Abstract
Methylene blue is an electroactive molecule that has been employed for the detection of the DNA hybridization event in electrochemical sensors. However, its use as a covalent label is very scarce and in most of the cases, non-covalent interactions (hydrophobic, electrostatic) are employed. Although it has advantages as simplicity and fewer number of procedure steps, the covalent attachment is less exploited in the development of these sensors. In this article, the electrochemical behavior of methylene blue attached to different DNA-strands is studied. Several lengths (15- and 30-mer) and different degree of DNA modification (MB-DNA, MB-DNA-MB and MB-DNA-SH) have been studied. The highest signals were obtained for longer strands with two MB molecules. In all the cases the signal is enhanced by CNT-nanostructuration of the electrode. Adsorption on these modified screen-printed electrodes allowed the amplification by employing an accumulation time. In this way, a sensitivity of −0.2864 μA μM−1 and a limit of detection of 800 nM for a 120 s accumulation time were obtained.
Keywords: Methylene blue, Covalent electroactive labels, Genosensors, Carbon nanotubes, Nanostructured electrodes, Screen-printed gold electrodes
1. Introduction
Various techniques as fluorescent [1], surface plasmon resonance spectroscopy [2], quartz crystal microbalance [3] and electrochemical methods [4] have been proposed for DNA hybridization detection. Electrochemical methods for DNA hybridization detection possess the advantage of simplicity, low cost, high sensitivity [5], [6], [7] and rapid and direct detection [8] of specific DNA sequences. A variety of alternative ways of electrochemically detecting DNA have been reported in the literature in the last years. Direct, indirect methods and the use of marks are the three formats which have been employed in the genosensors development. The intrinsic electroactivity of guanine or adenine bases can be used for the direct measurement of nucleic acids in a label-free assay [9], [10], but their poor sensitivity is their principal disadvantage. The electroactive determination of intercalate [11] or associate with double-stranded DNA (dsDNA) indicators involves the indirect methods of DNA determination. The bad sensitivity can be enhancing with the use of enzymes due to their inherent amplification. Finally, the use of marks, as fluorescent molecules [12], has been very used in the DNA sensors development. However, the use of electroactive species has been much less common [13], [14].
Methylene blue (MB), 3,7-bis(dimethylamino)phenothiazine-5-ium-chloride, an organic dye which belongs to the phenothiazine family, had been used as electroactive marker in biosensors to detect the hybridization event [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28]. The process is performed by different interactions: (i) by electrostatic interaction between cationic MB and anionic DNA [19], [20], [21], (ii) by intercalation of MB in the DNA double helix between alternating G–C base sequences [22], [23], [24], [25] and (iii) by covalent bound to the end of the single-stranded target DNA (ssDNA) [26], [27], [28], the least exploited of all. The first reference of a covalently modified MB-DNA is due to Plaxco et al. [26]. This work supports the utility of a DNA sensor with MB conjugated to the 3′ end of the DNA as probe sequence as a rapid, specific, and convenient method for the detection of DNA and RNA sequences. In this case, alternating current voltammetry (ACV) was employed for recording the electrochemical process. They found that signal generation on gold disk electrode is coupled to a hybridization-linked conformational change, rather than to only adsorption on the sensor surface. The covalent attachment presents, among others, the advantage that it is not necessary to perform a subsequent step of washing to remove unbound label. The use of ferrocene and their derivates [29], [30], [31], [32], gold complexes [13], metallic [33] or semiconductor [34] nanoparticles and osmium [35] or platinum [14] complexes covalently attachment to DNA strand had been used in the detection of the hybridization event.
Carbon nanotubes (CNTs) are widely considered to be among the most important materials of this century and can be applied to diverse fields including drug delivery [36], neuroengineering [37], gene therapy [38] and biosensor technology [39] among others. Several reviews of CNTs regarding the methods of synthesis [40], [41] and their mechanical, chemical and electrical properties [40], [42], [43], [44] are reported in the literature. Since their discovery and due to their unique properties, carbon nanotubes (CNTs) have been used in different electroanalytical applications [45], [46], [47]. Their adsorption capacity, the possibility of being functionalized, their ability to promote the electron transfer reactions of a great number of molecules [48], [49] and the possibility of use in miniaturized devices make CNTs a very attractive material for the further development of electrochemical biosensors. Different attempts have been made to improve the electroanalytical properties of disposable CNT-SPEs (screen-printed electrodes) biosensors, through their adsorption properties and the improvement in the reversibility of processes [50], [51].
In this work, a study of the electrochemical behavior of DNA-sequences of different lengths (15 and 30-mer) with MB covalently attached to one end of the strand was made. Sequences with two MB molecules, attached to both ends of the strand and sequences with one —SH group were tested too. All DNA sequences corresponded to a portion of the SARS genome. Severe Acute Respiratory Syndrome (SARS) is an emerging infectious disease emerged in Southern China and rapidly spread to different areas of the Far East, caused by a novel coronavirus (SARS-CoV), which has overwhelmed more than 30 countries claiming nearly 8400 cases with over 800 fatalities since late 2002 [52], [53]. To the best of our knowledge, this is the first time that a comparative study between DNA single strands, different in length and labeling degree is made on CNTs nanostructured gold SPEs.
2. Experimental
2.1. Reagents and solutions
Nafion® (perfluorinated ion-exchange resin, 5 wt.% solution in a mixture of lower aliphatic alcohols and water), methylene blue (MB, certified by the BSC) and boric acid were obtained from Sigma–Aldrich (St. Louis, MO, USA). Sulphuric acid (95–97% of purity) was purchased from Merck (Darmstadt, Germany). Finally, amine functionalized multi-wall carbon nanotubes (MWCNT-NH2) were purchased from Belgium Nanocyl (Auvelais, Belgium). Water was purified employing a Milli-Q directQS system from Millipore.
