Graphical abstract
Keywords: Shrimp, Covert mortality nodavirus, Reverse transcription loop-mediated isothermal amplification, RT-PCR, Litopenaeus vannamei
Highlights
-
•
A sensitive and specific detection method for covert mortality nodavirus was developed.
-
•
The detection threshold of the assay is 27 viral copies.
-
•
The developed assay is very promising for CMNV detection and quantification.
Abstract
A disease known as covert mortality disease has become an increasing problem in the shrimp farming industry in recent years in China and several countries of Southeast Asia, leading to serious losses in production. Litopenaeus vannamei (also known as Pacific white shrimp) is affected by this disease that leads to a range of clinical symptoms including hepatopancreas atrophy and necrosis, soft shell, slow growth, and abdominal muscle whitening and necrosis in the acute stage of disease. A new nodavirus, termed covert mortality nodavirus (CMNV), has been shown to be the etiological agent. In this study, we report a sensitive and specific real-time reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for the rapid and quantitative detection of CMNV. The optimal conditions for this newly developed RT-LAMP reaction were found to be 6 mM MgCl2 and 1.6 mM dNTPs, an incubation temperature of 65 °C and a reaction time of 50 min. The analytical sensitivity of the RT-LAMP assay was estimated to be 6.3 pg total RNA of CMNV-infected shrimp and 27 copies of the target plasmid. The diagnostic sensitivity and specificity of the newly developed assay versus the standard nested reverse transcription PCR (RT-PCR) assay was 96.4% and 94.4%, respectively. The reaction products were detected by visual inspection after staining with an in-tube DNA fluorescent dye, a measure taken to eliminate the risk of contamination. The quantitative RT-LAMP assay for CMNV showed high correlation coefficient (r2 = 0.9953) when the initial templates were above 1000 copies, however the correlation coefficient decreased when the initial templates were lower than 1000 copies. Test of viral load in shrimp indicated that the viral loads varied from 1.5 × 102 to 6.7 × 106 copies per mg of cephalothorax tissue. Thus, the CMNV RT-LAMP assay is a sensitive and specific new tool for the field detection and quantification of CMNV in the diagnosis and surveillance of covert mortality disease.
1. Introduction
Litopenaeus vannamei, also known as Pacific white shrimp, is an economically important variety of shrimp. However, production of L. vannamei is limited by its susceptibility to various diseases. In China, covert mortality disease (CMD) has been reported in pond cultures of L. vannamei recently (Xu and Ji, 2009, Zhang et al., 2014). The epidemiology study proved the prevalence of CMD in shrimp ponds in Thailand, Indonesia, India and Ecuador (Flegel, 2014). Typical clinical signs of CMD in the shrimp include hepatopancreas atrophy and necrosis, soft shell, slow growth, and abdominal muscle whitening and necrosis in the acute stage (Gu, 2012, Huang, 2012, Zhang, 2004, Zhang et al., 2014). The symptom of abdominal muscle whitening and necrosis is similar to the symptoms reported in the early stages of disease caused by infectious myonecrosis virus (IMNV) and Penaeus vannamei nodavirus (PvNV) in the shrimp. However, these diseases can be distinguished by RT-PCR (Zhang et al., 2014). CMD-related mortality has been found to be sporadic, and may be increased by environmental stress, such as high temperature. When the water temperature rises above 28 °C, survival in infected ponds decreases sharply and the cumulative mortality rate of shrimps infected by CMD has been reported to be up to 80% (Gu, 2012, Xing, 2004, Xu and Ji, 2009).
A range of techniques were employed to identify the etiological agent of CMD including histopathology, virion purification, sequence analysis, experimental infection, and fluorescence in situ hybridization. A new nodavirus, designated as covert mortality nodavirus (CMNV) (Zhang et al., 2014), was found to be the causative agent of CMD. Like other members of the genus Alphanodavirus, the genome of CMNV is a positive single-stranded RNA. Histological examination of the diseased L. vannamei shrimp revealed atrophied, structurally discorded, hepatopancreas tubules and coagulative necrosis of skeletal muscle. Eosinophilic inclusions were detected in the hepatopancreas and lymphoid organ (Zhang et al., 2014). Specimens of the main economic species of shrimp in China, Fenneropenaeus chinensis (Chinese white shrimp) and Marsupenaeus japonicus (Japanese tiger prawn), were also found to be infected by CMNV in an epidemiological investigation conducted using histological examination and RT-PCR for virus detection (Zhang et al., 2014). Therefore, the risk of a CMNV pandemic in both farmed and wild shrimp is substantial.
In an attempt to control the spread of CMNV, rapid and effective detection methods for CMNV are urgently required. In this study, we developed a rapid and sensitive reverse transcription loop-mediated isothermal amplification (RT-LAMP) method for the detection of CMNV in infected shrimp. The newly developed assay also has the advantage of quantifying CMNV in infected samples.
