Abstract
Dendritic cells loaded with antigenic peptides, because of their safety and robust immune stimulation, would be ideal for induction of immunity to protect against listeriosis. However, there is no currently accepted method to predict which peptides derived from the Listeria proteome might confer protection. While elution of peptides from MHC molecules after Listeria infection yields high-affinity immune-dominant epitopes, these individual epitopes did not reliably confer Listeria protection. Instead we applied bioinformatic predictions of MHC class I and II epitopes to generate antigenic peptides that were then formulated with Advax™, a novel polysaccharide particulate adjuvant able to enhance cross-presentation prior to being screened for their ability to induce protective T-cell responses. A combination of at least four intermediate strength MHC-I binding epitopes and one weak MHC-II binding epitope when expressed in a single peptide sequence and formulated with Advax adjuvant induced a potent T-cell response and high TNF-α and IL-12 production by dendritic cells resulting in robust listeriosis protection in susceptible mice. This T-cell vaccine approach might be useful for the design of vaccines to protect against listeriosis or other intracellular infections.
Abbreviations: DC, dendritic cell; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; i.p, intra-peritoneal; LLO, listeriolysin O; MHC, major histocompatibility complex
Keywords: Listeriosis, Dendritic cells, Vaccines, Listeriolysin O, Glyceraldehyde-3-phosfate-dehydrogenase
1. Introduction
Dendritic cells (DCs) present antigenic peptides on MHC I and class II molecules resulting in stimulation of CD8+ and CD4+ T cells, respectively. This ability to activate both CD4 and CD8 T cell responses is the basis for the effectiveness of DC-based vaccines in prophylaxis against infections including HIV, Streptococcus pneumonia, Chlamydia trachomatis or Mycobacterium (Vacas-Cordoba et al., 2013, Cohen et al., 2012, Rey-Ladino et al., 2014, Kawasaki et al., 2014, Fromen et al., 2015).
Groups at risk of listeriosis include pregnant women, neonates, the elderly and immune-compromised patients. DC vaccines loaded ex vivo with peptides and injected back into the host, are highly effective at inducing cellular immunity and thereby could be useful for Listeria immunization (Wood and Paterson, 2014).
The objective of this study was to find a simple method to predict the minimum epitope requirements for a DC vaccine against human listeriosis and involved testing DC vaccines in mice susceptible or resistant to Listeria. C57BL/6 mice have a Listeria resistant allele at the Hc locus on chromosome 2 and show reduced susceptibility to intravenous, i.v., intragastric, i.g. or intraperitoneal, i.p, challenge with Listeria when compared to susceptible Balb/c mice (Mainou-Fowler et al., 1988, Poulsen et al., 2011). Therefore, we used C57BL/6 mice to mimic the population at low risk of listeriosis, while Balb/c mice were used to mimic high-risk groups. Two Listeria antigens were selected as the focus of our study, listeriolysin O (LLO) and the glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) as they have previously been shown to elicit strong CD4+ and CD8+ T cell responses (Alvarez-Dominguez et al., 2008, Calderon-Gonzalez et al., 2014, Calderon-Gonzalez et al., 2015). This was important as human listeriosis is not well characterised and antigens other than LLO or GAPDH that induce cellular immunity have yet to be found (Angelakopoulos et al., 2002; CAD and RCG unpublished results).
Bioinformatic methods that predict T cell epitopes via their ability to bind MHC molecules provide a faster and less costly alternative to strategies for selection of T-cell vaccine candidates such as MHC peptide elution or assays to map T-cell epitopes using complete sets of overlapping peptides (Geginat et al., 2001, Skoberne and Geginat, 2002). This is especially relevant given the limited current data on Listeria epitopes (Yu et al., 2002).
Another challenge when developing T-cell vaccines is the restricted ability of soluble antigens to stimulate strong T-cell responses, which at least in part reflects the lack of MHC class I cross-presentation of such antigens. Hence to maximise T-cell responses against Listeria an adjuvant is needed to up-regulate presentation of antigen on MHC class I and class II molecules on the surface of antigen presenting cells. Advax™ is a novel polysaccharide adjuvant derived from microparticles of delta β-d-[2-1]poly(fructo-furanosyl)α-d-glucose (delta inulin) (Cooper and Petrovsky, 2011, Cooper et al., 2013, Cooper et al., 2014, Cooper et al., 2015). Advantages of Advax include the fact that it has already been shown to be well tolerated, effective and safe for human use (Gordon et al., 2012, Gordon et al., 2014), but most importantly for the current study, its demonstrated ability to induce potent CD4+ and CD8+ T cell responses against a wide variety of exogenous antigens including proteins from Japanese encephalitis, influenza, hepatitis B, HIV, and SARS coronavirus (Lobgis et al., 2010, Cristillo et al., 2011, Saade et al., 2013, Honda-Okubo et al., 2015). Hence, Advax adjuvant had ideal properties for use in our Listeria T-cell epitope screening programme.
