Abstract
The complete genomic DNA of a novel papillomavirus (PV) was isolated from a basosquamous carcinoma on the wing of an Egyptian fruit bat (Rousettus aegyptiacus). Initial short sequences of the E1 and L1 genes of this virus were retrieved by PCR with degenerate papillomavirus-specific primers, and the entire R. aegyptiacus papillomavirus type 1 (RaPV-1) DNA was then amplified by long template PCR, cloned and sequenced with a transposon insertion method. The RaPV-1 genome counts 7970 basepairs and contains the typical papillomavirus open reading frames (ORF) (E1, E2, E4, E6, E7, L1 and L2). Based on a concatenated alignment of the E1, E2, L1 and L2 open reading frames of RaPV-1 and 46 other human and animal papillomavirus type species, a neighbor-joining phylogenetic tree was constructed. This phylogenetic analysis shows that RaPV-1 has a close-to-root position in the papillomavirus evolutionary tree. Since RaPV-1 is only distantly related to other papillomaviruses (with maximally 50% nucleotide sequence identity across the L1 open reading frame), it cannot be assigned to one of the existing papillomavirus genera and therefore represents the first member of a novel, as yet unnamed, close-to-root papillomavirus genus. This is the first time a papillomavirus has been isolated and characterized from a member of the Chiroptera order.
Keywords: Papillomavirus, Fruit bat, Rousettus aegyptiacus, Chiroptera, Carcinoma, Phylogenetic analysis
1. Introduction
The Papillomaviridae are a large family of small, non-enveloped, double stranded DNA viruses with an epithelial tropism. They can cause benign and malignant proliferations of the stratified squamous epithelium of the skin and of the mucosa in various higher vertebrate species. In general, papillomaviruses (PVs) are highly species-specific, and interspecies transmission is a very rare event that has only been observed between closely related species (Sundberg, 1987, Sundberg et al., 1997). The vast majority of PV genotypes have been isolated from humans, the only intensively studied host, and a wide genetic diversity of different human papillomavirus (HPV) types is associated with a broad range of genotype-specific clinical conditions, ranging from genital and cutaneous warts to invasive cervical carcinoma and skin cancer (Van Ranst et al., 1992b, Syrjänen and Syrjänen, 2000). The limited number of non-human PVs that has been characterized to date covers a broad range of host species, including mostly domestic and wild mammals and two bird species (Sundberg, 1987, Sundberg et al., 1997, Tachezy et al., 2002b). Taken together with the numerous partial sequences of putative novel non-human PVs that have been reported (Chan et al., 1997, Antonsson and Hansson, 2002, Ogawa et al., 2004) this suggest that every vertebrate species could carry its own set of species-specific PVs.
In February 2003, a lesion of possible papillomaviral origin was noticed on an Egyptian fruit bat (Rousettus aegyptiacus), which to our knowledge is the first report of lesions suspicious of PV infection in a member of the Chiroptera. This animal presented with a small raised pigmented mass on the lateral canthus of the left eye, and later developed additional masses on the skin of the left wing membranes, showing bone invasion and displaying histopathologic features characteristic of basosquamous carcinoma. The papillomaviral origin of these lesions was confirmed by positive immunohistochemical staining with anti-HPV antibodies, and PCR amplification of a short DNA fragment of the L1 open reading frame (ORF) of an unknown papillomavirus type (McKnight et al., 2006).
A major impediment to the characterization of novel PVs is the absence of a conventional cell culture system for in vitro viral propagation. The isolation of PVs could therefore in the past only be accomplished when the virus was abundantly present. The recent methods for identification of novel PVs are mostly PCR-based, and use consensus or degenerate primers that are developed based on sequence information of previously characterized PV types. These primers are usually located in the L1 and/or E1 gene, which are the most conserved regions of the PV genome. This approach allows amplification and characterization of part of an unknown PV genome, even when this is only present in minute quantities. Given the circular structure of the PV genome, the complete genomic sequence can subsequently be amplified by inverse or overlapping long template PCR (Terai and Burk, 2002, Rector et al., 2005b). We have used this strategy to characterize the complete genome of a novel close-to-root PV from the carcinomatous wing lesion of the Egyptian fruit bat: the R. aegyptiacus PV type 1 (RaPV-1), which is the first PV isolated from a member of the Chiroptera order.
