Clayton Kelling, University of Nebraska, Lincoln NE for providing NCL-1–ISRE–Luc–Hygro cells for IFN - α/β assay. The authors would also like to acknowledge Angela Klein for her editing assistance.
References
- 1.Beutler B. Toll-like receptors and their place in immunology. Where does the immune response to infection begin? Nat. Rev. Immunol. 2004;4 doi: 10.1038/nri1401. [DOI] [PubMed] [Google Scholar]
- 2.Mariathasan S., Monack D.M. Inflammasome adaptors and sensors: intracellular regulators of infection and inflammation. Nat. Rev. Immunol. 2007;7:31–40. doi: 10.1038/nri1997. [DOI] [PubMed] [Google Scholar]
- 3.Oppenheim J.J., Yang D. Alarmins: chemotactic activators of immune responses. Curr. Opin. Immunol. 2005;17:359–365. doi: 10.1016/j.coi.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Yang D., Oppenheim J.J. Antimicrobial proteins act as “alarmins” in joint immune defense. Arthritis Rheum. 2004;50(11):3401–3403. doi: 10.1002/art.20604. 2004 Nov. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X., Mosser D.M. Macrophage activation by endogenous danger signals. J. Pathol. 2008;214:161–178. doi: 10.1002/path.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolf R., Howard O.M., Dong H.F., Voscopoulos C., Boeshans K., Winston J., Divi R., Gunsior M., Goldsmith P., Ahvazi B., Chavakis T., Oppenheim J.J., Yuspa S.H. Chemotactic activity of S100A7 (Psoriasin) is mediated by the receptor for advanced glycation end products and potentiates inflammation with highly homologous but functionally distinct S100A15. J. Immunol. 2008;181:1499–1506. doi: 10.4049/jimmunol.181.2.1499. PMCID: PMC2435511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foell D., Roth J. Proinflammatory S100 proteins in arthritis and autoimmune disease. Arthritis Rheum. 2004;50:3762–3771. doi: 10.1002/art.20631. [DOI] [PubMed] [Google Scholar]
- 8.Foell D., Wittkowski H., Roth J. Mechanisms of disease: a 'DAMP' view of inflammatory arthritis. Nat. Clin. Pract. Rheumatol. 2007;3:382–390. doi: 10.1038/ncprheum0531. [DOI] [PubMed] [Google Scholar]
- 9.Foell D., Wittkowski H., Vogl T., Roth J. S100 proteins expressed in phagocytes: a novel group of damage-associated molecular pattern molecules. J. Leukoc. Biol. 2007;81:28–37. doi: 10.1189/jlb.0306170. [DOI] [PubMed] [Google Scholar]
- 10.Tsai S.Y., Segovia J.A., Chang T.H., Morris I.R., Berton M.T., Tessier P.A., Tardif M.R., Cesaro A., Bose A. DAMP molecule S100A9 acts as a molecular pattern to enhance inflammation during influenza A virus infection: role of DDX21-TRIF-TLR4-MyD88 pathway. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1003848. https://DOI.org/10.1371/journal.ppat.1003848 e1003848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L., Luo H., Chen X., Jiang Y., Huang Q. Functional characterization of S100A8 and S100A9 in altering monolayer permeability of human umbilical endothelial cells. PLoS One. 2014;9:e90472. doi: 10.1371/journal.pone.0090472. https://DOI.org/10.1371/journal.pone.009047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Averill M.M., Barnhart S., Becker L., Li X., Heinecke J.W., Leboeuf R.C., Hamerman J.A., Sorg C., Kerkhoff C., Bornfeldt K.E. S100A9 differentially modifies phenotypic states of neutrophils, macrophages, and dendritic cells: implications for atherosclerosis and adipose tissue inflammation. Circulation. 2011;123:1216–1226. doi: 10.1161/CIRCULATIONAHA.110.985523. Epub 2011 Mar 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogl T., Tenbrock K., Ludwig S., Leukert N., Ehrhardt C., Zoelen M.A.D., Nacken W., Dl Foel, Poll T., Sorg C., Roth J. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat. Med. 2007;13:1042–1049. doi: 10.1038/nm1638. [DOI] [PubMed] [Google Scholar]