Synthetic single-stranded DNA (ssDNA) was purchased from BioTez (Berlin, Germany). Oligomers different in length possessed the following base sequences:
| 15-mer: | MB-DNA: | 5′-MB-ACA GAG CCT AAA AAG-3′ |
| MB-DNA-MB: | 5′-MB-ACA GAG CCT AAA AAG-MB-3′ | |
| 30-mer: | MB-DNA: | 5′-MB-ACA GAG CCT AAA AAG GAC AAA AAG AAA AAG-3′ |
| MB-DNA-MB: | 5′-MB-ACA GAG CCT AAA AAG GAC AAA AAG AAA AAG-MB-3′ | |
| MB-DNA-SH: | 5′-MB-ACA GAG CCT AAA AAG GAC AAA AAG AAA AAG-SH-(CH2)3-3′ |
The sequence chosen is included in the 29751-base genome of the SARS-associated coronavirus [54]. This is the causative agent of an outbreak of atypical pneumonia. The sequence corresponds to a gene that encodes the nucleocapsid protein (422 amino acids), specifically a short lysine-rich region that appears to be unique to SARS and suggestive of a nuclear localization signal. A 30-mer oligonucleotide with bases comprised between numbers 29218 and 29247, both included, and a 15-mer oligonucleotide was chosen.
Oligonucleotide solutions were prepared in TBE buffer pH 8.0 (0.1 M Tris–boric acid buffer solution, 1 mM in EDTA). Aliquots were prepared and stored at −20 °C. Working solutions of ssDNA were prepared from aliquots in the buffer solution (0.1 M Tris–H2SO4 pH 8.0) [55] and stored at 4 °C.
2.2. Apparatus and instruments
Voltammetric measurements were performed with an Autolab PGSTAT 10 (ECO Chemie) potentiostat interfaced to a Pentium 120 computer system and controlled by Autolab GPES software version 4.8. A JEOL JSM-5600 Scanning Electron Microscope (30 kV) was used to characterize the gold working electrodes modified with carbon nanotubes solutions. An Elma ultrasonic bath, a Nahita centrifuge with interchangeable car, a Mettler Toledo (AB54) balance, a Crison Micro-pH 2001 pH-meter, a magnetic stirrer Asincro (J.P. Selecta), a Sanyo refrigerator and a Sanyo (MIR-162) incubator were also used.
2.3. Gold screen-printed electrodes
2.3.1. Unmodified electrodes (AuSPEs)
Gold screen-printed electrodes purchased from DropSens (Asturias, Spain) include a traditional three-electrode configuration printed on the same strip. The format of these SPEs includes a gold disk electrode (12.6 mm2) as working electrode, a silver pseudo-reference electrode and a gold counter electrode using the same ink than this of the working electrode. All of them are screen-printed on a ceramic substrate (3.4 cm × 1.0 cm × 0.05 cm) and subjected to high-temperature curing (ref. 220AT). An insulating layer serves to delimit the working area and electric contacts. The production characteristics of commercial SPEs are regarded by the manufacturers as proprietary information. A specific connector supplied also by DropSens allows their connection to the potentiostat.
2.3.2. Nanostructured electrodes (CNTs-AuSPEs)
The nanostructuration of screen-printed electrodes with CNTs was carried out by evaporation at room temperature of a 3-μL drop of MWCNTs-NH2 (dispersed in a 0.5% Nafion®/ethanol solution [51]) deposited on the working electrode. Before the measurements, CNTs-AuSPEs were washed with the supporting electrolyte (0.1 M Tris–H2SO4 pH 8.0) and dried at room temperature.
2.4. Electrochemical measurements
Drops of 40 of 20 μM MB, MB-DNA, MB-DNA-MB or MB-DNA-SH in buffer solution (0.1 M Tris–H2SO4 pH 8.0) were deposited for performing the electrochemical detection. The reduction signal of MB was recorded in triplicate (in three different SPEs) by scanning the potential between 0.0 and −0.7 V using cyclic voltammetry (CV) with a scan rate of 250 mV s−1, square wave voltammetry (SWV) with a step potential of 0.008 V, amplitude of 0.05 V and frequency of 50 Hz) and differential pulse voltammetry (DPV), with a step potential of 0.008 V, modulation amplitude of 0.05 V and scan rate of 16 mV s−1. All experiments were performed at room temperature.