2. Materials and methods
2.1. Case summary
In February 2003, a first lesion of possible papillomaviral origin was noticed on an Egyptian fruit bat at the Organization for Bat Conservation in Bloomfield Hills, MI, USA. This animal presented with a small raised pigmented mass on the lateral canthus of the left eye, gradually increasing in size over a period of a few months. In January 2004, this animal developed six additional raised, smooth to cauliflower-like masses of variable size on the skin of the left wing membranes. Two of the lesions partially resolved over the next 2 months, and the other masses were surgically removed. The wing lesions showed bone invasion and the histopathologic features were characteristic of basosquamous carcinoma.
2.2. PCR and DNA sequencing
A frozen tissue biopsy of the basosquamous wing carcinoma from this animal was submitted to the Laboratory of Clinical and Epidemiological Virology at the Rega Institute for PV detection. The biopsy material was finely minced with a scalpel, and total genomic DNA was isolated by using phenol/chloroform extraction and ethanol precipitation as described previously (Rector et al., 2004b). Polymerase chain reactions with degenerate PV-specific primers were performed on the isolated DNA, with the following degenerate primer pairs (with the expected length of the amplification product, based on the sequence of HPV-1a): AR-E1F2/AR-E1R3 (371 bp), AR-E1F2/AR-E1R4 (552 bp) and AR-L1F8/AR-L1R9 (704 bp) (Rector et al., 2004a, Rector et al., 2005b). PCR was carried out in a total volume of 50 μl, containing 200 μM of each dNTP, 0.75 μM of forward and reverse primer, 1 U of Taq DNA polymerase (Perkin-Elmer/Roche Molecular Systems, Belgium), and 2.5 mM MgCl2 [pH 8.5], with 1 μl of the extracted DNA as template. PCR conditions comprised 10 min denaturation at 94 °C, 45 cycles of 1 min at 94 °C, 1 min at 50 °C and 1 min at 72 °C, followed by 5 min of final elongation at 72 °C. Amplicons suggestive of PV-specific amplification were generated with all these primer pairs. The PCR products were purified through 3% agarose gel electrophoresis and extraction of the PV-specific bands from the gel (QIAquick Gel Extraction Kit, QIAgen, Westburg, Leusden, The Netherlands), and sequenced with the same degenerate primers as used for PCR. Similarity searches, performed with the NCBI Basic Local Alignment Search Tool (BLASTN 2.2.10) server on the GenBank DNA database release 145.0 (Altschul et al., 1990), showed that partial E1 and L1 sequences of a novel PV were amplified. Primers for (inverse) long template PCR were chosen in these partial E1 and L1 sequences in order to amplify the complete genome of the R. aegyptiacus RaPV-1 in three overlapping long PCR fragments: RaPV-long1 of approximately 3.5 kb, amplified with forward and reverse primers RaPV-longF1 and RaPV-longR1, fragment RaPV-long2 of approximately 5.5 kb, amplified with primers RaPV-longF2 and RaPV-longR2, and fragment RaPV-long3 of approximately 8 kb, amplified with primers RaPV-longF3 and RaPV-longR3 (primer sequences available from the corresponding author upon request) (Fig. 1 ). Long template PCR was performed with the Expand Long Template PCR System (Roche Diagnostics, Mannheim, Germany). The PCR products were purified on a 0.8% agarose gel with crystal violet staining, and isolated from the gel by using SNAP purification columns (TOPO XL PCR Cloning kit, Invitrogen, Carlsbad, USA). The fragments were ligated into a pCR-XL-TOPO vector, followed by transformation into One Shot TOP10 competent cells (TOPO XL PCR Cloning Kit). The bacteria were selectively grown on LB agar plates containing 50 μg/ml kanamycin, and one clone containing the 3.5 kb RaPV-long1 PCR fragment, one containing the 5.5 kb RaPV-long2 PCR fragment and one containing the 8 kb RaPV-long3 fragment were selected. The EZ::TN™ < TET-1 > Insertion Kit (Epicentre, Landgraaf, The Netherlands) was used to sequence the cloned RaPV-long3 PCR fragment. This kit uses the Tn5 transposase to randomly insert primer binding sites and a tetracycline resistance selection marker into target DNA in vitro. The reaction was performed according to the manufacturer's protocol. The transposon reaction product was used to transform One Shot MAX Efficiency DH5α-T1R competent cells (Invitrogen). Twenty-four colonies were selected and the provided primers were used to sequence the insertion clones bi-directionally from primer binding sites at the 5′- and 3′-ends of the inserted transposon. The remaining gaps in the sequences were determined by primer-walking on the PCR-clones of the RaPV-long1, -long2 and -long3 fragments. Sequencing was performed on an ABI Prism 3100 Genetic Analyzer (Perkin-Elmer Applied Biosystems, Foster City, USA), chromatogram sequencing files were inspected with Chromas 2.2 (Technelysium, Helensvale, Australia), and contigs were prepared using SeqMan II (DNASTAR, Madison, USA). The complete nucleotide sequence of the R. aegyptiacus PV type 1 (RaPV-1) genome counts 7970 bp, and was deposited in GenBank under accession number DQ366842.