- 14.Nomura I., Goleva E., Howell M.D., Hamid Q.A., Ong P.Y., Hall C.F., Darst M.A., Gao B., Boguniewicz M., Travers J.B., Leung D.Y. Cytokine milieu of atopic dermatitis, as compared to psoriasis, skin prevents induction of innate immune response genes. J. Immunol. 2003;171:3262–3269. doi: 10.4049/jimmunol.171.6.3262. PMID: 12960356. [DOI] [PubMed] [Google Scholar]
- 15.Chase C.C., Hurley D.J., Reber A.J. Neonatal immune development in the calf and its impact on vaccine response. Vet. Clin. Food Anim. Pract. 2008;24:87–104. doi: 10.1016/j.cvfa.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chase C.C., Thakur N., Darweesh M.F., Morarie-Kane S.E., Rajput M.K. Immune response to bovine viral diarrhea virus–looking at newly defined targets. Anim. Health Res. Rev. 2015;16(1):4–14. doi: 10.1017/S1466252315000110. [DOI] [PubMed] [Google Scholar]
- 17.Darweesh M.D., Rajput M., Lebedev M., Young A., Braun L., Chase C. Bovine natural killer cells are infected by BVDV and the infection results in changes in phenotype and activation markers (P6128) J. Immunol. 2013;190(1 Supplement) doi: 10.1016/j.virusres.2014.09.015. 128.115-128.115. [DOI] [Google Scholar]
- 18.Rajput M.K., Darweesh M.F., Park K., Braun L.J., Mwangi W., Young A.J., Chase C.C. The effect of bovine viral diarrhea virus (BVDV) strains on bovine monocyte-derived dendritic cells (Mo-DC) phenotype and capacity to produce BVDV. Virol. J. 2014;11:44. doi: 10.1186/1743-422X-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harada T., Tautz N., Thiel H.J. E2-p7 region of the bovine viral diarrhea virus polyprotein: processing and functional studies. J. Virol. 2000;74(20):9498–9506. doi: 10.1128/jvi.74.20.9498-9506.2000. PMID: 11000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harada T., Tautz N., Thiel H.J. E2-p7 region of the bovine viral diarrhea virus polyprotein: processing and functional studies. J. Virol. 2000;74(20):9498–9506. doi: 10.1016/0042-6822(88)90672%5f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckels K.H., Putnak R. Formalin-inactivated whole virus and recombinant subunit flavivirus vaccines. Adv. Virus Res. 2003;61:395–418. doi: 10.1016/S0065-3527(03. [DOI] [PubMed] [Google Scholar]
- 22.Buckwold V.E., Beer B.E., Donis R.O. Bovine viral diarrhea virus as a surrogate model of hepatitis C virus for the evaluation of antiviral agents. Antivir. Res. 2003;60(1):1–15. doi: 10.1016/S0166-3542(03. [DOI] [PubMed] [Google Scholar]
- 23.Gil L.H., Ansari I.H., Vassilev V., Liang D., Lai V.C., Zhong W., Hong Z., Dubovi E.J., Donis R.O. The amino-terminal domain of bovine viral diarrhea virus Npro protein is necessary for alpha/beta interferon antagonism. J. Virol. 2006;80:900–911. doi: 10.1128/JVI.80.2.900-911.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rumenapf T., Stark R., Heimann M., Thiel H.J. N-terminal protease of pestiviruses: identification of putative catalytic residues by site-directed mutagenesis. J. Virol. 1998;72:2544–2547. doi: 10.1128/jvi.72.3.2544-2547.1998. PMCID: PMC109561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tratschin J.D., Moser C., Ruggli N., Hofmann M.A. Classical swine fever virus leader proteinase Npro is not required for viral replication in cell culture. J. Virol. 