3. Results and discussion
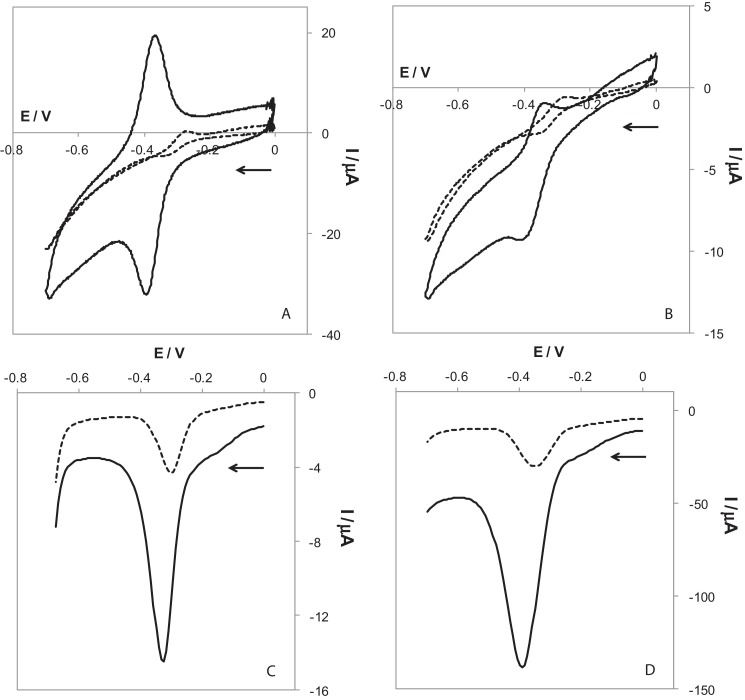
As commented in Section 1, direct electrochemical labeling, this made with electroactive molecules will simplify enormously the procedures of bioassays. However, highly detectable molecules as well as sensitive principles of detection are needed for replacing the amplification power of enzymes. Methylene blue is a molecule that presents a well-defined two-electron redox process [56] with cathodic (conversion to leucomethylene blue (LB)) and anodic (reoxidation to MB) peaks. The electrochemical behavior of this molecule converts it into a suitable candidate for covalent electrochemical biolabel. Since a covalent attachment does not produce a high label density (one or two MB molecules per DNA strand), the transducer has to favored electron transfer. In a previous work, it has been demonstrated that nanostructuration with MWCNTs-NH2 produces the adsorption of MB on the surface allowing a very substantial increase in the analytical signal by employing an accumulation time [50]. In Fig. 1A a cyclic voltammogram of MB on bare and nanostructured screen-printed electrodes is presented. After the nanostructuration, the increase in the reversibility of the electrochemical process allows the employ of sensitivity-enhancing electrochemical techniques such as SWV or DPV, producing a seven-time higher sensitivity. In this case, the next step is the study of the electrochemical behavior of MB covalently attached to a single-stranded DNA and comparison with that of the MB molecule. With this aim, several single MB-conjugated DNA strands have been evaluated. Examples of the cyclic, differential pulse and square wave voltammograms obtained are presented in Fig. 1B–D to show the effect that CNTs have on the recorded analytical signals. Although ACV was employed in the bibliography [26], in this case CV, DPV and SWV since they are less complex and more commonly found in commercial potentiostats. DPV was also used for recording the signal of MB covalently labeled to DNA in non-nanostructured conventional gold electrodes [27], [28]. Moreover, SWV is a fast electrochemical technique, which is very important when several assays are going to be performed.
Fig. 1.
Voltammograms recorded by CV (100 mV s−1) for 20 μM free MB (A) and 20 μM MB-DNA-MB 30-mer (B) and, by DPV (A = 0.05 V, s = 0.008 V) (C) and SWV (A = 0.05 V, s = 0.008 V, f = 50 Hz) (D) for 20 μM MB-DNA-MB 30-mer, obtained on bare (⋯) and nanostructured (—) electrodes.
3.1. Electrochemical signals of MB-DNA conjugate
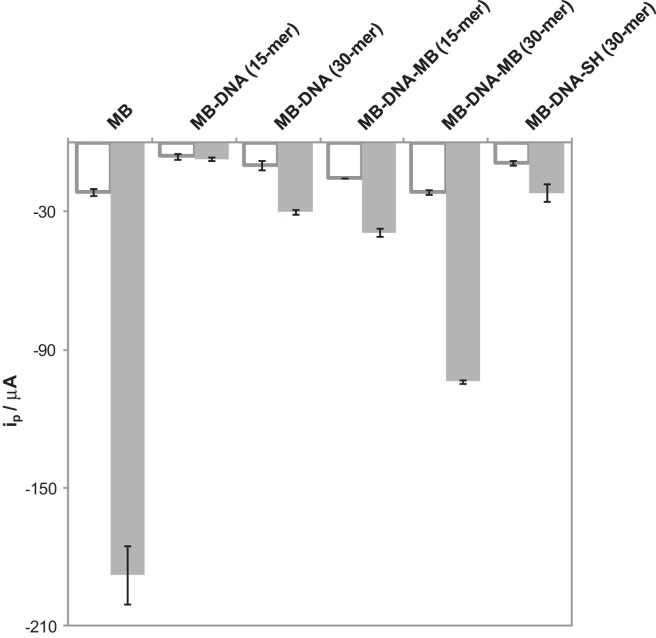
The electrochemical behavior of DNA strands with different length (15 and 30-mer) labeled with MB (either with 1 or 2 molecules per strand) was studied. Comparison between bare and nanostructured electrodes was performed by recording DP and SW voltammograms. Results obtained for SWV are shown in the diagram of bars of Fig. 2 , where the intensity corresponding to the label itself is included for the sake of comparison. Those corresponding to DPV are included in the Supplementary Data (Figure S1). The error bars are calculated for the measurements recorded on three different SPEs.
Fig. 2.
Bar diagrams obtained for different DNA-sequences (20 μM) by SWV (A = 0.05 V, s = 0.008 V, f = 50 Hz) on bare (colorless) and nanostructured (colored) electrodes.