Fig. 1.
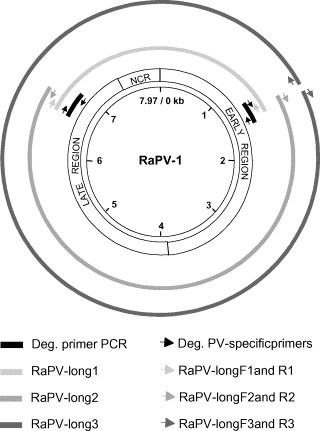
Schematic representation of the strategy for amplification of the complete genomic DNA of RaPV-1. NCR, non-coding region.
2.3. DNA and protein sequence analysis
Open reading frame analysis was performed with the ORF Finder tool on the NCBI server of the National Institutes of Health (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The molecular weight of the putative proteins was calculated using the ExPASy (Expert Protein Analysis System) Compute pI/Mw tool (http://www.expasy.org/tools/pi_tool.html). The sequence similarities between RaPV-1 and the porpoise PsPV-1 (GenBank accession number NC_003348), equine EcPV-1 (NC_003748), bovine BPV-1 (X02346), human HPV-1a (NC_001356), HPV-5 (NC_001531), HPV-16 (NC_001526) and parrot PePV (NC_003973) were investigated by pairwise sequence alignments of the different ORFs, performed at the amino acid level with the ClustalW program in DAMBE version 4.2.7 (Thompson et al., 1994, Xia and Xie, 2001), after which the nucleotide sequences were aligned accordingly. For multiple nucleotide sequence alignments, the sequence of RaPV-1 and 46 other PVs (type species of the different PV genera and species) were imported in DAMBE version 4.2.7, aligned at the amino acid level using ClustalW, after which the nucleotide sequences were aligned according to the aligned amino acid sequences. This was done separately for the different ORFs, and the unambiguously alignable parts of the E1, E2, L2 and L1 ORFs were pasted together in one compiled alignment of a total of 2700 nucleotides (nt). Nucleotide positions that were included in the alignment are nt 1691–1807, 1904–2119, 2156-2542 and 2573–2701 in E1, nt 2787–3143, nt 3165–3209 and 3231–3269 in E2, nt 3976–4011, 4018–4161, 4903–4920 and 5044–5142 in L2, and 5630–5746, 5783–5992, 6029–6112, 6164–6412, 6500–6652, 6695–6901 and 6932–7024 in L1, relative to the RaPV-1 sequence. Based on this alignment, a phylogenetic tree was constructed using the neighbor-joining method in MEGA version 3.1 (Kumar et al., 2004). Bootstrap support values were obtained for 1000 replicates.
3. Results
3.1. RaPV-1 complete genomic sequence
The complete genome of the RaPV-1 counts 7970 basepairs, with a GC content of 51.1%. As is the case for all PVs characterized to date, all ORFs of the RaPV-1 genome are located on the same strand of its circular double stranded DNA. The position of the first nucleotide of the RaPV-1 genome corresponds to the start of E6, the first ORF in the early protein region.
3.2. Open reading frame organization of early and late genes
The RaPV-1 genome contains the seven classical PV major ORFs, coding for five early (E) proteins (E1, E2, E4, E6 and E7) and two late (L) capsid proteins (L1 and L2). The exact location of the RaPV-1 ORFs, and the predicted molecular weights of the corresponding proteins are indicated in Fig. 2 .
Fig. 2.
