Abstract
MRNA2 of the arteriviruses lactate dehydrogenase-elevating virus (LDV) and equine arteritis virus (EAV) encodes two proteins that are read in different frames, an about 26 kDa minor envelope glycoprotein and an about 8 kDa protein that lacks N-glycosylation sites and a signal peptide, but possesses a central hydrophobic segment. Recent studies have shown that both proteins of EAV are translated from mRNA 2 in EAV infected BHK cells, that the 8 kDa protein is membrane associated and that small amounts of it are recovered in purified virions (Snijder, E.J., van Tol, H., Pederson, K.W., Raamsman, M.J.B., de Vries, A.A.F., 1999. Identification of a novel structural protein of arteriviruses. J. Virol. 73, 6335–6345). The authors concluded that the 8 kDa protein is another arterivirus envelope protein and designated it E protein. However, we have not detected a significant level of an 8 kDa protein in LDV virions and thus conclude that it is not a structural virion component.
Keywords: Lactate dehydrogenase elevating virus, ORF2a protein, Virion envelope proteins
The family Arteriviridae (Cavanagh, 1997) presently consists of equine arteritis virus (EAV), murine lactate dehydrogenase-elevating virus (LDV), porcine reproductive and respiratory syndrome virus (PRRSV) and simian hemorrhagic fever virus (SHFV). Arterivirus virions possess a cubical core nucleocapsid with a positive stranded RNA genome of 12.7–15.7 kb and the nucleocapsid protein (N; 12–13 kDa). This nucleocapsid is surrounded by an envelope which primarily contains disulfide-linked heterodimers of the matrix protein (M; 16–20 kDa) and the primary envelope glycoprotein, designated VP-3P, GL, and E for LDV, EAV, and PRRSV, respectively (Plagemann, 1996, Snijder and Meulenberg, 1998). In these viruses, VP-3P/GL/E, M, and N are encoded by ORFs 5, 6, and 7, respectively. In addition, the genomes of these viruses encode three other glycoproteins encoded by ORFs 2, 3 and 4. The ORF 2 protein seems to be a very minor envelope glycoprotein (De Vries et al., 1992, Faaberg and Plagemann, 1995, Meulenberg and Petersen-den Besten, 1996), but its function and those of the ORF 3 and ORF 4 glycoproteins are not clear. The ORF 3 protein of LDV is an antigenic, non-structural protein (Faaberg and Plagemann, 1997). ORFs 2–7 are expressed via a nested set of six subgenomic mRNAs (mRNAs 2–7) which possess a common leader derived from the 5′ end of the viral genome. The leader is fused to the bodies of the subgenomic mRNAs at closely related subgenomic mRNA transcription regulatory sequences (TRSs).
During sequencing of the genome of one LDV quasispecies, LDV-P, we noticed that mRNA 2 besides coding for the 26 kDa ORF 2 glycoprotein contained another smaller ORF read in a different frame that potentially encoded another protein with 70 amino acids (∼8 kDa; Fig. 1 , bold face; Chen et al., 1993). The initiation codon for the latter protein overlaps by one nucleotide with the termination codon of the ORF 1b replicase gene, is in relatively favorable context for translation initiation (Kozak, 1989) and precedes that for the 26 kDa ORF 2 protein by 41 nucleotides (Fig. 1). If both proteins were synthesized, they would be translated from a bicystronic mRNA since only a single TRS has been found to precede both AUG initiation codons (Fig. 1) and only a single mRNA 2 was detected (Chen et al., 1993).
Fig. 1.
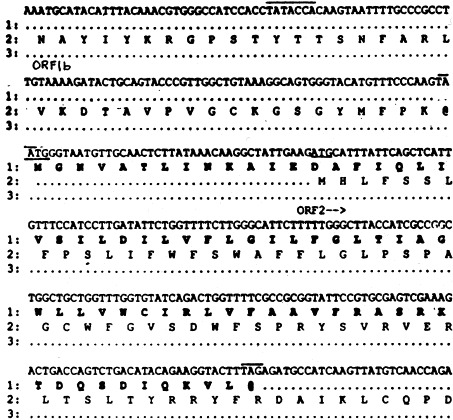
Nucleotide sequence of a segment of the LDV-P genome encompassing the 3′-end of ORF 1b, the 5′-end of ORF 2 and a small additional ORF that overlaps with ORF 2 and whose translation initiation codon overlaps by one nucleotide with the termination codon of ORF 1b. The 70 amino acids of the predicted protein product (about 8 kDa) encoded by the latter ORF are shown in bold face letters. Also shown are the amino acids of the segments of the ORF 1b and ORF 2 proteins encoded by the given genome segment. The AUG translation initiation codons are underlined and the termination codons and the TRS or mRNA 2 are overlined.
