Abstract
Diarrhea is a leading cause of sickness and death of beef and dairy calves in their first month of life. Field investigations of outbreaks of neonatal calf diarrhea should be conducted for the purposes of (1) reducing the losses associated with existing cases and (2) preventing new cases from occurring. The appropriate corrective actions are determined by taking a population approach to diagnostics and prevention. It can be difficult to provide solutions to disease outbreaks, but success is more likely if an organized, epidemiologic approach to outbreak investigation is followed.
Keywords: Calf, Diarrhea, Epidemiology, Outbreak, Investigation, Diagnostics
Key Points.
-
•
Diarrhea is a leading cause of sickness and death of beef and dairy calves in their first month of life.
-
•
Field investigations of outbreaks of neonatal calf diarrhea should be conducted for the purposes of (1) reducing the losses associated with existing cases and (2) preventing new cases from occurring.
-
•
Veterinarians investigating outbreaks of neonatal calf diarrhea must first make recommendations for treating affected calves and then take action to protect susceptible and unborn calves from ongoing illness.
-
•
Knowing the etiologic agent may provide an explanation for the proximal cause of a calf’s illness or death, but that knowledge rarely explains the outbreak or provides a solution for treatment, control, or prevention.
-
•
The goal is to conduct a useful outbreak investigation—one that leads to a solution to the problem.
Introduction
One of the most likely reasons for young beef or dairy calves to become sick or die is diarrhea.1 The disease is a detriment to calf health and well-being.2, 3 Also, neonatal calf diarrhea can be economically important to cattle producers because of poor calf performance, death, and the expense of medications and labor to treat sick calves. The act of catching and treating young calves puts farmers and ranchers at risk of physical injury by the calf or dam, and many producers are disheartened by seeing calves become sick or die at only 1 or 2 weeks of age. Finally, there may be public health risks associated with some outbreaks of neonatal calf diarrhea because (1) some diarrhea pathogens of calves also make people sick and (2) an important reason for using antibiotics on many beef and dairy farms is to treat young calves for diarrhea.
Until a serious outbreak occurs, veterinarians may not be aware that cattle producers are dealing with neonatal calf diarrhea problems. Veterinarians investigating outbreaks of neonatal calf diarrhea must first make recommendations for treating affected calves and then take action to protect susceptible and unborn calves from ongoing illness. Finally, attention should focus on determining what future actions might prevent the disease in subsequent calving periods or seasons.4 Although knowledge of the pathogens involved is often useful information, outbreak investigations sometimes become sidetracked in the sole pursuit of an etiologic agent rather than identifying more useful explanations for the outbreak. Knowing the etiologic agent may provide an explanation for the proximal cause of a calf’s illness or death, but that knowledge rarely explains the outbreak or provides a solution for treatment, control, or prevention. The goal, then, is to conduct a useful outbreak investigation—one that leads to a solution to the problem.
Principles of disease outbreak investigation
Veterinarians are often asked to investigate outbreaks of animal death or disease, including those related to neonatal calf diarrhea. The objectives of these investigations are commonly to (1) reduce the losses associated with existing cases and (2) prevent new cases from occurring. These objectives are met by (1) confirming the clinical diagnosis so that affected individuals are treated appropriately, (2) making an appropriate population diagnosis to determine the reasons for the outbreak, (3) identifying actions that can be taken to prevent new cases or future outbreaks, and (4) effectively communicating the plan of action. It can be difficult to provide solutions to disease outbreaks, but success is more likely if an organized, epidemiologic approach to outbreak investigation is followed.5, 6
Disease outbreaks (epidemics) are typically defined as an unexpected increase in disease or death, typically occurring during a brief period of time. Outbreaks may occur because of a common (point-source) exposure (eg, the potato salad at the church picnic). These types of outbreaks are typically rapid in development and resolution or they may become propagated epidemics (eg, you got sick from the church supper and then your family got sick and passed it on to friends at school). Propagated epidemics are typically characterized by less rapid but ongoing transmission from animal to animal. The distinction between these two types of outbreaks sometimes becomes blurred, but often they are distinguishable by observing the epidemic curve. Methods to prevent new cases may differ depending on the type of epidemic (eg, removing the potato salad or isolating sick individuals).7
Outbreak investigations involve an orderly process to characterize the outbreak:
-
•
Interview key individuals (eg, owners, caretakers, veterinarians, nutritionists, and other stakeholders)
-
•
Verify the clinical diagnosis and assure that treatments are appropriate
-
•Identify the factors responsible for the outbreak: make a population diagnosis
-
○Establish a system for collecting information
-
○Define what a case is and what a case is not
-
○Define the population at risk
-
○Characterize the outbreak by time, space, and unit (animal, pen, herd, region)
-
○Determine the nature of the outbreak (point source, propagated)
-
○Develop hypotheses about possible causal factors
-
○Test hypotheses using epidemiologic principles
-
○
-
•Develop strategies to prevent new cases and future outbreaks
-
○Identify control points and corrective actions based on the knowledge of causal factors and key determinants
-
○
-
•Communicate observations and recommendations with the key individuals
-
○Validate that recommendations are being performed
-
○Monitor progress
-
○
Investigating outbreaks of neonatal calf diarrhea
Identify the key individuals early in the course of the investigation. Although you may be reporting to the owner, other individuals may be able to provide history or other details unknown to the owner. Key individuals include those with direct financial interests (eg, the owner); employee caretakers who may know the most about day-to-day care and practice; and other parties, such as other veterinarians, nutritional consultants, or other advisors that may have special knowledge of the circumstances. Capture names and contact information at the start of the investigation.
Verify the Diagnosis and Assure that Treatments are Appropriate
Early in the outbreak investigation it is vital to conduct at least a preliminary examination of affected animals. This process includes a walk-through of the herd and facilities, completing physical examinations or postmortem examinations, and submitting samples to the diagnostic laboratory (see article by Blanchard in this issue) to verify the clinical diagnosis.5, 8 Assure that affected calves are receiving medically appropriate treatment, so that ongoing losses are minimized while the outbreak is being investigated. Determine through history and records analysis that this is indeed an outbreak (defined as an increased incidence of disease compared with expected or historical rates). Sometimes an outbreak is really a sudden awareness on the owner’s or caretaker’s part of a disease process that has been ongoing for some time.
Identify the Factors Responsible for the Outbreak: Make a Population Diagnosis
Establish a system for recording information
Few cow-calf and dairy operations collect animal health data in a format that is easily analyzed. If records exist at all, they may be on paper (eg, in pocket-sized ranch books) or in an electronic format with free text fields, none of which allow the veterinarian or the producer to quickly and easily discover animal health relationships. Some electronic record-keeping systems capture health information and may present information in standard graphs or summaries, but do not allow easy querying of health relationships. Free text fields are difficult or impossible to analyze because of misspelled words, multiple names, or descriptors for the same disease (eg, diarrhea, scours, loose, enteric) and multiple pieces of information in the same field (eg, scours, T103.8, bolus and fluids, retreat afternoon, watch out for the cow). The lack of a simple record-keeping system has hindered many disease outbreak investigations. It simply becomes too costly to wade through inefficient record systems to get the needed information from which questions can be asked. A desirable system for investigating neonatal calf diarrhea is one that captures individual calf information for all calves at risk, not just affected calves. Useful information includes the calf’s identification, the identification of the dam, the age of the dam, the calf’s birth date, the date of onset of the illness, the date of onset of other illnesses, and information describing various risk factors of interest.