When bare electrodes were used, much higher intensities were obtained for MB when compared with those form MB-conjugated strands. This can be explained because although the label is also MB, conjugation to DNA decreases the diffusion coefficient and therefore the peak intensities [57], [58]. On the other hand, the electrode surface can be reduced by the presence of DNA strands, decreasing then the effective area for the electron transfer. An increase in the signal is obtained for those strands with two MB molecules (MB-DNA-MB, 15 and 30-mer) attached at both ends, when compared to the respective MB-DNA of the same length. Although all the strands have the same concentration, this of MB doubles and can be an adequate approach for increasing the signals. Slightly higher values are obtained for 30-mer strands. The SWV intensities obtained were −6 ± 1 and −10 ± 2 μA for MB conjugated 15 and 30-mer respectively. These became −15.5 ± 0.1 and −21.6 ± 0.9 μA for the corresponding doubly conjugated 15 and 30-mer strands. Then, it seems that when the same number of label molecules is attached, a higher signal is obtained for longer sequences. (All data from SWV intensities were collected in Table S1 in the Supplementary Data.)
Finally, when the signal of a 30-mer MB-DNA sequence is compared to that similar but modified in the position 3′ by a thiol group (MB-DNA-SH), similar intensities were observed (−9 ± 1 μA). Similar results (reported in the Supplementary Data, Table S2) were obtained by DPV.
When SPEs were nanostructured with CNTs, intensities values were incremented considerably in all the cases. The enhancement for the molecule of MB is very notorious (more than 8.5 and 12.1 times for SWV and DPV, respectively). The same behavior than that observed for bare electrodes is here seen, that is to say, an increase with the number of labels for the same strand length, and also for the same number of labels an increase with the strand length. The highest value was obtained for the 30-mer sequence with two MB molecules (−103.9 ± 0.6 μA), meanwhile the lowest intensity was for the shortest sequence with one MB (−7 ± 1 μA (MB-DNA, 15-mer)). In the case of the DPV (Table S2 in the Supplementary Data) these values were −10 ± 1 and −1.8 ± 0.1 μA for the highest and lowest values respectively. In all the cases and similarly to bare electrodes, intensities obtained were lower than the intensity obtained for free MB, −188 ± 13 (SWV) and −34 ± 4 μA (DPV).
It can be deduced from these studies that the labeling of strands with MB reduces the analytical signal but this can be increased since higher signals were obtained for doubly labeled longer strands on nanostructured electrodes.
3.2. Nature of the MB process in DNA strands
Free MB presents a process that is controlled by diffusion on bare AuSPEs. However, MB becomes adsorbed when AuSPEs are nanostructured with CNTs and this allows a very substantial increase of the analytical signal [50]. In order to know this behavior for DNA strands, voltammograms were recorded at different scan rates ranging from 10 to 500 mV s−1 for MB-DNA (15-mer) on bare and nanostructured electrodes. On the other hand, to determine the effect of the thiol group present in DNA sequences this study was also performed for the MB-DNA-SH (30-mer). In Table 1 parameters of the linear regression from both DNA-sequences over bare and nanostructured electrodes are reported.
Table 1.
Analytical parameters obtained from measurements performed on bare and nanostructured electrodes with 20 μM ssDNA and different scan rates, from 10 to 500 mV s−1.
| DNA sequence | Electrode | ip/μA vs. | Slope | Intercept | r | n |
|---|---|---|---|---|---|---|
| MB-DNA (15-mer) | AuSPE | v1/2 | −3.4880 | 0.6398 | 0.9810 | 6 |
| v | −3.9858 | 0.0876 | 0.9983 | 6 | ||
| CNTs-AuSPE | v1/2 | −1.5210 | 0.7315 | 0.9838 | 6 | |
| v | −1.0066 | 0.1997 | 0.9998 | 6 | ||
| MB-DNA-SH (30-mer) | AuSPE | v1/2 | −3.4589 | 0.6683 | 0.9757 | 7 |
| v | −4.1164 | 0.1170 | 0.9986 | 7 | ||
| CNTs-AuSPE | v1/2 | −0.5420 | 0.0376 | 0.9857 | 7 | |
| v | −0.6699 | −0.0394 | 0.9952 | 7 | ||
v in mV s−1.
In all the cases assayed, DNA-sequences with and without thiol group, the study of scan rate shows that the electrochemical process is governed by an adsorption phenomenon because scan rate and intensities were directly related over both electrodes, unmodified as modified electrodes. Therefore, even for AuSPEs electrodes and without thiol group present, the MB process becomes adsorptive in nature.
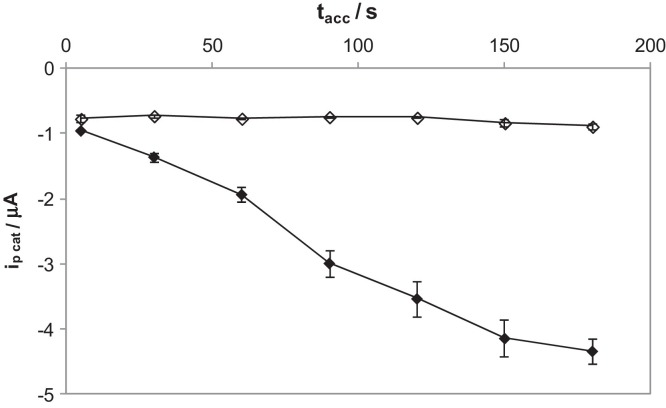
Since this adsorption process increased notoriously the sensitivity in the case of the MB molecule, by performing an accumulation step, accumulation curves were performed for the DNA that presented the higher signal, the MB-DNA-MB, 30-mer strand. This was performed on bare and nanostructured electrodes for the sake of comparison. The corresponding accumulation curves are presented in Fig. 3 . Surprisingly, it can be seen that over bare electrodes, no adsorption process is observed. It is known that there is a critical effect of the size of the oligonucleotide on the adsorption at solid electrodes that may explain this. Oligonucleotide size directly affects the adsorption process [59], so that easier adsorption of smaller molecules is produced. Meanwhile, larger molecules such as the 30-mer oligonucleotide, give weaker adsorption phenomena.