The smaller ORF of mRNA 2 is highly conserved among arteriviruses (Snijder et al., 1999) and their strains. For example, it is present in an identical position in the genomes of all three identified quasispecies of LDV isolated from laboratory mice carrying various transplantable tumors (LDV-P, LDV-vx, LDV-C; GenBank accession numbers: U15146, AF092283 and L13298, respectively; Godeny et al., 1993, Palmer et al., 1995, Li et al., 1999) as well as in the genomes of four LDVs isolated from wild house mice in the USA. (Li et al., 2000). Nucleotide divergence between the small ORFs 2 of LDV-P and LDV-vx and LDV-P and LDV-C are 3.3 and 17.1%, respectively. Most of the nucleotide differences are translationally silent (synonymous). Amino acid differences between the predicted proteins of LDV-P and LDV-vx and LDV-P and LDV-C are 1.4 and 15.6%, respectively, and involve largely val, ileu, leu, and cys residues. Similar small additional proteins are encoded by mRNA 2 of EAV and PRRSV and mRNA 4 of SHFV (Snijder et al., 1999). All the predicted proteins lack N-glycosylation sites and a signal peptide, but possess a central hydrophobic core that could function as a membrane anchor and a potential N-myristoylation site (Snijder et al., 1999). Recently, it has been shown that the additional ORF 2 protein of EAV of about 8 kDa is synthesized in EAV-infected BHK-21 cells, that it is associated with intracellular membranes, that it is required for the generation of infectious virions and that small amounts of the protein are recovered in sucrose density gradient purified EAV (Snijder et al., 1999). The authors suggested to designate the small and large ORFs of mRNA 2 of EAV and LDV as ORF 2a and ORF 2b, respectively. Furthermore, because the 8 kDa protein is apparently a membrane protein and recovered in small amounts in virions, they suggested the name envelope or E protein for the 8 kDa arterivirus protein and speculated that this protein, like the E proteins of coronaviruses (De Haan et al., 2000), might play a role in arterivirus morphogenesis. However, we have not detected significant amounts of an 8 kDa protein in LDV virions. In one experiment, an LDV-infected mouse macrophage culture was incubated with [3H]leu from 2 to 24 h post infection (p.i.). Progeny virus was purified from the culture fluid by isopycnic centrifugation in a 0.5–1.5 M sucrose density gradient and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; Brinton-Darnell and Plagemann, 1975). The gel was cut into 1-mm slices which were analyzed for radioactivity (Fig. 2 A). The three main structural proteins of LDV, N, M and VP-3P, were clearly separated, but there was no indication of the presence of a protein of about 8 kDa, which should be located in fractions 117–122. Leucine labeling is the most sensitive approach for detecting the presence of the small ORF 2a protein in LDV virions since its leucine content is 16%, whereas the leucine content of N, M, and VP-3P is 7.3, 11.7 and 11.6%, respectively. These results are consistent with those of another study of [3H]leu-labeled LDV virions (Michaelidis and Schlesinger, 1973).
Fig. 2.
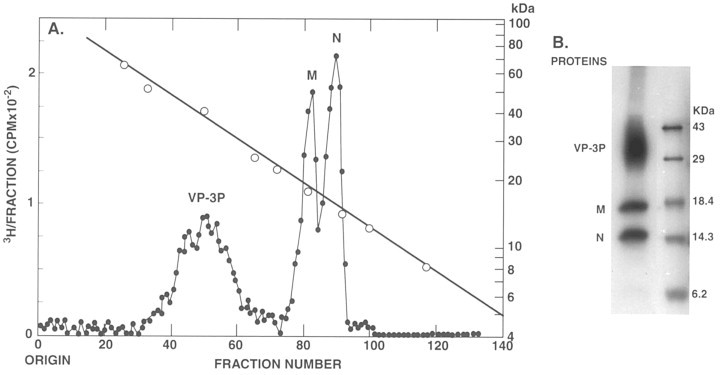
SDS-PAGE of [3H]leu-labeled (A) and 35S-labeled (B) LDV virion proteins. LDV-P infected mouse macrophage cultures were incubated beginning at 3 h p.i. with [3H]leu or [35S]met/[35S]cyc, respectively. At 20 or 24 h p.i., the virions were harvested from the culture fluid, semipurified and analyzed by SDS-PAGE as described by Brinton-Darnell and Plagemann (1975) and Faaberg and Plagemann (1995), respectively. In A, the gel was cut into slices and the latter were analyzed for radioactivity by liquid scintillation counting. The co-electrophoresed protein standards were: bovine serum albumin, 67 kDa; heavy chain of IgG, 50 kDa; creatine phosphokinase, 40 kDa; chymoytrypsin, 25 kDa; trypsin, 23 kDa; myoglobulin, 17.8 kDa; lysozyme, 14 kDa; cytochrome c, 12.4 kDa and West Nile virus M protein, 8.3 kDa. In B, the gel was autoradiographed. The protein standards were: ovalbumin, 43 kDa; carbonic anhydrase, 29 kDa; β lactoglobulin, 18.4 kDa; lysozyme, 14.3 kDa and bovine trypsin inhibitor, 6.2 kDa.