Define a case
A critical, but too often ignored, component of any disease outbreak investigation is specifying what a case is and what it is not. Neonatal calf diarrhea is no exception. Diarrhea is a subjective diagnosis and accompanying clinical signs (eg, dehydration, fever, and level of depression) may or may not be relevant to the specific syndrome associated with the outbreak of interest. For example, the complaint may be about high mortality in calves developing diarrhea at 1 to 3 weeks’ of age. So, do you count as cases calves with fever or dehydration but no observed diarrhea? What about calves with diarrhea at 60 days’ of age or calves with distended abdomens and pasty stools? The definition of a case is not always obvious but it is important. Having a reasonable level of specificity in the case definition is necessary to avoid the confusion of looking for a single solution to more than one problem. However, overspecifying the case definition may cause confusion because only part of the problem is being investigated.
Define the population at risk
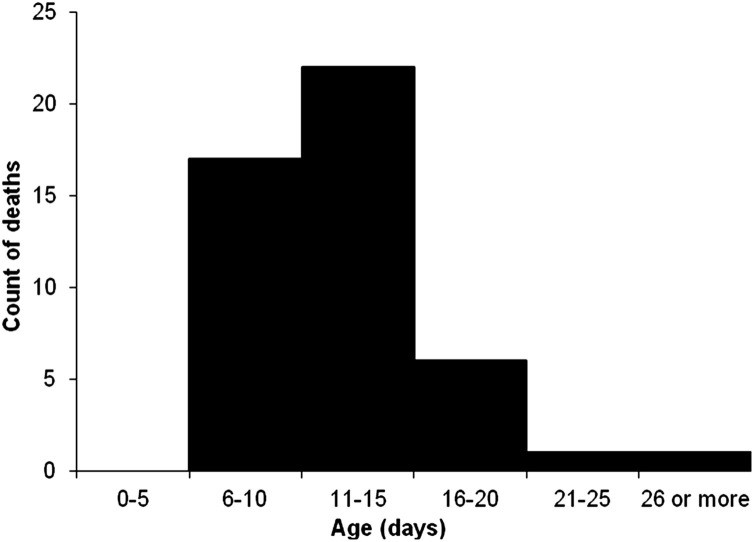
Incidence is a measure of the driving force of disease: the rate at which new cases of disease are occurring. The population at risk is the denominator for incidence and by itself is a measure of importance because it may describe a dynamic population. The process of gathering history, conducting physical examinations, reviewing the literature, and developing a case definition should help clarify who is at risk for the disease. Only a subset of animals in a population may be at risk for a given disease based on various conditions or characteristics, including age, gender, breed, physical location, and so forth (eg, only females are at risk for pregnancy and only pregnant cows are at risk for abortion). Neonatal calf diarrhea typically occurs at a narrow range in age, so, for example, it may be appropriate to define the population at risk as calves less than 4 weeks’ of age (Fig. 1 ).
Fig. 1.
Frequency distribution by age in days of beef ranch calves (n = 47) that died of neonatal calf diarrhea among a population of 402 calves.
(From Smith DR, Grotelueschen DM, Knott T, et al. Population dynamics of undifferentiated neonatal calf diarrhea among ranch beef calves. Bovine Practitioner 2008;42(8):1–8; with permission.)
Characterize the outbreak by time, space, and unit (eg, animal, pen, or herd)
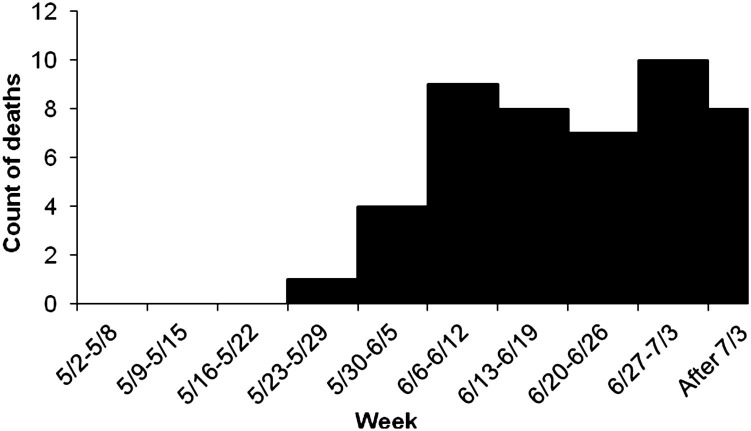
Often a disease outbreak investigation is initiated long after cases have occurred. It may be possible to reconstruct the outbreak by asking questions, analyzing health records, or preferably both. The most common way of doing this would be to characterize the outbreak by who got sick, when, and where. The who may be specifically identified individuals about whom you may have additional information or it may be group-level information (eg, the pens or groups that have been affected). This process may be iterative because there may be refinements in the case definition and the definition of the population at risk as the outbreak is reconstructed. It may be useful to graphically portray this information, for example, in the form of frequency histograms. A frequency histogram of particular interest is the count of cases plotted by time, also known as an epidemic curve (Fig. 2 ).
Fig. 2.
Epidemic curve of deaths occurring each week of the calving season caused by neonatal calf diarrhea among beef ranch calves.
(From Smith DR, Grotelueschen DM, Knott T, et al. Population dynamics of undifferentiated neonatal calf diarrhea among ranch beef calves. Bovine Practitioner 2008;42(8):1–8; with permission.)
Determine the nature of the outbreak
The shape of the epidemic curve may provide clues about the nature of the disease process and factors that have favored the occurrence of clinical signs.7 For example, outbreaks that occur as a point-source epidemic (eg, because of a sudden exposure to a pathogen, the sudden loss of immunity, or something that suddenly facilitates pathogen transmission) may be evidenced as an epidemic curve with a high peak in a relatively short period of time. When the outbreak is propagated (eg, when the disease process is one of ongoing transmission or there is a continuous presence of risk factors), the epidemic curve may appear flatter over a longer period of time. Sometimes outbreaks begin as a point-source exposure but are followed by secondary transmission and a propagated epidemic.
Develop hypotheses about possible causal risk factors
The causes of neonatal diarrhea in calves include more than the etiologic agent. In fact, it may be rare that knowledge of the presence of an agent by itself leads to a solution to outbreaks of neonatal calf diarrhea. To make effective recommendations for corrective actions following an outbreak, one often needs to understand how the pathogen interacts within the production system. For example, once you know the agent involved, it might still be useful to understand the various sources of the agent and routes of transmission on the farm.
Neonatal calf diarrhea is a complex, multifactorial, and temporally dynamic disease process.9, 10, 11 The factors that explain neonatal disease are the agent, factors related to host resistance or susceptibility to disease, and factors about the environment that favor the host or agent. These factors interact dynamically over time. To control the disease or prevent its occurrence, it is important to understand the relationships between these multiple factors and how they interact within the production system.12 It may be more useful to diagnose the failure of passive transfer or recognize a vehicle of pathogen transmission than to know the specific agent involved. Population diagnostics is concerned with correctly diagnosing or classifying the outbreak by agent, host, environment, and temporal factors.