Fig. 3.
Accumulation curves for 10 μM of MB-DNA-MB 30-mer on bare (colorless) and nanostructured (colored) AuSPEs by CV (v = 100 mV s−1). Accumulation time = 5, 30, 60, 90, 120, 150, 180 s.
However, when nanostructured electrodes were used, peak currents increases with accumulation time up to 150 s, following a linear relationship (i p/μA = −0.0227t acc/s − 0.803, r = 0.997). Therefore, with two MB molecules present in the DNA-sequence, the diffusion nature of free MB is maintained on bare electrodes meanwhile this becomes adsorptive with the presence of carbon nanotubes on the electrode. This was confirmed with the relationships among peak current and scan rate. A linear relationship exists between i p and v 1/2 (i p/μA = −8.6919v 1/2/(mV s−1)1/2 − 0.0003, r = 0.9994) for bare electrodes and between i p and v for nanostructured ones (i p/μA = −5.3662v/mV s−1 − 0.2852, r = 0.9989).
3.3. Calibration plots
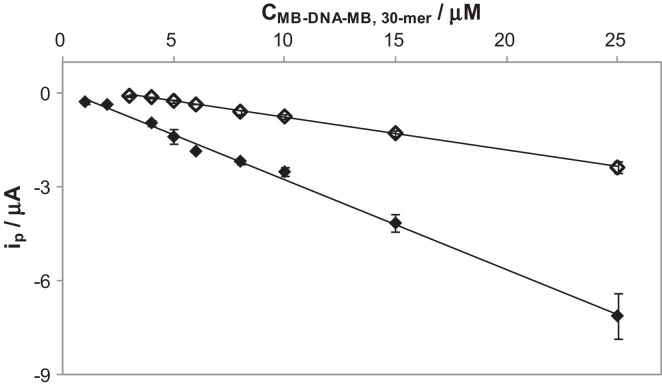
Calibration plots for the strand with the highest signal, MB-DNA-MB, 30-mer, were made by DPV on nanostructured electrodes. DPV was chosen because narrower and well-defined reduction peaks were obtained (127.0 and 87.3 mV as the half-height width for SWV and DPV, respectively). Since the simultaneous use of AuSPEs and CNTs originate a process governed by adsorption, this produces an enhancement of the analytical signal simply by the application of a short accumulation time. In this case 120 s was employed before recording the voltammogram and the calibration plot was compared to this obtained without accumulation. Graphs of the calibration plots are presented in Fig. 4 . In Table 2 are reported the parameters obtained from the linear relationship between i p (μA) and C (μM), performing three repeated observations at each concentration value. The accumulation gave a higher slope (sensitivity) with a value of −0.2864 μA μM−1, which meant an enhancement of almost 3 times. Similarly, lower detection limit (calculated as the concentration corresponding to three times the standard deviation of the intercept), 800 nM, was obtained by using accumulation time.
Fig. 4.
Calibration plots for MB-DNA-MB 30-mer obtained by DPV (A = 0.05 V, s = 0.008 V) on nanostructured electrodes without (colorless) and with 120 s (colored) of accumulation time.
Table 2.
Analytical parameters obtained from DPV (s = 0.008 V, A = 0.05 V, v = 16 mV s−1) calibration plots performed on nanostructured electrodes for the MB-DNA-MB, 30-mer strand.
| Accumulation time/s | Slope/μA μM−1 | Intercept/μA | r | n | LDR/μM | LD/μM |
|---|---|---|---|---|---|---|
| 0 | −0.1051 | 0.2745 | 0.9994 | 9 | 3–25 | 1.4 |
| 120 | −0.2864 | 0.1107 | 0.9979 | 9 | 1–25 | 0.8 |
4. Conclusions
A study of the electrochemical behavior of methylene blue covalently labeled to different SARS DNA-strands has been performed. Highly detectable labels and sensitive techniques are required. Although the signal of MB decreases due to the conjugation, labeling with two molecules and recording the signal on nanostructured electrodes allows an increase in the intensities. Nanostructuration increased the signal for all the strand evaluated (MB-DNA and MB-DNA-MB for 15 and 30-mer, and also for MB-DNA modified with a thiol group). On the other hand, higher analytical signals were obtained for longer DNA sequences.
The nature of the electrodic process was also studied and thus, when bare-AuSPEs were used the electrochemical process of MB covalently attached to DNA or DNA-SH sequences is controlled by adsorption. Only when two MB molecules are present, this process is governed by diffusion. The modification of AuSPEs with CNTs, made the electrochemical process of MB-DNA, MB-DNA-SH and MB-DNA-MB controlled by adsorption. This can be exploited for the enhancement of the sensitivity. The calibration plots for MB-DNA-MB employing a 120 s accumulation provided a limit of detection of 800 nM and an enhancement in the sensitivity of almost 3 times.
Work is in progress in order to perform a sensitive DNA sensor employing covalently labeled electroactive molecules on nanostructured electrodes.
Acknowledgment
This work has been supported by the Spanish Ministry of Science and Innovation (MICINN) under project CTQ-2011-25814.
Biographies
Raquel García-González obtained her B.Sc. degree in 2006 with the work “Electrochemical characterization of different screen printed gold electrodes by cyclic voltammetry”. Two years later she has obtained her M.Sc. entitled “Screen printed gold electrodes and their nanostructuration with carbon nanotubes: Application to methylene blue detection”. Her research is focused on electrochemistry and development of new electrodic surfaces as sensor platforms.