2. Materials and methods
2.1. Shrimp samples and the preparation of template RNA
Fifty-two living L. vannamei samples, ranging from 8 to 10 g in weight, were obtained from ponds of shrimp farms in Hebei, Shandong, Jiangsu and Zhejiang Provinces and 16 samples were determined as CMNV positive by using the previously described RT-PCR method (Zhang et al., 2014). The rest of 36 samples were CMNV negative by the RT-PCR method. In addition, to validate of the newly developed RT-LAMP method, 12 experimentally infected shrimp individuals with typical syndrome of CMD were collected. Total RNA of the shrimp was extracted using the RNAprep™ Pure Tissue Kit (Tiangen, Beijing, China) following the manufacturer’s protocol and was stored at −80 °C prior to use. For RT-PCR, CMNV cDNA was synthesized using the Reverse Transcription System (Promega, Madison, WI, USA) and random primers.
2.2. Primers and the RT-LAMP assay
A partial sequence of the RNA-dependent RNA polymerase gene in the RNA1 genome of CMNV (GenBank accession number KM112247) was used as a template to design the RT-LAMP primer set using the Primer Explorer software, version 4.0 (http://primerexplorer.jp/elamp4.0.0/index.html). The details of the primers are shown in Table 1 .
Table 1.
Primers used for reverse transcriptase loop-mediated isothermal amplification.
| Primer | Sequence |
|---|---|
| F3 | TGCCAAGCAAATACGAGCT |
| B3 | CATCAGCGATGTCACGGC |
| FIP | GTCGTCGACGGTTAGGTTGCGTTTTCCAAGCACTTCCCGACAA |
| (F1c + TTTT + F2) | |
| BIP | CGTCCAAAAGGACCTCCGCATTTTTGGAGACCTTGGTCACGC |
| (B1c + TTTT + B2) | |
| LF | GCTCACGGCTTTGGATACC |
| LB | GATTGCATGCGTCAACCTCA |
The RT-LAMP reaction was carried out in a 25-μL reaction mixture containing: 1.6 μM of each of the FIP and BIP primers; 0.2 μM of each of the F3 and B3 primers; 1× isothermal amplification buffer (20 mM Tris–HCl, 10 mM (NH4)2SO4, 50 mM KCl, 2 mM MgSO4, 0.1% Tween 20, pH 8.8); 6 mM MgCl2; 1 M betaine; 1.4 mM deoxynucleoside triphosphates (dNTPs); 0.2 mM MnCl2; 20 μM calcein; 2 U M-MLV reverse transcriptase (Promega); 8 U Bst DNA polymerase (New England Biolabs, Beverly, MA, USA) and 1 μL template RNA. Calcein was used as a fluorescent indicator which yields strong fluorescence by forming complexes with divalent magnesium ions in LAMP reaction as report by Tomita et al. (2008). The mixture was prepared on ice and then immediately incubated for 50 min (one circle per minute) in a gradient PCR machine at 59, 62, 65 or 68 °C, followed by 80 °C for 20 min to terminate the reaction. Changes in fluorescence were monitored every min at 520 nm. To determine the optimum concentration of MgCl2 and dNTPs, the RT-LAMP reaction was optimized by Taguchi’s L16 (2(4)) orthogonal design with two elements (dNTPs and MgCl2) at four concentration levels (Table 2 ).
Table 2.
The result of optimization of concentration of Mg2+ and dNTP.
| Concentration of MgCl2 (mM) | Concentration of dNTP (mM) | Meana of Ct value | SDa of Ct value |
|---|---|---|---|
| 4 | 1.2 | 23.40 | 1.09 |
| 4 | 1.4 | 22.38 | 0.65 |
| 4 | 1.6 | 25.25 | 0.16 |
| 4 | 1.8 | 28.51 | 1.38 |
| 6 | 1.2 | 26.63 | 0.36 |
| 6 | 1.4 | 23.06 | 0.74 |
| 6 | 1.6 | 23.31 | 0.04 |
| 6 | 1.8 | 27.31 | 0.59 |
| 8 | 1.2 | 31.29 | 0.59 |
| 8 | 1.4 | 29.02 | 0.38 |
| 8 | 1.6 | 25.90 | 0.32 |
| 8 | 1.8 | 25.09 | 0.28 |
| 10 | 1.2 | 33.81 | 1.12 |
| 10 | 1.4 | 29.86 | 0.35 |
| 10 | 1.6 | 28.23 | 0.08 |
| 10 | 1.8 | 27.49 | 0.17 |
Mean and SD were produced from the repeats of RT-LAMP assay in triplicate.