Agent factors
Numerous infectious agents have been recovered from calves with neonatal diarrhea.9, 10, 12, 13, 14, 15, 16, 17, 18, 19 The most common agents of neonatal calf diarrhea include bacteria, such as Escherichia coli and Salmonella; viruses, such as rotavirus and coronavirus; and protozoa, such as cryptosporidia. Bovine rotavirus, bovine coronavirus, and cryptosporidia can be found in most cattle populations and can be recovered from calves in herds not experiencing calf diarrhea.12 It is not unusual to be able to find multiple agents in herds experiencing outbreaks of calf diarrhea, suggesting that even during outbreaks, more than one agent may be involved. The adult cow herd commonly serves as the reservoir of pathogens from one year to the next.20, 21, 22, 23, 24, 25
Often outbreak investigations take place long after cases have occurred, so opportunities to identify pathogens have past. If neonatal diarrhea cases are ongoing, pathogen diagnostics may be helpful to the investigation (see article by Blanchard in this issue). For example, it may be useful to know if the pathologic condition is caused by bacterial or viral pathogens because that knowledge may guide treatment decisions, lead to hypotheses about routes and sources of exposure, or create awareness of zoonotic potential. Also, serologic responses or even recovery of the agent are not necessarily clear indications that the pathogen is a primary pathogen in the condition being investigated. The recovery of one pathogen may not mean that there are not other players, and the relative prevalence of agents recovered from neonatal calf diarrhea cases may simply reflect the relative ease of detecting the agent. Finally, diagnostics for the purpose of outbreak investigation are almost always conducted to answer population-based questions. Rarely is the question about what pathogen is causing diarrhea in a particular calf. More often the question is whether to and how to adequately test for pathogens that are affecting the health of the herd.
The investigator might first determine if knowledge of the agents involved will change the way current cases are treated or future cases will be prevented. If it would be useful to know the pathogens involved, it is reasonable to want the testing process to accurately reflect the status of the herd.26, 27 Ineffective herd-level testing might either fail to identify important pathogens present in the herd or falsely classify a herd as having a pathogen it does not, either way, misdirecting the investigation.
To determine if herd-level testing will have adequate negative or positive predictive value you must know: (1) what samples and test methods are appropriate for diagnosing the presence of the pathogens of interest, (2) how well those tests perform, (3) if samples can be collected from representative cases or appropriate environmental sources, (4) if an adequate number of cases can be sampled, and (5) how many test-positive results are necessary to classify the herd as infected. The number of samples needed to make a herd-level diagnosis depends on (1) the expected prevalence of the pathogen within the herd, (2) the performance of the test on individual samples (eg, sensitivity and specificity), and (3) the likelihood of the herd having the pathogen.26 When a diagnosis is rare, then the herd-level positive predictive value may be low, especially if the herd-level specificity is low. When a diagnosis is common, the herd-level negative predictive value may be low, especially if the herd-level sensitivity is low.
In conducting population (eg, herd-level) diagnostics, the herd is often classified by a process of testing many individuals. In this situation, the herd-level sensitivity improves because of the many opportunities to find the agent; however, the herd-level specificity decreases because there are more opportunities to find a false-positive result. Therefore, confirmatory testing using a test of high specificity (eg, serial testing) may be important.26 For example, a single bovine viral diarrhea virus (BVDV)-positive enzyme-linked immunosorbent assay test result might lead the investigator to think BVDV is circulating in the herd, taking the investigation and proposed corrective actions down a particular (and possibly costly) path. Confirmatory testing (eg, using immunohistochemistry) might be prudent before going down that path. The issues of predictive value of a test are equally relevant to diagnostics of host, environmental, and temporal risk factors.
Host factors
Calves obtain passive immunity against common agents of calf diarrhea after absorbing antibodies from colostrum or colostrum supplements shortly after birth.28, 29, 30 The quantity of antibodies absorbed is determined by the quality and quantity of colostrum the calf ingests and how soon after birth it is ingested. The presence of maternal antibodies against specific agents in colostrum requires that the dam had prior exposure to antigens of the agent. Vaccines are used to immunize the dam against specific agents, and some commercially available colostrum supplements contain polyclonal or monoclonal antibodies directed against specific agents. Unfortunately, the use of vaccines or colostrum supplements does not always prevent neonatal calf diarrhea.
Calves commonly become ill or die of neonatal diarrhea within 1 to 2 weeks’ of age.9, 13, 15, 31, 32 The relatively narrow age range for neonatal calf diarrhea is not explained solely by the incubation period of the agents. Diarrhea may be observed in colostrum-deprived and gnotobiotic calves within a few days of pathogen challenge regardless of age.33, 34, 35 Calves may have an age-specific susceptibility to neonatal diarrhea that occurs as maternal immunity is waning and before the calf is capable of an active immune response.28
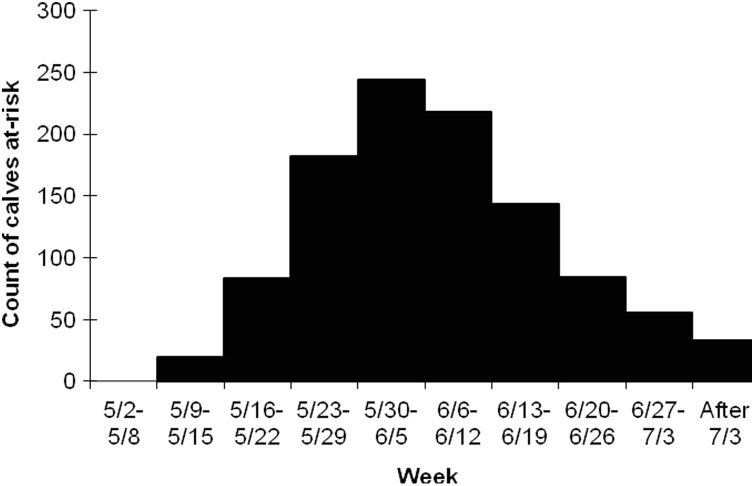
Regardless of the reason for the age-specificity of neonatal calf diarrhea, this period defines the age of calf susceptibility as well as the age calves are most likely to become infective and shed the agents in their feces.25, 36, 37, 38, 39, 40 Age-specificity of susceptibility and infectivity has important implications for preventing the transmission of the pathogens of neonatal diarrhea because, in some calving systems, the number of susceptible and infective calves can change dynamically with time (Fig. 3 ). At times, the number of potentially infective calves may greatly outnumber the number of susceptible calves resulting in an overwhelming opportunity for effective contacts.
Fig. 3.
Frequency distribution of the number of ranch calves considered at risk for neonatal calf diarrhea for each week of the calving season. The at-risk population was defined as the number of calves born in the previous 3 weeks and alive at the beginning of that week.
(From Smith DR, Grotelueschen DM, Knott T, et al. Population dynamics of undifferentiated neonatal calf diarrhea among ranch beef calves. Bovine Practitioner 2008;42(8):1–8; with permission.)