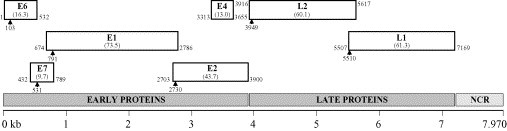
Linear representation of the open reading frames (ORFs) of the RaPV-1 genome (with the molecular mass of the predicted proteins in kilodaltons). Numbers show the nucleotide positions of the start of the ORF (first nucleotide after the last stop codon in the same reading frame) and of the stop codon. For each ORF, the position of the start codon is indicated (arrowhead), except for E4 which does not contain a start codon and therefore hypothetically begins at the first in-frame amino acid. NCR, non-coding region.
The E6 and E7 ORFs code for the major PV transforming proteins, together capable of immortalizing keratinocytes. A characteristic feature of E6 is the presence of two conserved zinc binding domains (CXXCX29CXXC), separated by 36 amino acids, whereas the E7 contains one such domain. In the putative RaPV-1 E6, the two zinc binding domains are separated by 37 amino acids instead of 36, and the second domain is slightly modified (CXXCX30CXXC). An identical modified motif is present in the RaPV-1 E7 protein. A putative retinoblastoma tumor suppressor binding domain, in which an LXCXE motif is considered critical for pRB binding (Moran, 1993, Chan et al., 2001), is present in the E7 protein sequence (DLHCDESLDQEAE). The E1 ORF codes for the largest RaPV-1 protein (665 amino acids), and contains the conserved ATP-binding site of the ATP-dependent helicase (consensus sequence GXXXXGK(T/S), GPPDTGKS in RaPV-1) (Titolo et al., 1999) in its carboxyterminal part. The PV E2 ORF codes for a DNA-binding protein that functions as a regulator of viral transcription and replication, and usually contains a leucine zipper domain, consisting of a periodic repetition of leucine residues at every seventh position over a distance covering eight helical turns (consensus sequence LX6LX6LX6L), which is involved in DNA-binding and dimerization. Although a canonical leucine zipper domain is not present in RaPV-1, we did locate a putative modified motif (IX6LX6LX6H) at the corresponding position in E2. The RaPV-1 E4 ORF is completely contained within the E2 gene, and as is the case in most other PVs it does not contain a start codon. E4 is usually expressed from two abundant mRNA species, an E1^E4 mRNA and an E1^E4-L1 mRNA. Although PV E4 proteins usually have a high proline content (15–20% on average), this is not the case for RaPV-1 (only 5% prolines). The late region of the genome contains the major (L1) and minor (L2) capsid protein genes, which both have a nuclear localization signal at their 3′-end.
In RaPV-1, the non-coding region (NCR) or upstream regulatory region (URR) between the stop codon of L1 and the first ATG of E6 is 901 bp long (nt 7172 to nt 102). To activate the origin of replication, an E1/E2 complex has to bind to the URR. PVs usually contain an E1-recognition site in the middle of two E2-binding sites (E2BS). Two typical palindromic E2BS with the consensus sequence ACCN6GGT are present in the RaPV-1 URR, at positions nt 7921 and nt 53, located equidistant to a putative E1 recognition site (TAATAGTTGCCAACAAC) at position nt 7965. The URR also contains at its 5′-end a polyadenylation site (AATAAA; nt 7614), located 31 nucleotides 5′ of a CA dinuleotide and G/T cluster, for processing of the L1 and L2 capsid mRNA transcripts, and the TATA box of the E6 promotor at its 3′-end (nt 72).
3.3. Phylogenetic analysis
To make an optimal sequence alignment of 47 PV types (all type species of the different PV genera and species), separate nucleotide sequence alignments were constructed for the different ORFs, based on the corresponding amino acid alignments. Only the PV core ORFs E1, E2, L1 and L2 were included, since only these ORFs are present in all characterized PVs. Unambiguously aligned regions were compiled in one concatenated alignment of 2700 nucleotides. The resulting neighbor-joining phylogenetic tree (Fig. 3 ) clusters the PVs in the 18 different PV genera, from Alpha- to Sigmapapillomavirus that have been defined to date (de Villiers et al., 2004, Rector et al., 2004a, Rector et al., 2005a). In this tree, RaPV-1 appears as a close-to-root PV, being the sole PV type on a branch that originates near the unresolved centre (as is indicated by the low bootstrap support values for nodes close to centre, all below 80%) of the PV evolutionary tree.