1998;72:7681–7684. doi: 10.1128/jvi.72.9.7681-7684.1998. PMID: 9696875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruggli N., Bird B.H., Liu L., Bauhofer O., Tratschin J.D., Hofmann M.A. N(pro) of classical swine fever virus is an antagonist of double-stranded RNA-mediated apoptosis and IFN-alpha/beta induction. Virology. 2005;340:265–276. doi: 10.1016/j.virol.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 27.Ruggli N., Tratschin J.D., Schweizer M., McCullough K.C., Hofmann M.A., Summerfield A. Classical swine fever virus interferes with cellular antiviral defense: evidence for a novel function of N(pro) J. Virol. 2003;77:7645–7654. doi: 10.1128/JVI.77.13.7645-7654.2003. PMCID: PMC164809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayer D., Hofmann M.A., Tratschin J.D. Attenuation of classical swine fever virus by deletion of the viral N(pro) gene. Vaccine. 2004;22:317–328. doi: 10.1016/j.vaccine.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Meyers G., Ege A., Fetzer C., von Freyburg M., Elbers K., Carr V., Prentice H., Charleston B., Schürmann E.M. Bovine viral diarrhea virus: prevention of persistent fetal infection by a combination of two mutations affecting Erns RNase and Npro protease. J. Virol. 2007;81:3327–3338. doi: 10.1128/JVI.02372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai V.C., Zhong W., Skelton A., Ingravallo P., Vassilev V., Donis R.O., Hong Z., Lau J.Y. Generation and characterization of a hepatitis C virus NS3 protease-dependent bovine viral diarrhea virus. J. Virol. 2000;74:6339–6347. doi: 10.1128/jvi.74.14.6339-6347.2000. PMID: 10864644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gabriele L., Ozato K. The role of the interferon regulatory factor (IRF) family in dendritic cell development and function. Cytokine Growth Factor Rev. 2007;18:503–510. doi: 10.1016/j.cytogfr.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 32.Hiscott J., Pitha P., Genin P., Nguyen H., Heylbroeck C., Mamane Y., Algarte M., Lin R. Triggering the interferon response: the role of IRF-3 transcription factor. J. Interferon Cytokine Res. 1999;19:1–13. doi: 10.1089/107999099314360. [DOI] [PubMed] [Google Scholar]
- 33.Horscroft N., Bellows D., Ansari I., Lai V.C., Dempsey S., Liang D., Donis R., Zhong W., Hong Z. Establishment of a subgenomic replicon for bovine viral diarrhea virus in Huh-7 cells and modulation of interferon-regulated factor 3-mediated antiviral response. J. Virol. 2005;79:2788–2796. doi: 10.1128/JVI.79.5.2788-2796.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madin S.H., Darby N.B., Jr. Established kidney cell lines of normal adult bovine and ovine origin. Proc Soc Exp Biol Med. 1958;98:574–576. doi: 10.3181/00379727-98-24111. PMID: 13567776. [DOI] [PubMed] [Google Scholar]
- 35.Schweizer M., Peterhans E. Noncytopathic bovine viral diarrhea virus inhibits double-stranded RNA-induced apoptosis and interferon synthesis. J. Virol. 2001;75(10):4692–4698. doi: 10.1128/JVI.75.10.4692-4698.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ridpath J.F., Neill J.D., Vilcek S., Dubovi E.J., Carman S. Multiple outbreaks of severe acute BVDV in North America occurring between 1993 and 1995 linked to the same BVDV2 strain. Vet. Microbiol. 2006;114(3–4):196–204. doi: 10.1016/j.vetmic.2005.11.059. [DOI] [PubMed] [Google Scholar]