The dam’s age is important to a calf’s risk for undifferentiated neonatal diarrhea. Calves born to heifers are at a higher risk for neonatal diarrhea and have lower maternal antibody levels than calves born to older cows.41 Calves born to heifers are probably more susceptible to disease because, compared with older cows, heifers produce less volume and lower quality of colostrum, may have poorer mothering skills, and are more likely to experience dystocia.42, 43
Environmental factors
The rate and magnitude of pathogen exposure as well as the ability of the calf to resist disease may be affected by environmental conditions. Exposure to pathogens may occur through direct contact with other cattle or via contact with contaminated environmental surfaces. Environmental hygiene has long been recognized as important for controlling neonatal calf diarrhea,44, 45 but providing effective hygiene has often been a challenge. An effective contact is an exposure to pathogens of a dose load or duration sufficient to cause disease. Crowded conditions increase opportunities for effective contacts with infected animals or contaminated surfaces. Ambient temperature (eg, excessive heat or cold) and moisture (eg, mud or snow) are important stressors that impair the ability of the calf to resist disease and may influence pathogen numbers as well as opportunities for oral ingestion.
Temporal factors
Risk factors related to time can be associated with calendar time (ie, chronologic order [eg, where a factor occurs on the calendar]) or relative time (ie, time measured from a zero point to an event [eg, the time that has elapsed since birth: age]).32
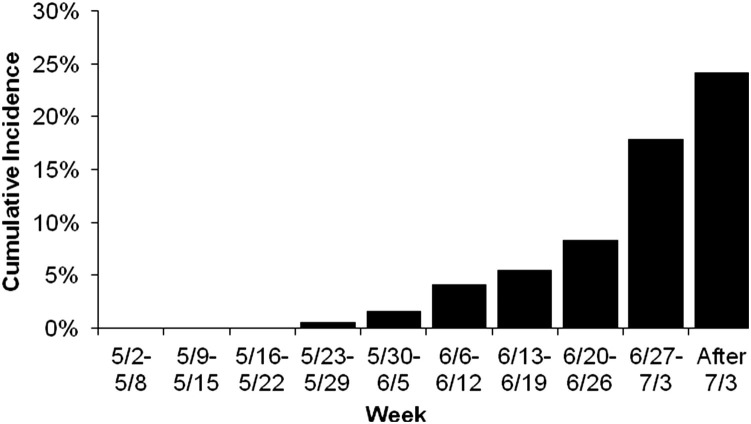
Host susceptibility, pathogen exposure, and pathogen transmission occur dynamically over calendar time within the calving season in beef herds and across seasons in dairies.12 Although the adult cow herd likely serves as the reservoir of neonatal diarrhea pathogens from year to year,20, 21, 22, 23, 24, 25 the average dose load of pathogen exposure to calves is likely to increase over time within a calving season because calves infected earlier serve as pathogen multipliers and become the primary source of exposure to younger susceptible calves. This multiplier effect can result in high calf infectivity and widespread environmental contamination with pathogens.46 Each calf serves as growth media for pathogen production, amplifying the dose load of the pathogen it received.35, 36, 37 Therefore, calves born later in a calving season may receive larger dose loads of pathogens and, in turn, may become relatively more infective by growing even greater numbers of agents. Eventually, the dose load of pathogens overwhelms the calf’s ability to resist disease. These factors alone or in combination may explain observations that calves born later in the calving season are at greater risk for disease or death (Fig. 4 ).31, 32
Fig. 4.
Cumulative incidence of death caused by neonatal calf diarrhea for each week of the calving season in beef ranch calves. Cumulative incidence was calculated as the number of deaths occurring each week divided by the number of calves at risk for undifferentiated neonatal calf diarrhea.
(From Smith DR, Grotelueschen DM, Knott T, et al. Population dynamics of undifferentiated neonatal calf diarrhea among ranch beef calves. Bovine Practitioner 2008;42(8):1–8; with permission.)
Test causal hypotheses using epidemiologic principles
The specific epidemiologic methods used to identify causes for disease outbreak will vary depending on the nature of the outbreak, time and resources available, and how well cases can be identified and exposures quantified. In disease causal theory, each factor that contributes to the development of disease is a component cause. Disease is observed when various component causes add up to complete a sufficient cause, which explains why some component causes are observed in the absence of disease (eg, we might recover Cryptosporidia from the feces of calves without diarrhea). This concept also explains why the manager of a herd that does a good job of assuring passive transfer might blame calf diarrhea on bad weather, whereas the manager of a herd in a moderate climate might observe diarrhea when they have been lax about assuring adequate colostrum intake. Removing one component cause means that the sufficient cause is not completed and, thus, disease is not observed.47 For example, one sufficient cause for neonatal calf diarrhea might include component causes of (1) exposure to rotavirus, (2) failure of passive transfer, and (3) weather stress. In the absence of other completed sufficient causes, the removal of any one of these factors would keep calves from developing diarrhea. In general, the objective of the investigation is to determine which possible component causes are contributing to the completion of a sufficient cause and determine which component causes (also known as causal factors or risk factors) are key determinants. Key determinants are those causal factors that can be modified. In the simple example described earlier, we might find it difficult to prevent exposure to rotavirus or correct the weather, so improving passive transfer might be the key determinant.
Gay48 recently reviewed causal thinking about animal diseases. Koch,49 Hill,50 Susser,51 and Evans52 have described important causal criteria for health investigations. Each has helped guide our thinking about causes of disease.48 Each suggests making a case for a causal relationship by finding the proposed cause (1) preceding the effect, (2) having a strong association with the effect, (3) being associated with the effect in more than one study, (4) demonstrating a dose effect, and (5) being consistent with current knowledge. These criteria can help guide the outbreak investigation; although, it is important to recognize that none are necessary or sufficient for determining if a causal relationship exists. Also important is the recognition of the cognitive biases that may prevent veterinarians from drawing the correct conclusions from an investigation.48
Sometimes the primary causal question is this: What is different about the calves that got sick and those that did not? But more often the question is this: What is the difference between this herd and other herds not experiencing an outbreak? Some causal factors are characteristics of individuals; other causes are characteristics of groups or herds. If the important causal factors occur at the herd level, it will not be possible to measure this association by comparing (statistically or otherwise) affected and nonaffected calves within a single herd. For example, if an important causal factor is the presence of BVDV circulating among cattle in the herd, it is likely that all calves become exposed to the virus, whether they get sick or not. The inference that BVDV is contributing to disease comes from comparing its presence in this herd with what is expected or observed in other herds. On the other hand, in this same herd, it may be possible to determine that failure of passive protection is also a causal factor by comparing postsuckling immunoglobulin G levels between affected and nonaffected calves within the same herd.