2.3. Analytical specificity of CMNV RT-LAMP primers
The analytical specificity of the RT-LAMP primers was tested under the optimized conditions described above. Nucleic acids from other shrimp pathogenic viruses, such as Macrobrachium rosenbergii nodavirus (MrNV), Taura syndrome virus (TSV), yellow head virus (YHV) and white spot syndrome virus (WSSV), were used in this analysis. Alignment of the target gene sequence of 179 bp of CMNV against the corresponding sequences of PvNV (GenBank accession number HQ259079.1) and MrNV (GenBank accession number FJ751226.1) was performed for comparison using the software of BioEdit 7.0 and BLAST.
2.4. Analytical sensitivity of CMNV RT-LAMP
The detection limit of the RT-LAMP assay was analyzed using two different templates: (1) the total RNA extracted from the shrimp infected by CMNV, (2) a plasmid vector (pMD19-T) containing the target fragment from the RNA-dependent RNA polymerase gene of CMNV (designated pMD19-T-CMNV). A 10-fold serial dilution of the total RNA (6.2 pg × 105–10−1) from CMNV-infected shrimp and plasmid pMD19-T-CMNV (2.7 × 108–101 copies) was used as a template for RT-LAMP under the predetermined conditions.
2.5. Evaluation of the CMNV RT-LAMP assay on clinical samples
The validity of the CMNV RT-LAMP was determined by testing 64 clinical samples, including: (1) 52 cephalothoraxes tissue samples collected from spontaneously infected L. vannamei from shrimp farms in 2014, (2) 12 cephalothoraxes tissue samples collected from artificially infected L. vannamei in the laboratory. Total RNA of the samples was extracted and the optimized CMNV RT-LAMP was performed as described in Section 2.2. RT-PCR assays were also performed on the same total RNA samples according to Zhang et al. (2014). The diagnostic sensitivity (DSe) and diagnostic specificity (DSp) of the two methods, as defined by the World Organization for Animal Health (2011), were calculated according to Zhang et al. (2013).
2.6. Detection of CMNV RT-LAMP products with fluorescent dyes
The CMNV RT-LAMP was performed according to the optimal protocol described in Section 2.2, except that calcein and MnCl2 were omitted. One microliter of fluorescent dye GeneFinder™ (Bio-V, Xiamen, China) was sealed into the cap of the reaction tube in advance using paraffin wax. At the end of the reaction, fluorescence development was initiated by incubating the tube at 95 °C for 5 min and then immediately mixing the GeneFinder™ with the reaction mixture. Different fluorescent colors appeared in the reaction mixtures of the positive and negative samples.
2.7. Quantitative CMNV RT-LAMP assay
To determine viral load in the shrimp tissues, the RNA extracted from the cephalothorax of the 64 shrimp clinical samples described in Section 2.5 was amplified by quantitative RT-LAMP (qRT-LAMP). The RT-LAMP assay was quantified using 10-fold dilutions of CMNV plasmid (pMD19-T-CMNV) as standards, in a final reaction volume of 25 μL. For real-time monitoring, the qRT-LAMP reactions were incubated at 65 °C for 50 cycles (1 min per cycle) with a CFX Connect™ Real-Time PCR Detection System (Bio-Rad, Foster City, CA, USA). For CMNV quantitative detection of samples, a standard curve was generated for CMNV qRT-LAMP by plotting a graph between different concentrations of pMD19-T-CMNV plasmids, ranging from 108 to 100 copy numbers, to cycle threshold (Ct) values, obtained through real-time monitoring of the amplification.
3. Results
3.1. Optimization of the CMNV RT-LAMP reaction conditions
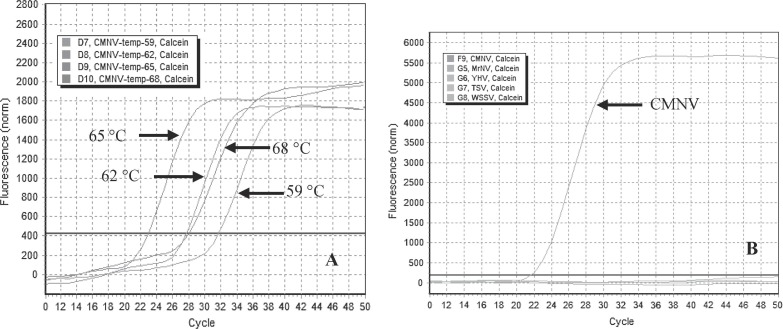
RT-LAMP reactions were performed using 4 mM MgCl2 and 1.5 mM dNTPs for 50 min at 56, 59, 62, 65 or 68 °C to determine the optical reaction temperature. The results showed that the amplification occurred earlier when the reaction was incubated at 65 °C compared with the other incubation temperatures (Fig. 1 A). Experiments to optimize the concentrations of MgCl2 and dNTPs showed that the smallest average Ct value (22.38) was generated when the concentrations of MgCl2 and dNTPs were 4.0 mM and 1.4 mM, respectively. However, the smallest average Ct value was accompanied by a standard error of 0.65, indicating substantial fluctuations in amplification efficiency. Meanwhile, the third smallest Ct value (23.31), with the smallest standard error of 0.04, was obtained when the concentrations of MgCl2 and dNTPs were 6.0 mM and 1.6 mM, respectively. Therefore, the optimal concentrations of MgCl2 and dNTPs were determined to be 6.0 mM and 1.6 mM, respectively. The real-time kinetics of the RT-LAMP reaction, with or without loop primers, was analyzed with 6.0 mM of MgCl2, 1.6 mM of dNTPs and incubation at 65 °C. The results indicated that the time required for initiation of amplification was 29.6 min with loop primers or 46.5 min without loop primers, which indicated that using loop primers accelerated the amplification, thereby reducing the detection time, compared with RT-LAMP without loop primers. Based on these findings, further RT-LAMP assays were incubated for 50 min at 65 °C with 6 mM MgCl2 and 1.6 mM dNTPs.