Agustín Costa-García obtained his B.Sc. degree in chemistry, focus in analytical chemistry, in 1974 (University of Oviedo) and the Ph.D. in chemistry in 1977 (University of Oviedo). Since February 2000 he is professor in analytical chemistry (University of Oviedo). He leads the Immunoelectroanalytical Research Group of the University of Oviedo and has been supervisor of several research projects developed at the electrochemistry laboratories of the Department of Physical and Analytical Chemistry of the University of Oviedo. Nowadays his research is focused on the development of nanostructured electrodic surfaces and its use as transducers for electrochemical immunosensors and genosensors employing electrochemical labels.
M. Teresa Fernández-Abedul received her Ph.D. in Chemistry in 1995 at University of Oviedo, Spain. Since 2002 is working as Senior Lecturer in Analytical Chemistry at the University of Oviedo. Her current research interests are the development of immunosensors and genosensors employing nanostructured transducers as well as the development of miniaturized analytical devices (microchip electrophoresis) for the sensitive electrochemical detection of analytes of interest, even those non-electroactive through adequate electroactive labeling systems.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.snb.2013.10.037.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Zhao X.J., Dytioco R.T., Tan W.H. Ultrasensitive DNA detection using highly fluorescent bioconjugated nanoparticles. Journal of the American Chemical Society. 2003;125:11474–11475. doi: 10.1021/ja0358854. [DOI] [PubMed] [Google Scholar]
- 2.Peterlinz K.A., Georgiadis R.M., Herne T.M., Tarlov M.J. Observation of hybridization and dehybridization of thiol-tethered DNA using two-color surface plasmon resonance spectroscopy. Journal of the American Chemical Society. 1997;119:3401–3402. [Google Scholar]
- 3.Okahata Y., Kawase M., Niikura K., Ohtake F., Furasawa H., Ebara Y. Kinetic measurements of DNA hybridization on an oligonucleotide-immobilized 27-MHz quartz crystal microbalance. Analytical Chemistry. 1998;70:1288–1296. doi: 10.1021/ac970584w. [DOI] [PubMed] [Google Scholar]
- 4.Wang J., Liu G.D., Merkoçi A. Electrochemical coding technology for simultaneous detection of multiple DNA targets. Journal of the American Chemical Society. 2003;125:3214–3215. doi: 10.1021/ja029668z. [DOI] [PubMed] [Google Scholar]
- 5.Xie H., Zhang C.Y., Gao Z.Q. Amperometric detection of nucleic acid at femtomolar levels with a nucleic acid/electrochemical activator bilayer on gold electrode. Analytical Chemistry. 2004;76:1611–1617. doi: 10.1021/ac0350965. [DOI] [PubMed] [Google Scholar]
- 6.Cai H., Cao X., Jiang Y., He P.G., Fang Y.Z. Carbon nanotube-enhanced electrochemical DNA biosensor for DNA hybridization detection. Analytical and Bioanalytical Chemistry. 2003;375:287–293. doi: 10.1007/s00216-002-1652-9. [DOI] [PubMed] [Google Scholar]
- 7.Wang J. Electroanalysis and biosensors. Analytical Chemistry. 1999;71:328–332. doi: 10.1021/a1999905e. [DOI] [PubMed] [Google Scholar]
- 8.Mikkelson S.R. Electrochemical biosensors for DNA sequence detection. Electroanalysis. 1996;8:15–19. [Google Scholar]
- 9.Lucarelli F., Marrazza G., Palchetti I., Cesaretti S., Mascini M. Coupling of an indicator-free electrochemical DNA biosensor with polymerase chain reaction for the detection of DNA sequences related to the apolipoprotein E. Analytica Chimica Acta. 2002;469:93–99. [Google Scholar]
- 10.Kerman K., Morita Y., Takamura Y., Tamiya E. Escherichia coli single-strand binding protein–DNA interactions on carbon nanotube-modified electrodes from a label-free electrochemical hybridization sensor. Analytical and Bioanalytical Chemistry. 2005;381:1114–1121. doi: 10.1007/s00216-004-3007-1. [DOI] [PubMed] [Google Scholar]
- 11.Arias P., Ferreyra N.F., Rivas G.A., Bollo S. Glassy carbon electrodes modified with CNT dispersed in chitosan: analytical applications for sensing DNA–methylene blue interaction. Journal of Electroanalytical Chemistry. 2009;634:123–126. [Google Scholar]
- 12.Martínez-Paredes G., González-García M.B., Costa-García A. Genosensor for detection of four pneumoniae bacteria using gold nanostructured screen-printed carbon electrodes as transducers. Sensors and Actuators B: Chemical. 2010;149:329–335. [Google Scholar]
- 13.de la Escosura-Muñiz A., González-García M.B., Costa-García A. DNA hybridization sensor based on aurothiomalate electroactive label on glassy carbon electrodes. Biosensors and Bioelectronics. 2007;22:1048–1054. doi: 10.1016/j.bios.2006.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernández-Santos D., González-García M.B., Costa-García A. Genosensor based on a platinum(II) complex as electrocatalytic label. Analytical Chemistry. 2005;77:2868–2874. doi: 10.1021/ac048091w. [DOI] [PubMed] [Google Scholar]