In another study, LDV-P-infected macrophage cultures were labeled with [35S]met and [35S]cys (TRANSLABEL) from 3 to 20 h p.i. (Faaberg and Plagemann, 1995). Progeny virus was collected by centrifugation through a 0.5 M sucrose cushion and analyzed by SDS-PAGE (Fig. 2B). Again, N, M, and VP-3P became heavily labeled, but no labeled protein was detected in the 6- to 10-kDa area (Fig. 2B). Results similar to those shown in Fig. 2B were obtained in another independent study in which LDV virions were isolated from macrophage cultures incubated with [35S]met alone (Cafruny et al., 1986).
The [3H]leu-labeling results (Fig. 2A) allow calculating the sensitivity of detecting an 8 kDa protein in LDV virions. On the basis of the number of amino acids of the LDV proteins and their leucine contents we estimated that for every 100 molecules of processed VP-3P (after removal of the signal peptide; Faaberg and Plagemann, 1995) LDV virions contained 52 molecules of M protein and less than 0.5 molecule of a potential 8 kDa protein. A similar analysis of the 35S-labeled virions was problematic since the amounts of radioactivity associated with each of the proteins cannot be accurately estimated from the SDS-PAGE profiles, and, indeed, no information on the relative amounts of the 8 kDa protein recovered in EAV virions was presented (Snijder et al., 1999). In order to obtain some comparative information on the presence of 8 kDa proteins in EAV and LDV virions we have scanned for density the SDS-PAGE profiles of 35S-labeled virion proteins presented in Fig. 7B of Snijder et al. (1999) and in Fig. 2B, respectively (Fig. 3 ). The overall density values for the LDV virion proteins (Fig. 3C) were comparable to those for the EAV virions (Fig. 3A,B), but no significant density was present in the 8 kDa region of LDV virion proteins, even though the scanned area was three times larger than those measured for the 8 kDa EAV protein. On the basis of the density values and the cys/met content of the LDV proteins we estimated that for every 100 molecules of processed VP-3P, there were 33 molecules of M protein and less than one molecule of a potential 8 kDa protein, values comparable to those estimated from [3H]leu labeling.
Fig. 3.
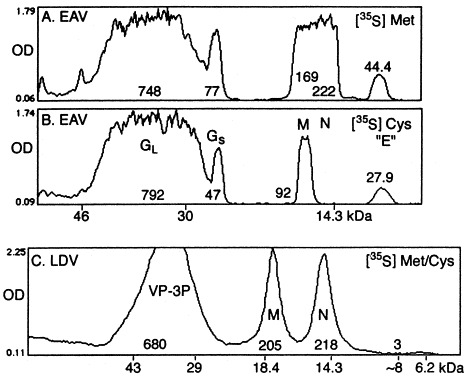
Density scanning of SDS-PAGE profiles of 35S-labeled proteins of EAV (A and B; Snijder et al., 1999; Fig. 7B; sucrose density gradient fractions 11 and of LDV; Fig. 2B). The profiles were scanned using NIH Image program 1.6.2. The density values listed with each protein are in arbitrary units=optical density (OD)×area.