Outbreak investigations of neonatal calf diarrhea are usually qualitative rather than quantitative because useful quantitative data (eg, from health records) are often not available for analysis. A qualitative investigation relies on more subjective observations, including partial records, memory, and perceptions of relationships. Causal inferences in qualitative investigations are largely based on systems of logic developed by John Stuart Mill in the nineteenth century.48 According to Mill, a causal factor might be identified by the method of agreement if the factor is common to multiple instances of the outcome when other factors are dissimilar (eg, finding that scours outbreaks are common to herds feeding a particular milk replacer, even though other management practices differ). A causal factor might also be identified by the method of difference if a particular factor differs, whereas others remain the same (eg, if calves receiving milk replacer from one bag develop diarrhea, but calves on the same farm receiving milk replacer from another bag do not). Finally, causal relationships may be revealed by the method of concomitant variations if the risk for the outcome changes with the level of the risk factor, all other factors being the same (eg, the more time that calves remain in the maternity pen, the greater the incidence of diarrhea).
When data are available, a quantitative approach is often more useful for discovering causal relationships and evaluating the effectiveness of interventions. The best study design for evaluating causal relationships depends on the circumstances. There are 3 basic observational study designs: (1) case-control, (2) cohort or longitudinal, and (3) cross-sectional. Case-control studies compare odds of exposure among cases to the odds of exposure among noncases. Case-control studies excel when the disease is rare and when there are many potential exposures to test. Cohort and longitudinal studies compare incidence of disease among subjects with an exposure to the incidence of those without the exposure. Cohort and longitudinal studies are best when data exist to follow subjects over time, either prospectively or retrospectively. Cross-sectional studies look at the relationship between disease and exposure prevalence at a point in time.53
The statistical measure of association is important because it helps relate the strength of the relationship between the risk factor and the occurrence of disease. When the outcome is dichotomous (eg, diseased or not diseased), the measure of association is the odds ratio or (in some cases) relative risk (RR). These measures are measures of the odds, probability, or incidence rate of observing disease with an exposure level compared with another exposure level (Figs. 5 and 6 ). If the odds ratio has a value of 1, then the exposure is not associated with the disease. If the odds ratio is greater than 1, then that exposure is associated with the disease. If the odds ratio is less than 1, then the exposure is associated with the absence of disease (eg, it is protective from disease). The further the odds ratio is from 1, the stronger the association.
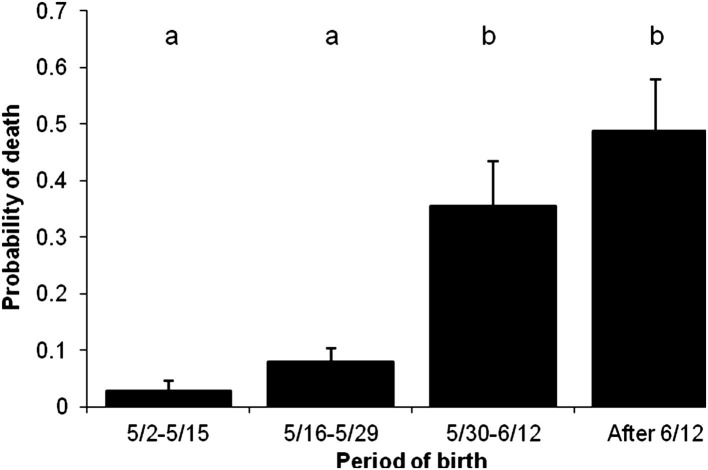
Fig. 5.
Model-adjusted probability for calves to die of neonatal calf diarrhea, depending on the period of time the calf was born during the calving season. Calves differed significantly in their risk for death depending on how late in the calving season they were born. Error bars represent the standard error of the mean. The outcomes of variables with different superscripts are statistically different (P≤ .05). Compared with calves born in the first 4 weeks, calves born in the last month of calving were 10 times more likely to die (relative risk = 10).
(From Smith DR, Grotelueschen DM, Knott T, et al. Population dynamics of undifferentiated neonatal calf diarrhea among ranch beef calves. Bovine Practitioner 2008;42(8):1–8; with permission.)
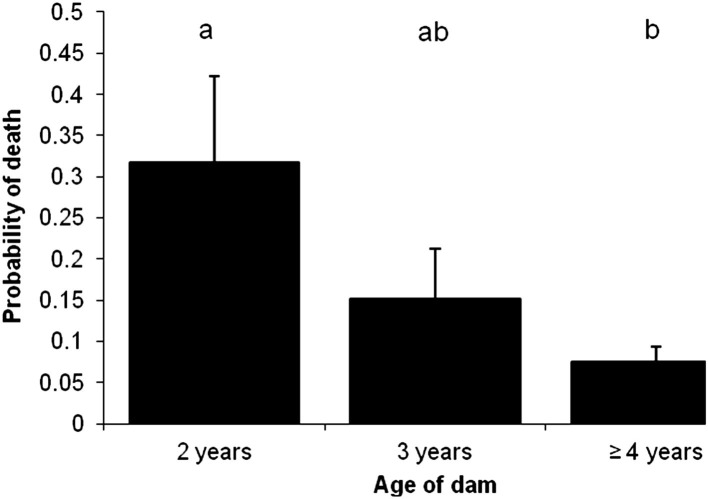
Fig. 6.
Model-adjusted probability for calves to die of neonatal calf diarrhea for calves born to dams of different ages. Error bars represent the standard error of the mean. The outcomes of variables with different superscripts are statistically different (P≤.05). Compared with calves born to mature cows, calves born from heifers were 4 times more likely to die (relative risk = 4).
(From Smith DR, Grotelueschen DM, Knott T, et al. Population dynamics of undifferentiated neonatal calf diarrhea among ranch beef calves. Bovine Practitioner 2008;42(8):1–8; with permission.)
Often quantitative analyses use statistical measures, such as the P value, to help make causal inferences.54 The P value helps us judge the role of chance in observing the measure of association. It is the probability of observing that relationship, or something more extreme, if in truth no relationship exists. It is a common convention in research to consider a finding statistical significant when the P value is equal to or less than 0.05. However, outbreak investigations often involve small sample sizes with low power to detect differences at that level of significance. In outbreak investigations, it is usually appropriate to interpret the P value liberally, if at all.
Develop Strategies to Prevent New Cases and Future Outbreaks
With some luck, the result of the outbreak investigation, whether qualitative or quantitative, is a list of potential causal factors that may relate to the agent, host immunity, or the environment. The art of the investigation is determining which factors are key determinants (also called control points) for preventing new cases and future outbreaks of neonatal calf diarrhea. The corrective actions should be (1) manageable (ie, within the capabilities of the caretakers), (2) economical (ie, not cost more than the disease), and (3) most likely to be effective (ie, to have the greatest impact).
Decisions on the relative importance of various causal factors in the outbreak should include consideration for the impact of the factor on the exposed (attributable risk of exposure) and on the population (population attributable risk). For example, attributable risk of exposure estimates the proportion of cases among the exposed that are actually caused by that exposure. Some calves infected with failure of passive transfer do not develop diarrhea and some develop diarrhea for other reasons, so for calves with failure of passive transfer, what proportion of cases are caused by failure of passive transfer? Attributable risk of exposure is a function of relative risk.
Attributable risk of exposure = (RR-1)/RR
where RR is relative risk.