- 15.Kerman K., Ozkan D., Kara P., Meric B., Gooding J.J., Ozsoz M. Voltammetric determination of DNA hybridization using methylene blue and self-assembled alkanethiol monolayer on gold electrodes. Analytica Chimica Acta. 2002;462:39–47. [Google Scholar]
- 16.Zhu N., Zhang A., Wang Q., He P., Fang Y. Electrochemical detection of DNA hybridization using methylene blue and electro-deposited zirconia thin films on gold electrodes. Analytica Chimica Acta. 2004;510:163–168. [Google Scholar]
- 17.Boon E.M., Barton J.K. DNA electrochemistry as a probe of base pair stacking in A-, B-, and Z-form DNA. Bioconjugate Chemistry. 2003;14:1140–1147. doi: 10.1021/bc034139l. [DOI] [PubMed] [Google Scholar]
- 18.Boon E.M., Jackson N.M., Wightman M.D., Kelley S.O., Hill M.G., Barton J.K. Intercalative stacking: a critical feature of DNA charge-transport electrochemistry. The Journal of Physical Chemistry B. 2003;107:11805–11812. [Google Scholar]
- 19.Pau D., Zuo X., Wan Y., Wang L., Zhang J., Song S., Fan C. Electrochemical interrogation of interactions between surface-confined DNA and methylene blue. Sensors. 2007;7:2671–2680. doi: 10.3390/s7112671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuite E., Norden B. Sequence-specific interactions of methylene blue with polynucleotides and DNA: a spectroscopic study. Journal of the American Chemical Society. 1994;116:7548–7556. [Google Scholar]
- 21.Wang Y., Zhou A. Spectroscopic studies on the binding of methylene blue with DNA by means of cyclodextrin supramolecular systems. Journal of Photochemistry and Photobiology A: Chemistry. 2007;190:121–127. [Google Scholar]
- 22.Farjami E., Clima L., Gothelf K.V., Ferapontova E.E. DNA interactions with a methylene blue redox indicator depend on the DNA length and are sequence specific. Analyst. 2010;135:1443–1448. doi: 10.1039/c0an00049c. [DOI] [PubMed] [Google Scholar]
- 23.Baranovskii S.F., Bolotin P.A., Evstigneev M.P., Chernyshev D.N. Complexation of heterocyclic ligands with DNA in aqueous solution. Journal of Applied Spectroscopy. 2008;75:251–260. [Google Scholar]
- 24.Rohs R., Sklenar H., Lavery R., Roder B. Methylene blue binding to DNA with alternating GC base sequence: a modeling study. Journal of the American Chemical Society. 2000;122:2860–2866. [Google Scholar]
- 25.Kelley S.O., Barton J.K., Jackson N.M., Hill M.G. Electrochemistry of methylene blue bound to a DNA-modified electrode. Bioconjugate Chemistry. 1997;8:31–37. doi: 10.1021/bc960070o. [DOI] [PubMed] [Google Scholar]
- 26.Lubin A.A., Lai R.Y., Baker B.R., Heeger A.J., Plaxco K.W. Sequence-specific, electronic detection of oligonucleotides in blood, soil, and foodstuffs with the reagentless, reusable E-DNA sensor. Analytical Chemistry. 2006;78:5671–5677. doi: 10.1021/ac0601819. [DOI] [PubMed] [Google Scholar]
- 27.Pänke O., Kirbs A., Lisdat F. Voltammetric detection of single base-pair mismatches and quantification of label-free target ssDNA using a competitive binding assay. Biosensors and Bioelectronics. 2007;22:2656–2662. doi: 10.1016/j.bios.2006.10.033. [DOI] [PubMed] [Google Scholar]
- 28.Kafka J., Pänke O., Abendroth B., Lisdat F. A label-free DNA sensor based on impedance spectroscopy. Electrochimica Acta. 2008;53:7467–7474. [Google Scholar]
- 29.Authier L., Grossiord C., Brossier P., Limoges B. Gold nanoparticle-based quantitative electrochemical detection of amplified human cytomegalovirus DNA using disposable microband electrodes. Analytical Chemistry. 2001;73:4450–4456. doi: 10.1021/ac0103221. [DOI] [PubMed] [Google Scholar]
- 30.Cai H., Su C., He P., Fang Y. Colloid Au-enhanced DNA immobilization for the electrochemical detection of sequence-specific DNA. Journal of Electroanalytical Chemistry. 2001;510:78–85. [Google Scholar]
- 31.Patolsky F., Weizmann Y., Willner I. Redox-active nucleic-acid replica for the amplified bioelectrocatalytic detection of viral DNA. Journal of the American Chemical Society. 2002;124:770–772. doi: 10.1021/ja0119752. [DOI] [PubMed] [Google Scholar]
- 32.Nakayama M., Ihara T., Nakano K., Maeda M. DNA sensors using a ferrocene-oligonucleotide conjugate. Talanta. 2002;56:857–866. doi: 10.1016/s0039-9140(01)00659-2. [DOI] [PubMed] [Google Scholar]
- 33.Wang J., Liu G., Merkoçi A. Particle-based detection of DNA hybridization using electrochemical stripping measurements of an iron tracer. Analytica Chimica Acta. 2003;482:149–155. [Google Scholar]
- 34.Castañeda M.T., Merkoçi A., Pumera M., Alegret S. Electrochemical genosensors for biomedical applications based on gold nanoparticles. Biosensors and Bioelectronics. 2007;22:1961–1967. doi: 10.1016/j.bios.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 35.Fotja M., Brázdilová P., Cahová K., Pečinka P. A single-surface electrochemical biosensor for the detection of DNA triplet repeat expansion. Electroanalysis. 2006;18:141–151. [Google Scholar]