- 37.Donofrio G., Bottarelli C.S., Flammini C.F. Expression of bovine viral diarrhea virus glycoprotein E2 as a soluble secreted form in a Mammalian cell line. Clin. Vaccine Immunol. 2006;13(6):698–701. doi: 10.1128/CVI.00071-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nogarol C., Decaro N., Bertolotti L., Colitti B., Iotti B., Petrini S., Lucente M.S., Elia G., Perona G., Profiti M., Buonavoglia C., Rosati S. Pestivirus infection in cattle dairy farms: E2 glycoprotein ELISA reveals the presence of bovine viral diarrhea virus type 2 in northwestern Italy. BMC Vet. Res. 2017;13(1):377. doi: 10.1186/s12917-017-1305-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. PMID: 14907713. [PubMed] [Google Scholar]
- 40.Free R.B., Hazelwood L.A., Sibley D.R. Identifying novel protein-protein interactions using co-immunoprecipitation and mass spectroscopy. Curr Protoc Neurosci. 2009 doi: 10.1002/0471142301.ns0528s46. 2009/01/27 ed. pp. Unit 5.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 42.Rubinstein S., Familletti P.C., Pestka S. Convenient assay for interferons. J. Virol. 1981;37:755–758. doi: 10.1128/jvi.37.2.755-758.1981. PMID: 6163873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voss A., Gescher K., Hensel A., Nacken W., Zanker K.S., Kerkhoff C. Double-stranded RNA induces S100 gene expression by a cycloheximide-sensitive factor. FEBS Lett. 2012;586:196–203. doi: 10.1016/j.febslet.2011.12.022. Epub 2011 Dec 29. [DOI] [PubMed] [Google Scholar]
- 44.Tugizov S., Berline J., Herrera R., Penaranda M.E., Nakagawa M., Palefsky J. Inhibition of human papillomavirus type 16 E7 phosphorylation by the S100 MRP-8/14 protein complex. J. Virol. 2005;79:1099–1112. doi: 10.1128/JVI.79.2.1099-1112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reghunathan R., Jayapal M., Hsu L.Y., Chng H.H., Tai D., Leung B.P., Melendez A.J. Expression profile of immune response genes in patients with Severe Acute Respiratory Syndrome. BMC Immunol. 2005;6(2) doi: 10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt A.M., Yan S.D., Yan S.F., Stern D.M. The multiligand receptor RAGE as a progression factor amplifying immune and inflammatory responses. J. Clin. Invest. 2001;108:949–955. doi: 10.1172/JCI14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rakoff-Nahoum S., Medzhitov R. Role of toll-like receptors in tissue repair and tumorigenesis. Biochemistry (Mosc.) 2008;73:555–561. doi: 10.1134/s0006297908050088. PMID: 18605980. [DOI] [PubMed] [Google Scholar]
- 48.Akira S., Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 49.Son D.S., Parl A.K., Rice V.M., Khabele D. Keratinocyte chemoattractant (KC)/human growth-regulated oncogene (GRO) chemokines and pro-inflammatory chemokine networks in mouse and human ovarian epithelial cancer cells. Canc. Biol. Ther. 2007;6:1302–1312. doi: 10.4161/cbt.6.8.4506. PMID: 17712227. [DOI] [PubMed] [Google Scholar]
- 50.Kagan J.C., Su T., Horng T., Chow A., Akira S., Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat. Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karaghiosoff M., Steinborn R., Kovarik P., Kriegshauser G., Baccarini M., Donabauer B., Reichart U., Kolbe T., Bogdan C., Leanderson T., Levy D., Decker T., Müller M. Central role for type I interferons and Tyk2 in lipopolysaccharide-induced endotoxin shock. Nat. Immunol. 2003;4:471–477. doi: 10.1038/ni910. [DOI] [PubMed] [Google Scholar]
- 52.Ikebe M., Kitaura Y., Nakamura M., Tanaka H., Yamasaki A., Nagai S., Wada J., Yanai K., Koga K., Sato N., Kubo M., Tanaka M., Onishi H., Katano M. Lipopolysaccharide (LPS) increases the invasive ability of pancreatic cancer cells through the TLR4/MyD88 signaling pathway. J. Surg. Oncol. 2009;100:725–731. doi: 10.1002/jso.21392. [DOI] [PubMed] [Google Scholar]