Not all calves in the population have the causal exposure but they may still develop diarrhea. For example, if failure of passive transfer is suspected as a causal factor in the outbreak, then the impact to the herd (population attributable risk) of taking corrective action to improve passive transfer depends on how strong the association is between failure of passive transfer and diarrhea and how commonly failure of passive transfer occurs.
Population attributable risk = PE × (RR-1)/(PE × [RR-1]) + 1)
where P E is probability of exposure (to failure of passive transfer) and RR is relative risk (of diarrhea for calves with failure of passive transfer compared to calves with adequate passive transfer).
For this example, the proportion of cases prevented in the herd (population attributable risk) by correcting failure of passive transfer is greater for herds with high rates of failure of passive transfer.
Biocontainment of neonatal calf diarrhea
Biosecurity is the sum of actions taken to prevent introducing a disease agent into a population (pen, herd, region), whereas biocontainment describes the actions taken to control or prevent the transmission of a pathogen already present in the population.55 In the context of disease, the term control refers to actions that reduce the prevalence of infected individuals (eg, actions to treat infected individuals to eliminate infection). The term prevention refers to actions that reduce the rate of pathogen transmission or disease occurrence (ie, reduce the incidence of new infections).
In theory, neonatal calf diarrhea could be prevented by (1) eliminating the pathogens from the herd and its environment (eg, create a biosecure herd), (2) improving calf resistance to disease, or (3) altering the production system to minimize opportunities for calves to be exposed to existing pathogens and transmitting the infection to others. However, the endemic nature of the common pathogens of neonatal calf diarrhea makes it unlikely that cattle populations could be made biosecure from these agents; therefore, a biocontainment approach to control neonatal calf diarrhea seems prudent and logical.55, 56 Biocontainment actions against neonatal diarrhea might include those that improve passive transfer of maternal antibodies or reducing effective contacts. Maternal immunity is important to calf resistance to enteric agents,11, 57 but maternal immunity wanes with time.28 Dairy producers might be able to improve maternal immunity by managing colostrum quality and quantity, but managers of extensive beef cattle systems have limited practical opportunities to improve rates of passive antibody transfer. In addition, vaccines are not available against all pathogens of calf diarrhea, may not induce sufficient cross-protection,40 and pathogens may evade the protection afforded by vaccination by evolving away from vaccine strains.58
Effective contacts can be prevented by (1) physically separating animals, (2) reducing the level of exposure (eg, through the use of sanitation or dilution over space), or (3) minimizing contact time. These principles have been successfully applied in calf hutch systems to control neonatal diseases in dairy calves.59 Various systems for biocontainment have been developed to prevent neonatal calf diarrhea in beef herds.60, 61, 62 Each of these are management strategies to prevent effective contacts by reducing the opportunities for exposure and subsequent transmission. For example, the Sandhills Calving System63 prevents effective contacts among beef calves by (1) segregating calves by age to prevent direct and indirect transmission of pathogens from older to younger calves and (2) scheduled movement of pregnant cows to clean calving pastures to minimize the pathogen dose load in the environment and minimizing contact time between calves and the larger portion of the cow herd.
Some herds are large enough to conduct their own in-house trials, or multiple herds might be included in a study to test the usefulness of a potential corrective action. If so, it is important to follow a valid experimental design with appropriate methods of data analysis.64
Communicate Your Observations and Findings with the Key Individuals
A written report should quickly follow the investigation. In the report, in simple language restate the problem, summarize the findings, explain your interpretation of the findings, and make recommendations to correct the problem. Use graphs, charts, and tables to supplement the report. Consider the risk for subsequent liability and be as complete as possible without overstating your case.
Validate that recommendations are being performed
The corrective actions recommended should be measureable. For example, measurable actions are that pregnant cows move to a new calving pasture every 7 days, calves are removed from the maternity barn within 4 hours of birth, or that 1 gal of colostrum is fed in the first 12 hours after birth. It is useful to establish a record-keeping system so that client compliance can be measured. For example, dates and times that procedures took place can be recorded with the initials of the individual responsible for the task.
Monitor progress
Outbreak investigations provide an educable moment for many cattle producers. This time may be right to convince herd managers of the value of an animal health and performance record-keeping system.65, 66 Records help validate that the corrective actions are working and a process control approach may catch problems before they become costly or harmful to calf health and well-being.67
Summary
Diarrhea is a leading cause of sickness and death of beef and dairy calves in their first month of life. Field investigations of outbreaks of neonatal calf diarrhea should be conducted for the purposes of (1) reducing the losses associated with existing cases and (2) preventing new cases from occurring.
Outbreak investigations for any infectious or toxicant-induced disease involve an orderly process of characterizing the outbreak and finding solutions using epidemiologic concepts that include the following:
-
•
Interviewing key individuals
-
•
Verifying the clinical diagnosis and assure that treatments are appropriate
-
•
Identifying the factors responsible for the outbreak using a rational diagnostic plan and qualitative or quantitative analytical methods
-
•
Developing strategies to prevent new cases and future outbreaks by identifying corrective actions likely to have the most impact without excessive cost
-
•
Communicating observations and recommendations with the key individuals, and using records to verify compliance and monitor progress.
Footnotes
The author has nothing to disclose.