- 36.Bianco A. Carbon nanotubes for the delivery of therapeutic molecules. Expert Opinion on Drug Delivery. 2004;1:57–65. doi: 10.1517/17425247.1.1.57. [DOI] [PubMed] [Google Scholar]
- 37.Malarkey E.B., Parpura V. Applications of carbon nanotubes in neurobiology. Neurodegenerative Diseases. 2007;4:292–299. doi: 10.1159/000101885. [DOI] [PubMed] [Google Scholar]
- 38.Yang R., Yang X., Zhang Z., Zhang Y., Wang S., Cai Z. Single-walled carbon nanotubes-mediated in vivo and in vitro delivery of siRNA into antigen-presenting cells. Gene Therapy. 2006;13:1714–1723. doi: 10.1038/sj.gt.3302808. [DOI] [PubMed] [Google Scholar]
- 39.Star A., Gabriel J.C.P., Bradley K., Gruner G. Electronic detection of specific protein binding using nanotube FET devices. Nanoletters. 2003;3:459–463. [Google Scholar]
- 40.Ajayan P.M. Nanotubes from carbon. Chemical Reviews. 1999;99:1787–1799. doi: 10.1021/cr970102g. [DOI] [PubMed] [Google Scholar]
- 41.Teo K.B.K., Singh C., Chhowalla M., Milne W.I. In: Encyclopedia of Nanoscience and Nanotechnology. Nalwa H.S., editor. American Scientific Publishers; Stevenson Ranch, CA: 2003. pp. 1–22. [Google Scholar]
- 42.Collins P.G., Avouris P. Nanotubes for electronics. Scientific American. 2000;283:62–69. doi: 10.1038/scientificamerican1200-62. [DOI] [PubMed] [Google Scholar]
- 43.Dresselhaus M.S., Dresselhaus G., Jorio J. Unusual properties and structure of carbon nanotubes. Annual Review of Materials Research. 2004;34:247–278. [Google Scholar]
- 44.Hollingsworth J.P., Bandaru P. Carbon nanotube based nonvolatile memory. Applied Physics Letters. 2005;87:233115–233117. [Google Scholar]
- 45.Merkoçi A., Pumera M., Llopis X., Pérez B., del Valle M., Alegret S. New materials for electrochemical sensing. VI: Carbon nanotubes. TrAC – Trends in Analytical Chemistry. 2005;24:826–838. [Google Scholar]
- 46.Wang J., Liu G., Jan M.R. Ultrasensitive electrical biosensing of proteins and DNA: carbon-nanotube derived amplification of the recognition and transduction events. Journal of the American Chemical Society. 2004;126:3010–3011. doi: 10.1021/ja031723w. [DOI] [PubMed] [Google Scholar]
- 47.Robertson J. Realistic applications of CNTs. Materials Today. 2004;7:46–52. [Google Scholar]
- 48.Luo H., Shi Z., Li N., Gu Z., Zhuang Q. Investigation of the electrochemical and electrocatalytic behavior of single-wall carbon nanotube film on a glassy carbon electrode. Analytical Chemistry. 2001;73:915–920. doi: 10.1021/ac000967l. [DOI] [PubMed] [Google Scholar]
- 49.Musameh M., Wang J., Merkoçi A., Lin Y. Low-potential stable NADH detection at carbon-nanotube-modified glassy carbon electrodes. Electrochemistry Communications. 2002;4:743–746. [Google Scholar]
- 50.García-González R., Costa-García A., Fernández-Abedul M.T. Sensors and Actuators B: Chemical. 2013 doi: 10.1016/j.snb.2013.10.037. (submitted for publication) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fanjul-Bolado P., Queipo P., Lamas-Ardisana P.J., Costa-García A. Manufacture and evaluation of carbon nanotube modified screen-printed electrodes as electrochemical tools. Talanta. 2007;74:427–433. doi: 10.1016/j.talanta.2007.07.035. [DOI] [PubMed] [Google Scholar]
- 52.Feng Y., Gao G.F. Towards our understanding of SARS-CoV, an emerging and devastating but quickly conquered virus. Comparative Immunology, Microbiology and Infectious Diseases. 2007;30:309–327. doi: 10.1016/j.cimid.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berger A., Drosten Ch., Doerr H.W., Stürmer M., Preiser W. Severe Acute Respiratory Syndrome (SARS) – paradigm of an emerging viral infection. Journal of Clinical Virology. 2004;29:13–22. doi: 10.1016/j.jcv.2003.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marra M.A., Jones S.J.M., Astell C.R., Holt R.A. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 55.García-González R., Fernández-La-Villa A., Costa-García A., Fernández-Abedul M.T. Dispersion studies of carboxyl, amine and thiol-functionalized carbon nanotubes for improving the electrochemical behavior of screen printed electrodes. Sensors and Actuators B: Chemical. 2013;181:353–360. [Google Scholar]
- 56.Boon E.M., Ceres D.M., Drummond T.G., Hill M.G., Barton J.K. Mutation detection by electrocatalysis at DNA-modified electrodes. Nature Biotechnology. 2000;18:1096–1100. doi: 10.1038/80301. [DOI] [PubMed] [Google Scholar]
- 57.Randles J.E. A cathode ray polarograph. Part II. The current–voltage curves. Transactions of the Faraday Society. 1948;44:327–338. [Google Scholar]
- 58.Sevcik A. Oscillographic polarography with periodical triangular voltage. Collection of Czechoslovak Chemical Communications. 1948;13:349–377. [Google Scholar]
- 59.Pedano M.L., Rivas G.A. Immobilization of DNA on glassy carbon electrodes for the development of affinity biosensors. Biosensors and Bioelectronics. 2003;18:269–277. doi: 10.1016/s0956-5663(02)00176-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.