Fig. 1.
Optimization of the CMNV qRT-LAMP assay. A. Effect of temperature on the qRT-LAMP reaction using 6 mM MgCl2. The following temperatures were tested: 65, 62, 68 and 59 °C (from left to right). B. Specificity of the qRT-LAMP assay for the detection of CMNV. Calcein indicated the fluorescent indicator used in the qRT-LAMP assay.
3.2. Analytical specificity of the CMNV RT-LAMP primers
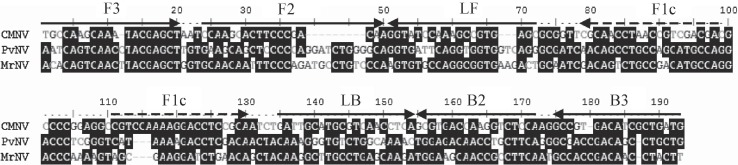
Alignment of the CMNV-LAMP target sequence (179 bp) with the corresponding sequences from the closely related viruses, PvNV and MrNV, indicated that the eight CMNV RT-LAMP primers covered 86 or more mutation sites in the corresponding sequences of PvNV and MrNV (Fig. 2 ). Amplification only occurred when the template was the RNA from CMNV; no amplification was observed when the template was the nucleic acid from MrNV, YHV, TSV or WSSV (Fig. 1B). Furthermore, both BLAST and EMBL searches revealed that the CMNV-LAMP target sequence did not align with any other viral sequences available in these databases, including all members of the Nodaviridae, such as the flock house virus (FHV), black beetle virus (BBV), MrNV and PvNV. Taken together, these results indicate that the RT-LAMP primer set is specific for amplification of CMNV nucleic acid.
Fig. 2.
Alignment of the CMNV target gene (KM112247) region with the corresponding sequences of closely related viruses, Penaeus vannamei nodavirus (PvNV) and Macrobrachium rosenbergii nodavirus (MrNV) (GenBank accession numbers HQ259079.1 and FJ751226.1). The eight CMNV RT-LAMP primers cover at least 86 mutation sites in the corresponding sequences of PvNV and MrNV. F: forward primer, B: backward primer, LF: loop-forward primer, LB: loop-backward primer.
3.3. Analytical sensitivities of the CMNV RT-LAMP assay
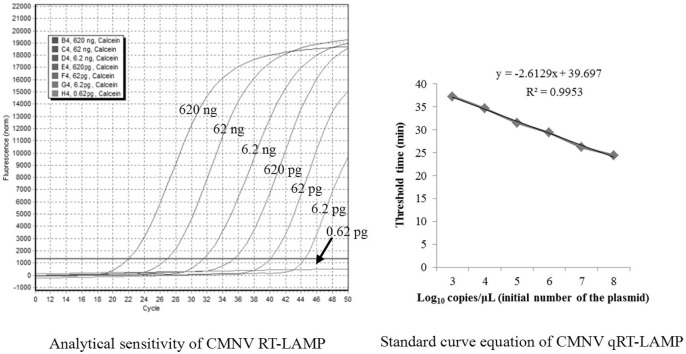
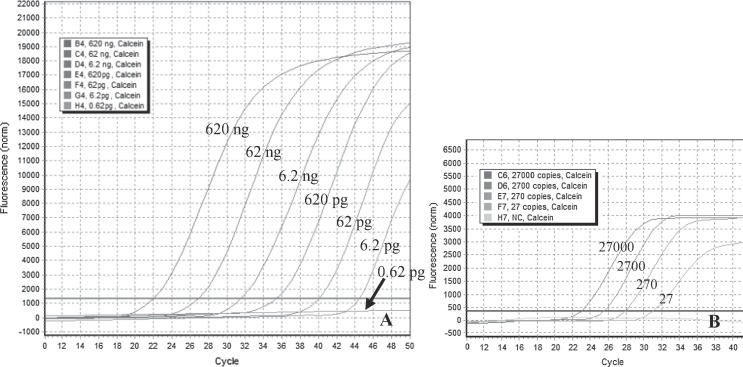
The lowest detection limit of the newly developed CMNV RT-LAMP method was 6.2 pg of total RNA when the reactions were tested using 1 μL of 10-fold serially diluted RNA from shrimp artificially infected with CMNV (Fig. 3 A). When the reaction was tested using 1 μL of 10-fold serially diluted pMD19-T-CMNV DNA (8.5 ng/μL equivalent to 2.7 × 109 copies/μL), the analytical sensitivity of the RT-LAMP method was estimated to be as low as 27 copies of the plasmid (Fig. 3B).