It could be argued that an 8 kDa ORF 2a protein was not detected in LDV virions because it is linked to another structural LDV protein. This seems very unlikely. The symmetry of the N and M peaks in Fig. 2A and of the N and M bands in Fig. 2B rule out any type of linkage of the ORF 2a protein to these proteins. Disulfide linkage of the 8 kDa protein to VP-3P is also ruled out since the virion proteins were analyzed under reducing conditions that have been shown to break the disulfide bond linking together the VP-3P-M heterodimer (Faaberg et al., 1995). Furthermore, the single cys residue in the 8 kDa ORF 2a protein of LDV-P is replaced by a val residue in those of LDV-C and LDV-74 (isolated from a wild house mouse; Li et al., 2000). Similarly, the 8 kDa ORF 2a protein of EAV was found not to form intermolecular or intramolecular disulfide bonds or to undergo other post-translational modifications (Snijder et al., 1999). It is possible that the 8 kDa ORF 2a protein is lost from LDV virions during purification, but not if it is an integral membrane protein as proposed for the EAV protein (Snijder et al., 1999). The reasons for the different results with LDV and EAV are not apparent since the experimental conditions for labeling the virus proteins and purification of the virions and their analysis by SDS-PAGE were comparable. The only difference was that labeling of the LDV virion proteins was conducted in the natural host cell of the virus, namely mouse macrophages, whereas labeled EAV was produced in an established cell line, BHK-21. Virus morphogenesis could differ in the two cell types. Although the results with EAV clearly indicated that the 8 kDa ORF 2a protein is synthesized in infected BHK cells, the possibility that small amounts of it remained non-specifically bound to virions during purification rather than being an integral part of the envelope, was not completely ruled out. It could also be possible that small amounts of the 8 kDa protein can be non-specifically incorporated into virions. The same difficulties in interpretation apply to the recovery of small amounts of the 26 kDa ORF 2 protein and of other viral proteins in virions (see later).
It is obvious that our results do not rule out that a 8 kDa protein is associated with LDV virions at an extremely low level, that is, for example, at one molecule/1000 VP-3P molecules, but such a low level seems hardly significant. In any case, such levels would not justify to designate the 8 kDa protein a structural envelope (E) protein unless it can be demonstrated that such trace amounts associated with virions play a functional role in virion structure or virus adsorption/penetration and do not result from an accidental interaction with virions.
This does not mean that the 8 kDa protein may not play an important role in virion morphogenesis. However, there is no information available on this point and comparison to the coronavirus E protein may not be valid, since the envelope structures of coronaviruses and arteriviruses differ greatly. The envelope of LDV largely contains disulfide bonded heterodimers of the envelope glycoprotein and the M protein (Faaberg and Plagemann, 1995) which seem to form ring-like structures in the envelope (Brinton-Darnell and Plagemann, 1975). In contrast, the envelope of coronaviruses consists of a dense matrix of lateral, non-covalently interacting M proteins into which the S glycoprotein becomes inserted (De Haan et al., 2000). The 8 kDa protein is synthesized in mouse hepatitis virus (MHV) infected cells along with all other viral proteins and most of it accumulates in masses of tubular, smooth convoluted membrane structures (Raamsman et al., 2000). Only trace amounts of it are recovered in MHV virions (De Haan et al., 2000), perhaps due to accidental incorporation into the envelope since the exclusion of foreign proteins from in vitro assembled virus-like particles is not perfect (De Haan et al., 2000). No tubular membrane structures resembling those containing the E protein observed in MHV-infected cells are detected in arterivirus-infected cells. The 8 kDa protein seems associated with ER/Golgi membranes (Snijder et al., 1999) into which free cubical nucleocapsids bud (Stueckemann et al., 1982) whereby most or all of the protein seems to become excluded from virions (Snijder et al., 1999).
Since the 8 kDa protein is not a significant structural component of LDV and no virion associated function has been identified for it, it might be preferable to provisionally designate the two proteins translated from mRNA 2 the ORF 2S and ORF 2L proteins, where S and L stand for small and large, respectively.
Acknowledgements
I thank Sara Veglahn and Patricia Nelson for competent secretarial assistance.