We present a method that combines bioinformatics and structural predictions of peptide binding to MHC class I and class II molecules with convenient T-cell readouts. Vaccine predictions were validated in vivo and immunological parameters checked to confirm vaccine efficiency.
2. Methods
2.1. Cells
Mice were 8–12 week-old female Balb/c from Charles River (France) or C57BL/6 from our animal facility. DCs were obtained from femur bone-marrow, differentiated with GM-CSF, and CD11c+ cells isolated with anti-mouse CD11c-coated magnetic beads and MACS™ separation columns (Miltenyi Biotech Inc., Auburn, CA) (Kono et al., 2012, Calderon-Gonzalez et al., 2014). DCs used were MHC-II+CD11c+CD40+CD11b+/−CD86+/−F4/80−Gr-1−.
2.2. Peptides and adjuvants
We used GAPDH1–15, GAPDH1–22, LLO91–99, LLO296–304, LLO189–201, LLO189–200 and LLO190–201 peptides. Peptides were synthesized at CNB (CSIC, Madrid) followed by HPLC and mass spectrometry using a MALDI-TOF Reflex™ IV mass spectrometer (Bruker Daltonics, Bremen, Germany). Peptide purity was > 95% after HPLC. Advax™ adjuvant (Adv1) was supplied by Vaxine (Adelaide, Australia) and was mixed with antigen by simple admixture immediately before immunization of mice or in vitro analysis.
2.3. Bacteria
Listeria monocytogenes 10403S strain (LMWT) was obtained from D.A. Portnoy (University of California, Berkeley, CA, USA).
2.4. Preparation of peptide pulsed DC vaccines
Cultured DCs were ex vivo loaded with 50 μg/ml of GAPDH1–15, GAPDH1–22, LLO91–99, LLO296–304, LLO189–200, LLO190–201 or LLO189–201 peptides for 24 h in the presence of 50 μg/ml of Advax. Supernatants were collected and stored at − 80 °C for cytokine measurements. Cells were analysed for cell surface markers by FACS.
2.5. Measurement of viability and apoptosis
Cultured DCs were also analysed for viability incubating cells with Trypan-blue. Cells stained with Trypan-blue were considered dead cells, while cells non-stained with the dye were viable. Cells were counted in a Neubauer haemocytometer and results were expressed as the mean of the percentages of viable cells ± SD. DCs were also examined for apoptosis by FACS analysis after double staining with 7-AAD and annexin-V (BD-Biosciences). In brief, DCs were incubated with 7-AAD and annexin-V-APC for 60 min at 4 °C, washed and analysed by flow cytometry. Double negative cells correspond to cells non-permeable to any reagent and therefore alive (Q3 region). Cells positive for annexin-V but negative for 7-AAD corresponded to early apoptotic cells (Q4 region). Double positive cells for annexin-V and 7-AAD corresponded to late apoptotic cells (Q2 region) and cells negative for annexin-V but positive for 7-AAD correspond to necrotic cells (Q1 region). Results are expressed as the mean of the percentages of late apoptotic cells (l-apoptosis) or early apoptotic cells (E-apoptosis) ± SD.
2.6. T-cell responses elicited by DC-LLO or DC-GAPDH vaccines
For delayed type hypersensitivity (DTH) analysis, Balb/c and C57BL/6 mice immunised i.p with LMWT (5 × 103 CFU) were inoculated 7 days later in left hind footpads with DC vaccines (106 cells/mice) in the presence or absence of Advax (50 μg/ml). Non-inoculated right hind footpads served as negative controls. Positive controls of this assay included DC pre-infected in vitro with wild type LM (DC-LMWT) as previously reported (Calderon-Gonzalez et al., 2015). After 24 h, we measured the footpad thickness with a calliper. T cell results in the footpads are expressed in millimetres as the mean of three different experiments ± SD.