Fig. 3.
Sensitivity of the RT-LAMP assay for the detection of CMNV RNA. A. Amplification plots 1–7 (from left to right), reaction conducted using 10-fold serial dilutions of RNA from shrimp infected by CMNV: 6.2 × 105, 6.2 × 104, 6.2 × 103, 6.2 × 102, 6.2 × 101, 6.2 and 0.62 pg of RNA, respectively. B. Sensitivity of RT-LAMP detection of pMD19-T-CMNV plasmid containing the target DNA fragments (the RNA-dependent RNA polymerase gene of CMNV). Amplification plots 1–5 (from left to right), reaction conducted using 10-fold serial dilutions of the plasmid (pMD19-T-CMNV): 2.7 × 104, 2.7 × 103, 2.7 × 102, 2.7 × 101 and 2.7 plasmid copies, respectively. Calcein indicated the fluorescent indicator used in the qRT-LAMP assay.
3.4. Evaluation of the newly developed CMNV RT-LAMP assay on clinical samples
RNA from 64 clinical samples was used to compare the newly developed RT-LAMP assay with conventional RT-PCR methods. RT-PCR results indicated that 28 of the 64 samples were positive for CMNV. The RT-LAMP assay showed that 27 samples from the 28 CMNV-positive samples determined by RT-PCR gave positive results. Moreover, two of the 36 samples that were CMNV negative by RT-PCR yielded a positive result with the RT-LAMP assay (Table 3 ). Therefore, the DSe and DSp values for the RT-LAMP method compared with the RT-PCR method were 96.4% and 94.4%, respectively.
Table 3.
The comparison of detection results of the newly developed RT-LAMP and RT-PCR.
| Detection result | RT-PCR positive | RT-PCR negative | Total |
|---|---|---|---|
| CMNV-RT-LAMP positive | 27 | 2 | 29 |
| CMNV-RT-LAMP negative | 1 | 34 | 35 |
| Total | 28 | 36 | 64 |
3.5. Detection of CMNV RT-LAMP products with fluorescent dyes
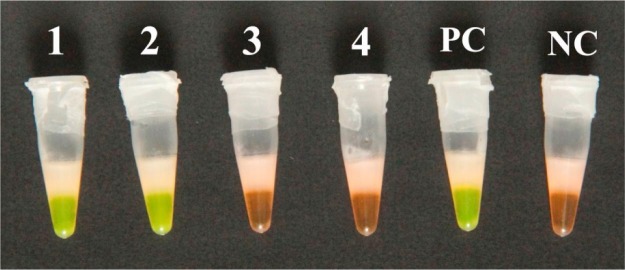
Visual inspection of CMNV RT-LAMP products was performed by adding a fluorescent dye to the reaction mixture. Green fluorescence was observed clearly with the naked eye in the reaction tubes of the CMNV-infected shrimp and the positive control, whereas an orange color was observed in the tubes of the negative control and the CMNV-free shrimp (Fig. 4 ).
Fig. 4.
Visual inspection of the CMNV RT-LAMP results. The amplification products (35 μL) in the reaction tubes were dyed using 1 μL of GeneFinder™ (1:35 dilution), which was sealed into the cap of the tube in advance using paraffin wax. Negative reactions appeared orange and positive reactions appeared green in daylight against a black background. Tubes 1 and 2: samples from spontaneously infected shrimps; tubes 3 and 4: samples from shrimps without signs of CMD; PC: positive control (pMD19-T-CMNV plasmid); NC: negative control (sterile water). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.6. CMNV quantitative RT-LAMP assay
A high correlation coefficient (r 2 = 0.9953) was obtained for the CMNV quantitative RT-LAMP (qRT-LAMP) assay when the initial template was above 1000 copies (Fig. 5 ). Quantitation of viral copies in the cephalothorax of the 64 clinical samples was calculated based on the Ct value of all RNA samples using the generated standard curve. The viral loads of the positive clinical samples varied within the range 1.5 × 102–6.7 × 106 copy number per mg of tissue.
Fig. 5.