Fig. 3.
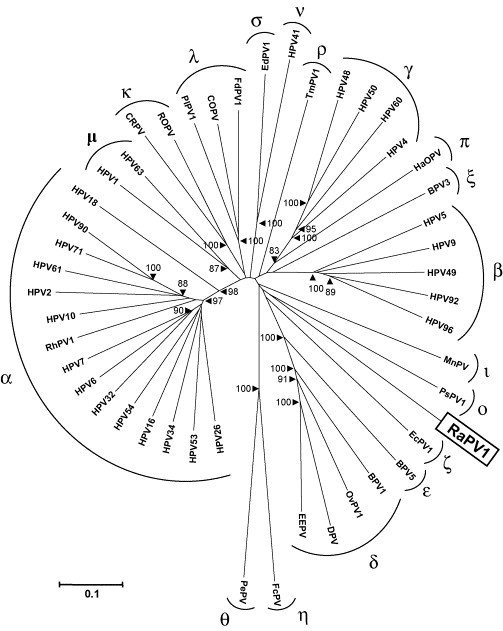
Neighbor-joining phylogenetic tree, based on a concatenated E1/E2/L2/L1 nucleotide sequence alignment of RaPV-1 and 46 other PV type species of the different PV genera and species. The PV-types included (with their GenBank accession numbers) were bovine BPV-1 (NC_001522), BPV-3 (NC_004197), BPV-5 (NC_004195), canine oral COPV (NC_001619), cottontail rabbit CRPV (NC_001541), deer DPV (NC_001523), Equus caballus EcPV-1 (NC_003748), Erethizon dorsatum EdPV-1 (NC_006951) European elk EEPV (NC_001524), Fringilla coelebs FcPV (NC_004068), Felis domesticus FdPV-1 (NC_004765), hamster oral HaOPV (E15111), HPV-1a (NC_001356), HPV-2a (NC_001352), HPV-4 (NC_001457), HPV-5 (NC_001531), HPV-6b (NC_001355), HPV-7 (NC_001595), HPV-9 (NC_001596), HPV-10 (NC_001576), HPV-16 (NC_001526), HPV-18 (NC_001357), HPV-26 (NC_001583), HPV-32 (NC_001586), HPV-34 (NC_001587), HPV-41 (NC_001354), HPV-48 (NC_001690), HPV-49 (NC_001591), HPV-50 (NC_001691), HPV-53 (NC_001593), HPV-54 (NC_001676), HPV-60 (NC_001693), HPV-61 (NC_001694), HPV-63 (NC_001458), HPV-71 (NC_002644), HPV-90 (NC_004104), HPV-92 (NC_004500), HPV-96 (NC_005134), Mastomys natalensis MnPV (NC_001605), ovine OvPV-1 (NC_001789), Psittacus erithacus PePV (NC_003973), Procyon lotor PlPV-1 (AY763115), Phocoena spinipinnis PsPV-1 (NC_003348), Rousettus aegyptiacus RaPV-1 (DQ366842), rhesus monkey RhPV-1 (NC_001678), rabbit oral ROPV (NC_002232), Trichechus manatus latirostris TmPV-1 (NC_006563). The PV genera are indicated with their Greek symbols. The numbers at the internal nodes represent the bootstrap probabilities (in percent), as determined for 1000 iterations by the neighbor-joining method. Only bootstrap values greater than 80% are shown. The scale bar indicates the genetic distance (nucleotide substitutions per site).
3.4. RaPV-1 sequence similarity to other papillomaviruses
We investigated the sequence similarity between RaPV-1 and PsPV-1 (which in our phylogenetic tree appeared as most closely related to RaPV-1), EcPV-1 and BPV-1 (representing the cluster that shares a common ancestral node with RaPV-1 and PsPV-1 in our tree), and the benign cutaneous HPV-1a, the epidermodysplasia verruciformis associated HPV-5, the prototype high-risk mucosal HPV-16 and the parrot PePV representing some of the major PV genera. The percentages similarity, calculated by pairwise alignments of the corresponding ORFs and their proteins, are provided in Table 1 . Although the RaPV-1 ORFs were found to be homologous to the corresponding ORFs of other PVs, only low percentages of similarity were found, and these values were comparable for all investigated PVs. The highest similarity scores (46–50%) were noted for the L1 ORF, which is characteristically the most conserved region of the PV genome. According to the recent classification criteria, PV types belong to different PV genera when they share less then 60% nucleotide sequence identity across the entire L1 ORF (de Villiers et al., 2004). Since the RaPV-1 L1 shares maximally 50% nucleotide identity with the L1 of other PVs, it cannot be placed in one of these existing genera. The RaPV-1 therefore represents the first member of a novel, as yet unnamed PV genus.