2.7. Vaccination protocols
Vaccines used were DC-LLO91–99, DC-LLO296–304, DC-LLO189–201, DC-LLO189–200, DC-LLO190–201, DC-GAPDH1–15 and DC-GAPDH1–22 (1 × 106 cells/mice). Mice were immunised into the peritoneal cavity (i.p) (n = 5) in the presence or absence of Advax (50 μg/ml) or left non-vaccinated (NV). Seven days after immunization, Balb/c and C57BL/6 mice were challenged i.p with 103 and 104 CFU of LMWT/mice, respectively, and then bled before termination (Lauer et al., 2008). Sera were stored at − 80 °C to measure cytokines by FACS analysis using the CBA kit that measures all Th1 cytokines of a sample at the same time (Becton Dickinson, Palo Alto, CA, USA). Data were analysed using the FlowJo software (Treestar, Ashland, OR). Samples were performed in triplicate and the results are the mean ± SD of two separate experiments. ANNOVA was used for statistics with the cytokine measurements. At termination spleens and livers were homogenized and CFUs counted in blood agar plates. To measure CD4+ frequencies producing intracellular IFN-γ, spleen cells were cultured in 96-well plates (5 × 106 cells/ml) and stimulated with GAPDH1–15 or GAPDH1–22 peptides (5 μg/ml each peptide) for 5 h in the presence of brefeldin A as previously reported (Calderon-Gonzalez et al., 2014). Next, cells were surface labelled for CD4, fixed and permeabilized with cytofix/cytoperm kit before staining for intracellular IFN-γ (BD Biosciences). FACS data were analysed using FlowJo software (Treestar, Ashland, OR). After sample acquisition, data were gated for CD4+ events, and the percentages of cells expressing IFN-γ determined according to the manufacturer's recommendations. Results were corrected according to the percentages of total CD4+ cells. Samples were tested in triplicate and the results are the mean ± SD of two separate experiments.
2.8. Frequencies of CD8+-LLO or GAPDH specific T cells
To confirm the frequency of LLO91–99, LLO296–304, GAPDH1–15 or GAPDH1–22 specific CD8 T cells producing IFN-γ, we used recombinant soluble dimeric mouse H-2Kb:Ig (for C57BL/6 mice) or H-2Ld:Ig (for Balb/c mice) fusion proteins following the manufacturer's instructions (DimerX I; BD Bioscience) and as previously described (Lauer et al., 2008, Calderon-Gonzalez et al., 2014).
2.9. ELISA for Listeria peptides
LLO91–99 or GAPDH1–22 peptides were prepared at 50 μg/ml in coating buffer (50 nM of Na2CO3 at pH 9.7) and used to coat Nunc MaxiSorp plates overnight at 4 °C. Next, plates were blocked with 1% BSA in PBS at RT for 30 min and mice sera diluted 1/200 in PBS-0.5% BSA and incubated for 1 h at RT. Mice IgM was detected with a goat anti-mouse IgM conjugated with horseradish peroxidase (0.8 μg/ml) and enzymatic reactions developed with tetramethylbenzidine in citric/acetate buffer and H2O2. Reactions were stopped with 50 μl of H2SO4 (0.8 M) and absorbances measured at 450 nm in the ELISA reader. Results were expressed as absorbance units of triplicates ± SD.
2.10. Statistical analysis
For statistical analysis, Student's t test was applied. ANOVA analysis was applied to cytokine measurements. P ≤ 0.05 was considered significant. GraphPad software was used for generation of graphs.
2.11. Ethics statement
This study was carried out in accordance with the Guide for the Care and Use of Laboratory Animals of the Spanish Ministry of Science, Research and Innovation. The Committee on the Ethics of Animal Experiments of the University of Cantabria approved the protocol (Permit Number: 2012/06) that follows the Spanish legislation (RD 1201/2005). All surgeries were performed under sodium pentobarbital anaesthesia, and all efforts were made to minimize suffering.
3. Results and discussion
3.1. Bioinformatics approach to classify Listeria epitopes
The objective of this study was to design a simple experimental approach that predicted the minimal epitope requirements to load and prepare DC vaccine formulations able to protect against listeriosis. We focussed on the Listeria antigens LLO and GAPDH as vaccine candidates since sera from patients recovering from listeriosis recognized various peptides from these antigens (Angelakopoulos et al., 2002; CAD and RCG unpublished results). Listeriosis is characterised by low humoral but robust T cell responses (Pamer, 2004). Previously T-cell epitopes for Listeria vaccine design have been identified by peptide elution from MHC molecules of Listeria-loaded macrophages (Geginat et al., 2001, Skoberne and Geginat, 2002). Since this approach is technically challenging, predominantly only identifies high affinity epitopes and is performed using macrophages which are less efficient at antigen presentation cells than DC, it may fail to identify many other relevant and protective Listeria T-cell epitopes. There is also no consensus of whether high, medium or low affinity epitopes may represent the best vaccine candidates. Bioinformatics approaches have been used in other infectious diseases to identify T-cell epitopes to include in vaccines with great success (Yu et al., 2002, Stephenson et al., 2015). Hence we used a bioinformatics approach to screen for potential T cell epitopes in LLO and GAPDH and followed this with a modified delayed-type-hypersensitivity (DTH) test to identify T-cell epitopes with potential to provide protection against human and veterinary listeriosis. As positive controls we used five known LLO epitopes previously identified by the elution method (Geginat et al., 2001). For T-cell epitope prediction we used IEDB, a bioinformatic prediction resource Consensus tool that combines predictions from artificial neural network (ANN), SMM and Comblib for GAPDH binding predictions (Nielsen et al., 2003, Peters and Sette, 2005, Sidney et al., 2008, Kim et al., 2012). IEDB analysis predicted that good MHC class I binders showed percentile ranks < 10 and weak binders percentile ranks < 100. Similarly, good MHC class II binders showed percentile thresholds < 50, intermediate binders < 500 and low binders < 5000.