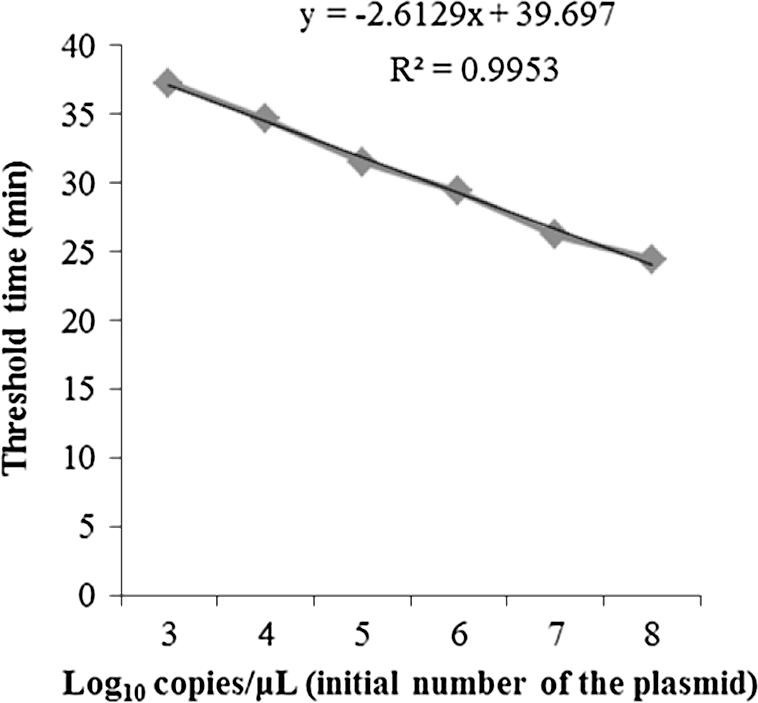
Standard curve equation for the CMNV-specific qRT-LAMP assay generated from the amplification plots for the 10-fold serially diluted pMD19-T-CMNV plasmid and the Ct value.
4. Discussion
The optimal reaction temperature was determined to be 65 °C and the optimal concentration of MgCl2 and dNTPs was determined to be 6 mM and 1.6 mM, respectively. These conditions are similar to those reported for LAMP assays for the detection of other organisms such as hepatitis B virus (Nagamine et al., 2001) and the toxic dinoflagellate Alexandrium (Wang et al., 2008). The results of the temperature optimization showed that Bst DNA polymerase effectively amplified the nucleic acid templates at temperatures from 62 to 68 °C, which is consistent with other classic LAMP assays (Mori et al., 2001, Mori et al., 2006, Notomi et al., 2000). The effectiveness of this assay across a wide temperature range would greatly benefit the future application of this method under field conditions. With regard to reaction time, when 21 ng of RNA (approximately 106 copies of the virus, as determined by quantitative RT-LAMP) from CMNV-infected shrimp was used as template, initiation of amplification took 29.6 min with loop primers and saved 16.9 min comparing with the reaction without loop primers, indicating that the presence of loop primers accelerated the LAMP reaction (Nagamine et al., 2002, Parida et al., 2004, Yang et al., 2012). The development of a rapid RT-LAMP assay for CMDV detection would be advantageous in the field.
To develop a detection assay with high specificity for the target species, it is important that the primers employed do not cross react with other viruses. In this study, we needed to ensure that the primers used for the RT-LAMP assay allowed for the detection of CMNV but did not cross react with other shrimp virus species, especially with other members of the genus Alphanodavirus. The 179-bp target sequence using for primer designing in this study shared no similarity with any other viral sequences available in the GenBank database. Specificity testing confirmed that amplification only occurred when nucleic acid from CMNV was used as template; the assay developed in this study can therefore be used for the specific detection of CMNV.
The analytical sensitivities of the RT-LAMP method were 6.2 pg of RNA from CMNV-infected shrimp. This degree of analytical sensitivity is similar to that reported by Hirayama et al. (2006) for sexing water buffalo, by Chen et al. (2010) for the detection of swine-transmissible gastroenteritis coronavirus and by Li and Ling (2014) for the detection of tomato necrotic stunt virus. In comparison to the reference method of nested RT-PCR, the DSe and DSp values for the CMNV RT-LAMP assay were 96.4% and 94.4%, respectively. One spontaneously infected L. vannamei sample was determined as negative of CMNV by RT-PCR; however it showed positive of CMNV by qRT-LAMP and was quantified as 6.2 × 105 viral copies. The inconsistent result tested by RT-PCR and qRT-LAMP might attribute to PCR inhibitors that could affect the Taq DNA polymerase used in conventional RT-PCR (Alhassan et al., 2007, Bakheit et al., 2008, Liang et al., 2009). In contrast, such inhibitors may not affect the Bst polymerase used in LAMP (Enosawa et al., 2003, Alhassan et al., 2007). This characteristic of Bst polymerase makes it be appropriate for field application.