Table 1.
Percentage nucleotide (amino acid) similarity of the different RaPV-1 ORFs with the ORFs of PsPV-1, EcPV-1, BPV-1, HPV-1a, HPV-5, HPV-16 and PePV
| RaPV-1 | PsPV | EcPV-1 | BPV-1 | HPV-1a | HPV-5 | HPV16 | PePV |
|---|---|---|---|---|---|---|---|
| E6 | 24 (15) | 35 (24) | 32 (27) | 38 (28) | 33 (27) | 35 (27) | No E6 |
| E7 | No E7 | 37 (31) | 25 (19) | 42 (30) | 41 (30) | 31 (25) | No E7 |
| E1 | 48 (41) | 49 (41) | 45 (36) | 46 (40) | 48 (40) | 46 (40) | 39 (28) |
| E2 | 43 (35) | 42 (36) | 35 (31) | 40 (32) | 34 (29) | 41 (33) | 37 (26) |
| E4 | 27 (15) | 34 (21) | 30 (17) | 30 (16) | 17 (11) | 27 (13) | 20 (10) |
| L2 | 37 (29) | 35 (26) | 34 (26) | 38 (31) | 37 (29) | 37 (29) | 31 (20) |
| L1 | 48 (42) | 48 (44) | 46 (43) | 49 (45) | 48 (45) | 50 (43) | 48 (41) |
4. Discussion
Papillomaviruses are very stable and slow-evolving viruses, with a mutation rate of only 0.73–1.2 × 10−8 (Tachezy et al., 2002a, Van Ranst et al., 1995). Their evolution mainly occurs through slow accumulation of point mutations, and recombination between the genomes of different PV types has to date not been documented. The general consensus on PV evolution is that different ancient PV lineages have co-evolved and co-speciated with their vertebrate host species. This assumption is supported by the observation that PVs of closely related host species are generally closely related themselves and cluster together in the PV phylogenetic tree, with dating of PV divergence largely coinciding with the divergence of their host species (Van Ranst et al., 1992a, Tachezy et al., 2002b). The fact that the genetic diversity of PVs in relation to their host species is complicated by a superimposed diversity within single host species (as is the case for human, artiodactyl and rodent PV types, which are divided over different PV genera) probably originates from the presence of different ancestral PV lineages within the ancestors of these species. Our phylogenetic analysis shows that the novel RaPV-1, isolated from a carcinomatous wing lesion of an Egyptian fruit bat, is only distantly related to other known PV sequences. This close-to-root position of RaPV-1 was to be expected from the co-evolution viewpoint, since no PVs from species of the same order as the fruit bat (Chiroptera) have been characterized to date, and the divergence between the Chiroptera and the closest related mammalian orders within the Laurasiatheria (i.e. the Cetartiodactyla, Perissodactyla, Carnivora and Pholidota) is dated as far back as 83 million years ago (Mya) (Springer et al., 2003). This is not much later than the time at which the placental mammals first started diverging (107 Mya), which accords with RaPV-1 branching off near the root of the evolutionary tree of PVs (which were all isolated from placental mammals except for the two bird PVs PePV and FcPV).
Given the species-specific nature of PVs, with interspecies transmission being very rare and only occurring between closely related species, it is unlikely that PVs were transmitted to fruit bats in recent history. Bats are probably latently infected with PV on their healthy skin, as is the case for many mammal species (Antonsson and Hansson, 2002, Van Doorslaer et al., 2006). In humans, cutaneous HPVs are highly prevalent in the general population, where they usually are contained by the immune system and result in subclinical infections. These infections however tend to become clinically apparent upon genetic, acquired, or iatrogenic deficiencies in cell-mediated immunity, and cutaneous HPVs have been linked to non-melanoma skin cancer in these patients (Leigh et al., 1999). Whether the occurrence of PV-associated carcinoma in the Egyptian fruit bat is also related to immunosuppression is not clear.