References
- 1.USDA. Part II: reference of 1997 beef cow-calf health and management practices. USDA, APHIS, VS, CEAH, National Animal Health Monitoring System; Fort Collins (CO): 1997. [Google Scholar]
- 2.Anderson D.C., Kress D.D., Bernardini M.M. The effect of scours on calf weaning weight. Prof Anim Sci. 2003;19:399–403. [Google Scholar]
- 3.Swift B.L., Nelms G.E., Coles R. The effect of neonatal diarrhea on subsequent weight gains in beef calves. Vet Med Small Anim Clin. 1976;71(9):1269–1272. [PubMed] [Google Scholar]
- 4.Smith D.R. Management of neonatal diarrhea in cow-calf herds. In: Anderson D.E., Rings D.M., editors. Current veterinary therapy food animal practice. 5th edition. Saunders Elsevier; St Louis (MO): 2009. pp. 599–602. [Google Scholar]
- 5.Waldner C.L., Campbell J.R. Disease outbreak investigation in food animal practice. Vet Clin North Am Food Anim Pract. 2006;22(1):75–101. doi: 10.1016/j.cvfa.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Hancock D.D., Wikse S.E. Investigation planning and data gathering. Vet Clin North Am Food Anim Pract. 1988;4:1–15. doi: 10.1016/s0749-0720(15)31072-0. [DOI] [PubMed] [Google Scholar]
- 7.Lessard P.R. The characterization of disease outbreaks. Vet Clin North Am Food Anim Pract. 1988;4:17–32. doi: 10.1016/s0749-0720(15)31073-2. [DOI] [PubMed] [Google Scholar]
- 8.Mason G.L., Madden D.J. Performing the field necropsy examination. Vet Clin North Am Food Anim Pract. 2007;23(3):503–526. doi: 10.1016/j.cvfa.2007.07.006. vi. [DOI] [PubMed] [Google Scholar]
- 9.Acres S.D., Laing C.J., Saunders J.R. Acute undifferentiated neonatal diarrhea in beef calves. I. Occurrence and distribution of infectious agents. Can J Comp Med. 1975;39(2):116–132. [PMC free article] [PubMed] [Google Scholar]
- 10.Acres S.D., Saunders J.R., Radostits O.M. Acute undifferentiated neonatal diarrhea of beef calves: the prevalence of enterotoxigenic E. coli, reo-like (rota) virus and other enteropathogens in cow-calf herds. Can Vet J. 1977;18(5):274–280. [PMC free article] [PubMed] [Google Scholar]
- 11.Saif L.J., Smith K.L. Enteric viral infections of calves and passive immunity. J Dairy Sci. 1985;68(1):206–228. doi: 10.3168/jds.S0022-0302(85)80813-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrington G.M., Gay J.M., Evermann J.F. Biosecurity for neonatal gastrointestinal diseases. Vet Clin North Am Food Anim Pract. 2002;18(1):7–34. doi: 10.1016/S0749-0720(02)00005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bulgin M.S., Anderson B.C., Ward A.C. Infectious agents associated with neonatal calf disease in southwestern Idaho and eastern Oregon. J Am Vet Med Assoc. 1982;180(10):1222–1226. [PubMed] [Google Scholar]
- 14.Mebus C.A., Stair E.L., Rhodes M.B. Neonatal calf diarrhea: propagation, attenuation, and characteristics of coronavirus-like agents. Am J Vet Res. 1973;34:145–150. [PubMed] [Google Scholar]
- 15.Trotz-Williams L.A., Jarvie B.D., Martin S.W. Prevalence of Cryptosporidium parvum infection in southwestern Ontario and its association with diarrhea in neonatal dairy calves. Can Vet J. 2005;46(4):349–351. [PMC free article] [PubMed] [Google Scholar]
- 16.Athanassious R., Marsollais G., Assaf R. Detection of bovine coronavirus and type A rotavirus in neonatal calf diarrhea and winter dysentery of cattle in Quebec: evaluation of three diagnostic methods. Can Vet J. 1994;35:163–169. [PMC free article] [PubMed] [Google Scholar]
- 17.Naciri M., Lefay M.P., Mancassola R. Role of Cryptosporidium parvum as a pathogen in neonatal diarrhoea complex in suckling and dairy calves in France. Vet Parasitol. 1999;85(4):245–257. doi: 10.1016/S0304-4017(99)00111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morin M., Lariviere S., Lallier R. Pathological and microbiological observations made on spontaneous cases of acute neonatal calf diarrhea. Can J Comp Med. 1976;40(3):228–240. [PMC free article] [PubMed] [Google Scholar]
- 19.Lucchelli A., Lance S.A., Bartlett P.B. Prevalence of bovine group A rotavirus shedding among dairy calves in Ohio. Am J Vet Res. 1992;53:169–174. [PubMed] [Google Scholar]
- 20.Crouch C.F., Bielefeldt Ohman H., Watts T.C. Chronic shedding of bovine enteric coronavirus antigen-antibody complexes by clinically normal cows. J Gen Virol. 1985;66:1489–1500. doi: 10.1099/0022-1317-66-7-1489. [DOI] [PubMed] [Google Scholar]
- 21.Collins J.K., Riegel C.A., Olson J.D. Shedding of enteric coronavirus in adult cattle. Am J Vet Res. 1987;48:361–365. [PubMed] [Google Scholar]
- 22.Crouch C.F., Acres S.D. Prevalence of rotavirus and coronavirus antigens in the feces of normal cows. Can J Comp Med. 1984;48:340–342. [PMC free article] [PubMed] [Google Scholar]
- 23.McAllister T.A., Olson M.E., Fletch A. Prevalence of Giardia and Cryptosporidium in beef cows in southern Ontario and in beef calves in southern British Columbia. Can Vet J. 2005;46(1):47–55. doi: 10.4141/cjas66-008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe Y., Yang C.H., Ooi H.K. Cryptosporidium infection in livestock and first identification of Cryptosporidium parvum genotype in cattle feces in Taiwan. Parasitol Res. 2005;97(3):238–241. doi: 10.1007/s00436-005-1428-1. [DOI] [PubMed] [Google Scholar]
- 25.Ralston B.J., McAllister T.A., Olson M.E. Prevalence and infection pattern of naturally acquired giardiasis and cryptosporidiosis in range beef calves and their dams. Vet Parasitol. 2003;114(2):113–122. doi: 10.1016/s0304-4017(03)00134-1. [DOI] [PubMed] [Google Scholar]
- 26.Smith D.R. Epidemiologic tools for biosecurity and biocontainment. Vet Clin North Am Food Anim Pract. 2002;18(1):157–175. doi: 10.1016/s0749-0720(02)00006-3. [DOI] [PubMed] [Google Scholar]
- 27.McKenna S.L., Dohoo I.R. Using and interpreting diagnostic tests. Vet Clin North Am Food Anim Pract. 2006;22(1):195–205. doi: 10.1016/j.cvfa.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Barrington G.M., Parish S.M. Bovine neonatal immunology. Vet Clin North Am Food Anim Pract. 2001;17(3):463–476. doi: 10.1016/S0749-0720(15)30001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Besser T.E., Gay C.C. The importance of colostrum to the health of the neonatal calf. Vet Clin North Am Food Anim Pract. 1994;10(1):107–117. doi: 10.1016/s0749-0720(15)30591-0. [DOI] [PubMed] [Google Scholar]
- 30.Besser T.E., Gay C.C., McGuire T.C. Passive immunity to bovine rotavirus infection associated with transfer of serum antibody into the intestinal lumen. J Virol. 1988;62(7):2238–2242. doi: 10.1128/jvi.62.7.2238-2242.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clement J.C., King M.E., Salman M.D. Use of epidemiologic principles to identify risk factors associated with the development of diarrhea in calves in five beef herds. J Am Vet Med Assoc. 1995;207(10):1334–1338. [PubMed] [Google Scholar]
- 32.Smith D.R., Grotelueschen D.M., Knott T. Population dynamics of undifferentiated neonatal calf diarrhea among ranch beef calves. Bovine Practitioner. 2008;42(8):1–8. [Google Scholar]