For visual inspection of the CMNV RT-LAMP assay result and to eliminate the risk of contamination caused by reopening the reaction tube, a method of DNA fluorescent dye pre-setting was developed in the study. At the end of the reaction, the dye was incorporated into the reaction mixture by shaking, allowing for the visual inspection of the RT-LAMP products. The use of a fluorescent dye can therefore substitute for gel electrophoresis or real-time PCR as a method of viral detection, simplifying the assay and reducing the contamination risk.
A standard curve was constructed using 10-fold serial dilutions of the pMD19-T-CMNV plasmid with reference to the Ct value. Based on the standard curve, an equation was calculated using regression analysis comparing Ct values with the standard copy number. In the range of 108–103 plasmid copies, the correlation coefficient was high (r 2 = 0.9953), which indicates that qRT-LAMP is appropriate as a quantitation tool. However, when the plasmid copy number decreased to less than 1000 copies, the correlation coefficient also decreased significantly, which confirmed the findings of previous reports that it is difficult to determine the exact correlation of the initial template quantity and the Ct value at very low concentrations of template (Mori et al., 2004, Suzuki et al., 2011, Wei et al., 2013).
In summary, this report describes a rapid, highly sensitive, highly specific, financially economical, quantitative RT-LAMP method for CMNV detection. This assay could therefore become the assay of choice for the routine detection and quantification of CMNV in the laboratory and in the field.
Acknowledgments
This work was supported by the following projects: Special Scientific Research Funds for Central Non-profit Institutes, the Chinese Academy of Fishery Sciences (2013A0601 and 2014A06XK01), the Special Fund for Agro-Scientific Research in the Public Interest (Grant: 201103034), the China Agriculture Research System (CARS-47), the Construction Programme for “Taishan Scholarship” of Shandong Province of China, and the Programme for Chinese Outstanding Talents in Agricultural Scientific Research.
Contributor Information
Qingli Zhang, Email: zhangql@live.com.
Shuang Liu, Email: liushuang8@163.com.
Haolin Yang, Email: yanghlwin@163.com.
Luoluo Zhu, Email: zhuluoluo666@163.com.
Xiaoyuan Wan, Email: wanxy@ysfri.ac.cn.
Jie Huang, Email: huangjie@ysfri.ac.cn.
References
- Alhassan A., Thekisoe O.M., Yokoyama N., Inoue N., Motloang M.Y., Mbati P.A., Yin H., Katayama Y., Anzai T., Sugimoto C., Igarashi I. Development of loop-mediated isothermal amplification (LAMP) method for diagnosis of equine piroplasmosis. Vet. Parasitol. 2007;143:155–160. doi: 10.1016/j.vetpar.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Bakheit M.A., Torra D., Palomino L.A., Thekisoe O.M., Mbati P.A., Ongerth J., Karanis P. Sensitive and specific detection of Cryptosporidium species in PCR-negative samples by loop-mediated isothermal DNA amplification and confirmation of generated LAMP products by sequencing. Vet. Parasitol. 2008;158:11–22. doi: 10.1016/j.vetpar.2008.09.012. [DOI] [PubMed] [Google Scholar]
- Chen Q., Li J., Fang X.E., Xiong W. Detection of swine transmissible gastroenteritis coronavirus using loop-mediated isothermal amplification. Virol. J. 2010;7:206. doi: 10.1186/1743-422X-7-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enosawa M., Kageyama S., Sawai K., Watanabe K., Notomi T., Onoe S., Mori Y., Yokomizo Y. Use of loop-mediated isothermal amplification of the IS900 sequence for rapid detection of cultured Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 2003;41:4359–4365. doi: 10.1128/JCM.41.9.4359-4365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegel, T.W., 2014. EMS/AHPND: a game changer for the future development of aquaculture. In: The 9th Symposium on Diseases in Asian Aquaculture (DAA9) in Ho Chi Minh City, Vietnam, November 24–28.
- Gu S.J. Analysis of causes of the covert mortality disease of Pacific white shrimp and its control strategies. (Chin. J.) Sci. Fish. Farming. 2012;8:62–63. (in Chinese with English abstract) [Google Scholar]
- Hirayama H., Kageyama S., Takahashi Y., Moriyasu S., Sawai K., Onoe S., Watanabe K., Kojiya S., Notomi T., Minamihashi A. Rapid sexing of water buffalo (Bubalus bubalis) embryos using loop-mediated isothermal amplification. Theriogenology. 2006;66:1249–1256. doi: 10.1016/j.theriogenology.2006.03.036. [DOI] [PubMed] [Google Scholar]
- Huang, J., 2012. Experience in EMS/AHPNS from China. In: NACA, (2012). The Asia Pacific Emergency Regional Consultation on the Emerging Shrimp Disease: Early Mortality Syndrome (EMS)/Acute Hepatopancreatic Necrosis Syndrome (AHPNS). Bangkok, Thailand, August 2012.