References
- Brinton-Darnell M., Plagemann P.G.W. Structure and chemical-physical characteristics of lactate dehydrogenase-elevating virus and its RNA. J. Virol. 1975;16:420–433. doi: 10.1128/jvi.16.2.420-433.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafruny W.A., Chan S.P.K., Harty J.T., Yousefi S., Kowalchyk K., McDonald D., Foreman B., Budweg G., Plagemann P.G.W. Antibody response of mice to lactate dehydrogenase-elevating virus during infection and immunization with inactivated virus. Virus Res. 1986;5:357–375. doi: 10.1016/0168-1702(86)90029-8. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;142/43:629–633. [PubMed] [Google Scholar]
- Chen Z., Kuo L., Rowland R.R.R., Even C., Faaberg K.S., Plagemann P.G.W. Sequence of 3′-end of genome and of 5′-end of ORF 1a of lactate dehydrogenase-elevating virus and common junction motifs between 5′-leader and bodies of seven subgenomic mRNAs. J. Gen. Virol. 1993;74:643–660. doi: 10.1099/0022-1317-74-4-643. [DOI] [PubMed] [Google Scholar]
- De Haan C.A.M., Vennema H., Rottier P.J.G. Assembly of coronavirus envelope: Homotypic interactions between the M proteins. J. Virol. 2000;74:4967–4978. doi: 10.1128/jvi.74.11.4967-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries A.A.F., Chirnside E.D., Horzinek M.C., Rottier P.J.M. Structural proteins of equine arteritis virus. J. Virol. 1992;66:6294–6303. doi: 10.1128/jvi.66.11.6294-6303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faaberg K.S., Plagemann P.G.W. The envelope proteins of lactate dehydrogenase-elevating virus and their membrane topography. Virology. 1995;212:512–525. doi: 10.1006/viro.1995.1509. [DOI] [PubMed] [Google Scholar]
- Faaberg K.S., Plagemann P.G.W. ORF3 of lactate dehydrogenase-elevating virus encodes a soluble, nonstructural, highly glycosylated, and antigenic protein. Virology. 1997;227:245–251. doi: 10.1006/viro.1996.8310. [DOI] [PubMed] [Google Scholar]
- Faaberg K.S., Even C., Palmer G.A., Plagemann P.G.W. Disulfide bonds between two envelope proteins of lactate dehydrogenase-elevating virus are essential for viral infectivity. J. Virol. 1995;69:613–617. doi: 10.1128/jvi.69.1.613-617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godeny E.K., Chen L., Kumar S.N., Methven S.L., Koonin E.V., Brinton M.A. Complete genomic sequence and phylogenetic analysis of the lactate dehydrogenase-elevating virus (LDV) Virology. 1993;194:585–617. doi: 10.1006/viro.1993.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J. Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Chen Z., Plagemann P.G.W. High frequency genetic recombination of an arterivirus, lactate dehydrogenase-elevating virus, in mice and evolution of neuropathogenic variants. Virology. 1999;258:73–83. doi: 10.1006/viro.1999.9660. [DOI] [PubMed] [Google Scholar]
- Li K., Schuler T., Chen Z., Glass G.E.G., Childs J.E., Plagemann P.G.W. Isolation of lactate dehydrogenase-elevating viruses from wild house mice and their biological and molecular characterization. Virus Res. 2000;67:153–162. doi: 10.1016/s0168-1702(00)00142-8. [DOI] [PubMed] [Google Scholar]
- Meulenberg J.J.M., Petersen-den Besten A. Identification and characterization of a sixth structural protein of Lelystad virus: the glycoprotein GP2 encoded by ORF2 is incorporated in virus particles. Virology. 1996;225:44–51. doi: 10.1006/viro.1996.0573. [DOI] [PubMed] [Google Scholar]
- Michaelidis M.C., Schlesinger S. Structural proteins of lactic dehydrogenase virus. Virology. 1973;55:211–217. doi: 10.1016/s0042-6822(73)81023-2. [DOI] [PubMed] [Google Scholar]
- Palmer G.A., Kuo L., Chen Z., Faaberg K., Plagemann P.G.W. Sequence of genome of lactate dehydrogenase-elevating virus. Heterogeneity between strains P and C. Virology. 1995;209:637–642. doi: 10.1006/viro.1995.1296. [DOI] [PubMed] [Google Scholar]
- Plagemann P.G.W. Lactate dehydrogenase-elevating virus and related viruses. In: Fields B.N., Knipe D.M., Howley P.M., editors. Virology. 3rd edn. Raven Press; New York: 1996. pp. 1105–1120. [Google Scholar]
- Raamsman M.J.B., Locker J.K., de Hooge A., de Vries A.A.F., Griffiths G., Vennena H., Rottier P.J.M. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 2000;74:2333–2342. doi: 10.1128/jvi.74.5.2333-2342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Meulenberg J.J.M. The molecular biology of arteriviruses. J. Gen. Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., van Tol H., Pederson K.W., Raamsman M.J.B., de Vries A.A.F. Identification of a novel structural protein of arteriviruses. J. Virol. 1999;73:6335–6345. doi: 10.1128/jvi.73.8.6335-6345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stueckemann J.A., Ritzi D.M., Holth M., Smith M.S., Swart W.J., Cafruny W.A., Plagemann P.G.W. Replication of lactate dehydrogenase-elevating virus in macrophages. 1. Evidence for cytocidal replication. J. Gen. Virol. 1982;59:245–262. doi: 10.1099/0022-1317-59-2-245. [DOI] [PubMed] [Google Scholar]