- 33.El-Kanawati Z.R., Tsunemitsu H., Smith D.R. Infection and cross-protection studies of winter dysentery and calf diarrhea bovine coronavirus strains in colostrum-deprived and gnotobiotic calves. Am J Vet Res. 1996;57:48–53. [PubMed] [Google Scholar]
- 34.Heckert R.A., Saif L.J., Mengel J.P. Mucosal and systemic antibody responses to bovine coronavirus structural proteins in experimentally challenge-exposed calves fed low or high amounts of colostral antibodies. Am J Vet Res. 1991;52(5):700–708. [PubMed] [Google Scholar]
- 35.Saif L.J., Redman D.R., Moorhead P.D. Experimentally induced coronavirus infections in calves: viral replication in the respiratory and intestinal tracts. Am J Vet Res. 1986;47:1426–1432. [PubMed] [Google Scholar]
- 36.Kapil S., Trent A.M., Goyal S.M. Excretion and persistence of bovine coronavirus in neonatal calves. Arch Virol. 1990;115:127–132. doi: 10.1007/BF01310629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uga S., Matsuo J., Kono E. Prevalence of Cryptosporidium parvum infection and pattern of oocyst shedding in calves in Japan. Vet Parasitol. 2000;94(1–2):27–32. doi: 10.1016/s0304-4017(00)00338-1. [DOI] [PubMed] [Google Scholar]
- 38.Nydam D.V., Wade S.E., Schaaf S.L. Number of Cryptosporidium parvum oocysts of Giardia spp. cysts by dairy calves after natural infection. Am J Vet Res. 2001;62(10):1612–1615. doi: 10.2460/ajvr.2001.62.1612. [DOI] [PubMed] [Google Scholar]
- 39.O'Handley R.M., Cockwill C., McAllister T.A. Duration of naturally acquired giardiasis and cryptosporidiosis in dairy calves and their association with diarrhea. J Am Vet Med Assoc. 1999;214(3):391–396. [PubMed] [Google Scholar]
- 40.Murakami Y., Nishioka N., Watanabe T. Prolonged excretion and failure of cross-protection between distinct serotypes of bovine rotavirus. Vet Microbiol. 1986;12(1):7–14. doi: 10.1016/0378-1135(86)90036-2. [DOI] [PubMed] [Google Scholar]
- 41.Schumann F.J., Townsend H.G., Naylor J.M. Risk factors for mortality from diarrhea in beef calves in Alberta. Can J Vet Res. 1990;54(3):366–372. [PMC free article] [PubMed] [Google Scholar]
- 42.Odde K.G. Reducing neonatal calf losses through selection, nutrition and management. Agri-Practice. 1996;17(3/4):12–15. [Google Scholar]
- 43.Odde K.G. Survival of the neonatal calf. Vet Clin North Am Food Anim Pract. 1988;4:501–508. doi: 10.1016/s0749-0720(15)31027-6. [DOI] [PubMed] [Google Scholar]
- 44.Law J. United States Department of Agriculture, Bureau of Animal Industry; Washington, DC: 1916. Diseases of young calves. Special report on diseases of cattle. p. 245–61. [Google Scholar]
- 45.Van Es L. John Wiley and Sons, Inc.; New York: 1932. White scours. The principles of animal hygiene and preventive veterinary medicine. p. 504–13. [Google Scholar]
- 46.Atwill E.R., Johnson E.M., Pereira M.G. Association of herd composition, stocking rate, and duration of calving season with fecal shedding of Cryptosporidium parvum oocysts in beef herds. J Am Vet Med Assoc. 1999;215(12):1833–1838. [PubMed] [Google Scholar]
- 47.Rothman K.J. Causes. Am J Epidemiol. 1976;104:587–592. doi: 10.1093/oxfordjournals.aje.a112335. [DOI] [PubMed] [Google Scholar]
- 48.Gay J.M. Determining cause and effect in herds. Vet Clin North Am Food Anim Pract. 2006;22(1):125–147. doi: 10.1016/j.cvfa.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 49.Koch R. Die aetiologie der tuberkulose. Mitt Kaiser Gesundh. 1884;2:1–88. [in German] [Google Scholar]
- 50.Hill A.B. The environment and disease: association or causation? Proc R Soc Med. 1965;58:295–300. doi: 10.1177/003591576505800503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Susser M. What is a cause and how do we know one? A grammar for pragmatic epidemiology. Am J Epidemiol. 1991;133:635–648. doi: 10.1093/oxfordjournals.aje.a115939. [DOI] [PubMed] [Google Scholar]
- 52.Evans A.S. Causation and disease: a chronological journey. Am J Epidemiol. 1978;108:249–258. doi: 10.1093/oxfordjournals.aje.a112617. [DOI] [PubMed] [Google Scholar]
- 53.Shott S. Designing studies that answer questions. J Am Vet Med Assoc. 2011;238(1):55–58. doi: 10.2460/javma.238.1.55. [DOI] [PubMed] [Google Scholar]
- 54.Slenning B.D. Hood of the truck statistics for food animal practitioners. Vet Clin North Am Food Anim Pract. 2006;22(1):149–170. doi: 10.1016/j.cvfa.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 55.Dargatz D.A., Garry F.B., Traub-Dargatz J.L. An introduction to biosecurity of cattle operations. Vet Clin North Am Food Anim Pract. 2002;18(1):1–5. doi: 10.1016/s0749-0720(02)00002-6. [DOI] [PubMed] [Google Scholar]
- 56.Larson R.L., Tyler J.W., Schultz L.G. Management strategies to decrease calf death losses in beef herds. J Am Vet Med Assoc. 2004;224(1):42–48. doi: 10.2460/javma.2004.224.42. [DOI] [PubMed] [Google Scholar]
- 57.Nocek J.E., Braund D.G., Warner R.G. Influence of neonatal colostrum administration, immunoglobulin, and continued feeding of colostrum on calf gain, health, and serum protein. J Dairy Sci. 1984;67(2):319–333. doi: 10.3168/jds.S0022-0302(84)81305-3. [DOI] [PubMed] [Google Scholar]
- 58.Lu W., Duhamel G.E., Benfield D.A. Serological and genotypic characterization of group A rotavirus reassortants from diarrheic calves born to dams vaccinated against rotavirus. Vet Microbiol. 1994;42(2–3):159–170. doi: 10.1016/0378-1135(94)90015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanders D.E. Field management of neonatal diarrhea. Vet Clin North Am Food Anim Pract. 1985;1(3):621–637. doi: 10.1016/s0749-0720(15)31307-4. [DOI] [PubMed] [Google Scholar]
- 60.Radostits O.M., Acres S.D. The control of acute undifferentiated diarrhea of newborn beef calves. Vet Clin North Am Large Anim Pract. 1983;5(1):143–155. doi: 10.1016/S0196-9846(17)30097-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomson J.U. Implementing biosecurity in beef and dairy herds. Proc Am Assoc Bov Pract. 1997;30:8–14. [Google Scholar]
- 62.Pence M., Robbe S., Thomson J. Reducing the incidence of neonatal calf diarrhea through evidence-based management. Compendium on Continuing Education for the Practicing Veterinarian. 2001;23:S73–S75. [Google Scholar]
- 63.Smith D.R., Grotelueschen D.M., Knott T. Prevention of neonatal calf diarrhea with the Sandhills calving system. Proc Am Assoc Bov Pract. 2004;37:166–168. [Google Scholar]
- 64.Sanderson M.W. Designing and running clinical trials on farms. Vet Clin North Am Food Anim Pract. 2006;22(1):103–123. doi: 10.1016/j.cvfa.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 65.Rae D.O. Assessing performance of cow-calf operations using epidemiology. Vet Clin North Am Food Anim Pract. 2006;22(1):53–74. doi: 10.1016/j.cvfa.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 66.Kelton D.F. Epidemiology: a foundation for dairy production medicine. Vet Clin North Am Food Anim Pract. 2006;22(1):21–33. doi: 10.1016/j.cvfa.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 67.Reneau J.K., Lukas J. Using statistical process control methods to improve herd performance. Vet Clin North Am Food Anim Pract. 2006;22(1):171–193. doi: 10.1016/j.cvfa.2005.11.006. [DOI] [PubMed] [Google Scholar]