- Li R., Ling K.S. Development of reverse transcription loop-mediated isothermal amplification assay for rapid detection of an emerging potyvirus: tomato necrotic stunt virus. J. Virol. Methods. 2014;200:35–40. doi: 10.1016/j.jviromet.2014.01.017. [DOI] [PubMed] [Google Scholar]
- Liang S.Y., Chan Y.H., Hsia K.T., Lee J.L., Kuo M.C., Hwa K.Y., Chan C.W., Chiang T.Y., Chen J.S., Wu F.T., Ji D.D. Development of loop-mediated isothermal amplification assay for detection of Entamoeba histolytica. J. Clin. Microbiol. 2009;47(6):1892–1895. doi: 10.1128/JCM.00105-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y., Kitao M., Tomita N., Notomi T. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J. Biochem. Biophys. Methods. 2004;59:145–157. doi: 10.1016/j.jbbm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Mori N., Motegi Y., Shimamura Y., Ezaki T., Natsumeda T., Yonekawa T., Ota Y., Notomi T., Nakayama T. Development of a new method for diagnosis of rubella virus infection by reverse transcription-loop-mediated isothermal amplification. J. Clin. Microbiol. 2006;44:3268–3273. doi: 10.1128/JCM.00803-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- Nagamine K., Watanabe K., Ohtsuka K., Hase T., Notomi T. Loop-mediated isothermal amplification reaction using a nondenatured template. Clin. Chem. 2001;47(9):1742–1743. [PubMed] [Google Scholar]
- Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M., Posadas G., Inoue S., Hasebe F., Morita K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J. Clin. Microbiol. 2004;42:257–263. doi: 10.1128/JCM.42.1.257-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Narimatsu S., Furukawa T., Iwakiri A., Miura M., Yamamoto S., Katayama H. Comparison of real-time reverse transcription loop-mediated isothermal amplification and real-time reverse transcription polymerase chain reaction for detection of noroviruses in municipal wastewater. J. Biosci. Bioeng. 2011;112:369–372. doi: 10.1016/j.jbiosc.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Tomita N., Mori Y., Kanda H., Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008;3(5):877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- Wang L., Li L., Alam M.J., Geng Y., Li Z., Yamasaki S., Shi L. Loop-mediated isothermal amplification method for rapid detection of the toxic dinoflagellate Alexandrium, which causes algal blooms and poisoning of shellfish. FEMS Microbiol. Lett. 2008;282:15–21. doi: 10.1111/j.1574-6968.2008.01074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Zeng J., Deng C., Zheng C., Zhang X., Ma D., Yi Y. A novel method of real-time reverse transcription loop-mediated isothermal amplification developed for rapid and quantitative detection of human astrovirus. J. Virol. Methods. 2013;188:126–131. doi: 10.1016/j.jviromet.2012.11.040. [DOI] [PubMed] [Google Scholar]
- World Organization for Animal Health (OIE), 2011. Principles and methods of validation of diagnostic assays for infectious diseases. In: Manual of Diagnostic Tests for Aquatic Animals. OIE, Paris, France (Chapter 1.1.2).
- Xing H. Discussion of the control measures for the “bottom death” (covert mortality disease) of Pacific white shrimp. China Fish. 2004;4:88–89. (in Chinese with English abstract) [Google Scholar]
- Xu Z.J., Ji F. Comprehensive control of the covert mortality disease of Pacific white shrimp. Chin. J.: Fish. Guide Rich. 2009;1:60. (in Chinese with English abstract) [Google Scholar]
- Yang J.L., Zhang S.H., Liu Z.H., Yang R., Huang Y., Wen M. Development and evaluation of a loop-mediated isothermal amplification assay for the rapid detection of porcine cytomegalovirus under field conditions. Virol. J. 2012;9:321. doi: 10.1186/1743-422X-9-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.H. To be cautious of “bottom death” in the intensive farming of Pacific white shrimp. (Chin. J.) Sci. Fish. Farming. 2004;10:48–49. (in Chinese with English abstract) [Google Scholar]
- Zhang Q.L., Liu Q., Liu S., Yang H., Liu S., Zhu L., Yang B., Jin J., Ding L., Wang X., Liang Y., Wang Q., Huang J. A new nodavirus is associated with covert mortality disease of shrimp. J. Gen. Virol. 2014;95:2700–2709. doi: 10.1099/vir.0.070078-0. [DOI] [PubMed] [Google Scholar]
- Zhang Q.L., Yan Y., Shen J.Y., Hao G.J., Shi C.Y., Wang Q.T., Liu H., Huang J. Development of a reverse transcription loop-mediated isothermal amplification assay for rapid detection of grass carp reovirus. J. Virol. Methods. 2013;187:384–389. doi: 10.1016/j.jviromet.2012.11.005. [DOI] [PubMed] [Google Scholar]