- 53.Chen R., Alvero A.B., Silasi D.A., Mor G. Inflammation, cancer and chemoresistance: taking advantage of the toll-like receptor signaling pathway. Am. J. Reprod. Immunol. 2007;57:93–107. doi: 10.1111/j.1600-0897.2006.00441.x. [DOI] [PubMed] [Google Scholar]
- 54.Koy M., Hambruch N., Hussen J., Pfarrer C., Seyfert H.M., Schuberth H.J. Recombinant bovine S100A8 and A9 enhance IL-1beta secretion of interferon-gamma primed monocytes. Vet. Immunol. Immunopathol. 2013;155:162–170. doi: 10.1016/j.vetimm.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 55.Hilton L., Moganeradj K., Zhang G., Chen Y.H., Randall R.E., McCauley W.J., Goodbourn S. The NPro product of bovine viral diarrhea virus inhibits DNA binding by interferon regulatory factor 3 and targets it for proteasomal degradation. J. Virol. 2006;80(23):11723–11732. doi: 10.1128/JVI.01145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Z., Rijnbrand R., Jangra R.K., Devaraj S.G., Qu L., Ma Y., Lemon S.M., Li K. Ubiquitination and proteasomal degradation of interferon regulatory factor-3 induced by Npro from a cytopathic bovine viral diarrhea virus. Virology. 2007;366(2):277–292. doi: 10.1016/j.virol.2007.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown G.B., Bolin S.R., Frank D.E., Roth J.A. Defective function of leukocytes from cattle persistently infected with bovine viral diarrhea virus, and the influence of recombinant cytokines. Am. J. Vet. Res. 1991;52:381–387. PMID: 1852143. [PubMed] [Google Scholar]
- 58.Roth J.A., Kaeberle M.L. Suppression of neutrophil and lymphocyte function induced by a vaccinal strain of bovine viral diarrhea virus with and without the administration of ACTH. Am. J. Vet. Res. 1983;44:2366–2372. PMID: 6318614. [PubMed] [Google Scholar]
- 59.Roth J.A., Kaeberle M.L., Griffith R.W. Effects of bovine viral diarrhea virus infection on bovine polymorphonuclear leukocyte function. Am. J. Vet. Res. 1981;42:244–250. PMID: 6266288. [PubMed] [Google Scholar]
- 60.Raquil M.A., Anceriz N., Rouleau P., Tessier P.A. Blockade of antimicrobial proteins S100A8 and S100A9 inhibits phagocyte migration to the alveoli in streptococcal pneumonia. J. Immunol. 2008;180:3366–3374. doi: 10.4049/jimmunol.180.5.3366. PMID: 18292562. [DOI] [PubMed] [Google Scholar]
- 61.Vogl T., Ludwig S., Goebeler M., Strey A., Thorey I.S., Reichelt R., Foell D., Gerke V., Manitz M.P., Nacken W., Werner S., Sorg C., Roth J. MRP8 and MRP14 control microtubule reorganization during transendothelial migration of phagocytes. Blood. 2004;104:4260–4268. doi: 10.1182/blood-2004-02-044. [DOI] [PubMed] [Google Scholar]
- 62.Markowitz J., Carson W.E., 3rd Review of S100A9 biology and its role in cancer. Biochim. Biophys. Acta. 2013;1835:100–109. doi: 10.1016/j.bbcan.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roth J.A., Kaeberle M.L. Effects of in vivo dexamethasone administration on in vitro bovine polymorphonuclear leukocyte function. Infect. Immun. 1981;33:434–441. doi: 10.1128/iai.33.2.434-441.1981. PMCID: PMC350716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beutler B., Rietschel E.T. Innate immune sensing and its roots: the story of endotoxin. Nat. Rev. Immunol. 2003;3:169–176. doi: 10.1038/nri1004. [DOI] [PubMed] [Google Scholar]
- 65.Payen D., Lukaszewicz A.C., Belikova I., Faivre V., Gelin C., Russwurm S., Launay J.M., Sevenet N. Gene profiling in human blood leucocytes during recovery from septic shock. Intensive Care Med. 2008;34:1371–1376. doi: 10.1007/s00134-008-1048-1. [DOI] [PubMed] [Google Scholar]