Virological investigations with respect to chiropteran species have mostly focused on their role as natural reservoir hosts of many viruses, and their implication in the emergence of a number of disease pathogens in the human population, such as rabies virus, Nipah and Hendra viruses, Ebola and Marburg viruses, and very recently also the SARS-coronavirus (Dobson, 2005). The RaPV-1 described in this paper was isolated from a basosquamous carcinoma on the wing of an Egyptian fruit bat. To our knowledge, this is the first chiropteran PV, and the first report of PV-associated carcinomatosis in bats. The close-to-root position of RaPV-1 in the PV evolutionary tree, as the sole member of a novel PV genus, indicates that the knowledge on PV genetic diversity is still very incomplete, with many of their host species being under-sampled or not sampled at all. Sampling and study of many more animal PVs is therefore a prerequisite to obtain a more detailed picture of PV phylogeny and evolution.
Acknowledgements
We thank the colleagues of the laboratory of Clinical Virology, Rega Institute, University of Leuven, for helpful comments and critical reading of the manuscript. This work was supported by the Flemish Fund for Scientific Research (Fonds voor Wetenschappelijk Onderzoek, FWO) grant G.0513.06 and by a postdoctoral fellowship of the Research Fund K.U. Leuven to Annabel Rector.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman P.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Antonsson A., Hansson B.G. Healthy skin of many animal species harbors papillomaviruses which are closely related to their human counterparts. J. Virol. 2002;76:12537–12542. doi: 10.1128/JVI.76.24.12537-12542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.Y., Bernard H.U., Ratterree M., Birkebak T.A., Faras A.J., Ostrow R.S. Genomic diversity and evolution of papillomaviruses in rhesus monkeys. J. Virol. 1997;71:4938–4943. doi: 10.1128/jvi.71.7.4938-4943.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H.M., Smith L., La Thangue N.B. Role of LXCXE motif-dependent interactions in the activity of the retinoblastoma protein. Oncogene. 2001;20:6152–6163. doi: 10.1038/sj.onc.1204793. [DOI] [PubMed] [Google Scholar]
- de Villiers E.M., Fauquet C., Broker T.R., Bernard H.U., zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Dobson A.P. Virology. What links bats to emerging infectious diseases? Science. 2005;310:628–629. doi: 10.1126/science.1120872. [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Leigh I.M., Buchanan J.A., Harwood C.A., Cerio R., Storey A. Role of human papillomaviruses in cutaneous and oral manifestations of immunosuppression. J. Acquir. Immune. Defic. Syndr. 1999;21:S49–S57. [PubMed] [Google Scholar]
- McKnight C.A., Wise A.G., Maes R.K., Howe C., Rector A., Van Ranst M., Kiupel M. Papillomavirus-associated basosquamous carcinoma in an Egyptian fruit bat (Rousettus aegyptiacus) J. Zoo Wildl. Med. 2006;37:193–196. doi: 10.1638/05-101.1. [DOI] [PubMed] [Google Scholar]
- Moran E. DNA tumor virus transforming proteins and the cell cycle. Curr. Opin. Genet. Dev. 1993;3:63–70. doi: 10.1016/s0959-437x(05)80342-9. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Tomita Y., Okada M., Shinozaki K., Kubonoya H., Kaiho I., Shirasawa H. Broad-spectrum detection of papillomaviruses in bovine teat papillomas and healthy teat skin. J. Gen. Virol. 2004;85:2191–2197. doi: 10.1099/vir.0.80086-0. [DOI] [PubMed] [Google Scholar]
- Rector A., Bossart G.D., Ghim S.J., Sundberg J.P., Jenson A.B., Van Ranst M. Characterization of a novel close-to-root papillomavirus from a Florida manatee by using multiply primed rolling-circle amplification: Trichechus manatus latirostris papillomavirus type 1. J. Virol. 2004;78:12698–12702. doi: 10.1128/JVI.78.22.12698-12702.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rector A., Tachezy R., Van Ranst M. A sequence-independent strategy for detection and cloning of circular DNA virus genomes by using multiply primed rolling-circle amplification. J. Virol. 2004;78:4993–4998. doi: 10.1128/JVI.78.10.4993-4998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rector A., Tachezy R., Van Doorslaer K., MacNamara T., Burk R.D., Sundberg J.P., Van Ranst M. Isolation and cloning of a papillomavirus from a North American porcupine by using multiply primed rolling-circle amplification: the Erethizon dorsatum papillomavirus type 1. Virology. 2005;331:449–456. doi: 10.1016/j.virol.2004.10.033. [DOI] [PubMed] [Google Scholar]
- Rector A., Van Doorslaer K., Bertelsen M., Barker I.K., Olberg R.-A., Lemey P., Sundberg J.P., Van Ranst M. Isolation and cloning of the raccoon (Procyon lotor) papillomavirus type 1 by using degenerate papillomavirus-specific primers. J. Gen. Virol. 2005;86:2029–2033. doi: 10.1099/vir.0.80874-0. [DOI] [PubMed] [Google Scholar]
- Springer M.S., Murphy W.J., Eizirik E., O’Brien S.J. Placental mammal diversification and the Cretaceous-tertiary boundary. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1056–1061. doi: 10.1073/pnas.0334222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg J.P. Papillomavirus infections in animals. In: Syrjänen K., Gissmann L., Koss L.G., editors. Papillomaviruses and Human Disease. Springer-Verlag; Berlin: 1987. pp. 40–103. [Google Scholar]
- Sundberg J.P., Van Ranst M., Burk R.D., Jenson A.B. The nonhuman (animal) papillomaviruses: host range, epitope conservation, and molecular diversity. In: Gross G., von Krogh G., editors. Human Papillomavirus Infections in Dermatovenereology. CRC Press; Boca Raton: 1997. pp. 47–68. [Google Scholar]
- Syrjänen K.J., Syrjänen S.M. John Wiley and Sons Ltd.; Chichester, UK: 2000. Papillomavirus Infections in Human Pathology. [Google Scholar]
- Tachezy R., Duson G., Rector A., Jenson A.B., Sundberg J.P., Van Ranst M. Cloning and genomic characterization of Felis domesticus papillomavirus type 1. Virology. 2002;301:313–321. doi: 10.1006/viro.2002.1566. [DOI] [PubMed] [Google Scholar]
- Tachezy R., Rector A., Havelkova M., Wollants E., Fiten P., Opdenakker G., Jenson B., Sundberg J., Van Ranst M. Avian papillomaviruses: the parrot Psittacus erithacus papillomavirus (PePV) genome has a unique organization of the early protein region and is phylogenetically related to the chaffinch papillomavirus. BMC Microbiol. 2002;2:19–27. doi: 10.1186/1471-2180-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai M., Burk R.D. Identification and characterization of 3 novel genital human papillomaviruses by overlapping polymerase chain reaction: candHPV89, candHPV90, and candHPV91. J. Infect. Dis. 2002;185:1794–1797. doi: 10.1086/340824. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titolo S., Pelletier A., Sauve F., Brault K., Wardrop E., White P.W., Amin A., Cordingley M.G., Archambault J. Role of the ATP-binding domain of the human papillomavirus type 11 E1 helicase in E2-dependent binding to the origin. J. Virol. 1999;73:5282–5293. doi: 10.1128/jvi.73.7.5282-5293.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorslaer K., Rector A., Vos P., Van Ranst M. The genetic characterization of the Capra hircus papillomavirus; a novel close to root Artiodactyl papillomavirus. Virus Res. 2006;118:164–169. doi: 10.1016/j.virusres.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Van Ranst M., Fuse A., Fiten P., Beuken E., Pfister H., Burk R.D., Opdenakker G. Human papillomavirus type 13 and pygmy chimpanzee papillomavirus type 1: comparison of the genome organizations. Virology. 1992;190:587–596. doi: 10.1016/0042-6822(92)90896-w. [DOI] [PubMed] [Google Scholar]
- Van Ranst M., Kaplan J.B., Burk R.D. Phylogenetic classification of human papillomaviruses: correlation with clinical manifestations. J. Gen. Virol. 1992;73:2653–2660. doi: 10.1099/0022-1317-73-10-2653. [DOI] [PubMed] [Google Scholar]
- Van Ranst M., Kaplan J.B., Sundberg J.P., Burk R.D. Molecular evolution of papillomaviruses. In: Gibbs A., Calisher C.H., Garcia-Arenal F., editors. Molecular Basis of Virus Evolution. Cambridge University Press; Cambridge: 1995. pp. 455–476. [Google Scholar]
- Xia X., Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]