GAPDH1–15 contained four epitopes predicted to be good binders for H-2Kb, H-2Kd and H-2Ld MHC-I molecules, and one predicted intermediate binding epitope for IAb and low binder to IAd (lower right image in Fig. 1A). GAPDH1–22 contained six epitopes predicted to be good binders to H-2Kd and H-2Ld, six epitopes predicted to be intermediate binders to IAb and one epitope predicted to be a low binder to IAd (GAPDH binding values in Fig. 1A). These data indicated that GAPDH1–15 and GAPDH1–22 epitopes were able to bind to different MHC-I and MHC-II molecules of Balb/c and C57BL/6 mice strains.
Fig. 1.
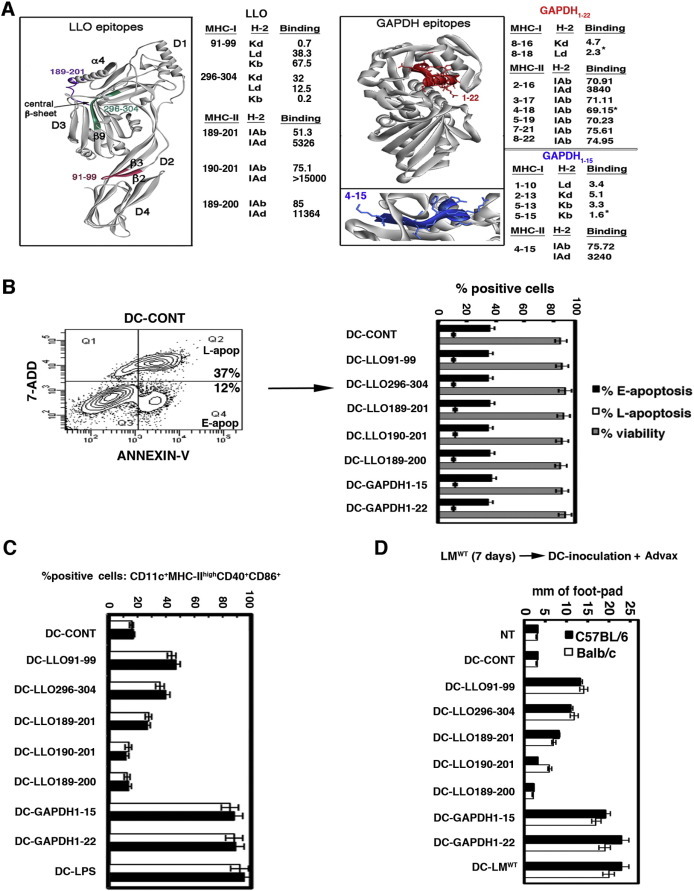
Methodology to predict efficient vaccine epitopes for DC vaccine formulations. (A) diagram showing the bioinformatic strategy for epitope prediction including the 3D-structure of LLO based on the recently reported crystal structure (Köster et al., 2014) and the relevant MHC-I and MHC-II epitopes in Balb/c and C57BL/6 mice. The purple sequence corresponds to LLO189–201 peptide that contains LLO189–200 and LLO190–201 MHC-II epitopes and the pink sequence corresponds to LLO91–99 MHC-I epitope. The green sequence corresponds to the LLO296–394 MHC-I epitope. On the right, the predicted 3D structure of GAPDH is shown with the GAPDH1–22 peptide highlighted in red. The lower image shows an enlargement of amino acids 3–15 of GAPDH1–15 epitope. The sequences included on the right correspond to MHC binding predictions performed with IEDB Consensus tool. (B) DCs were incubated with 50 μg/ml of peptides for 24 h and then stained with Trypan blue to assay cytotoxicity (grey bars) or incubated with 7-ADD and annexin-V to analyse apoptosis by FACS (black and white bars). An example of apoptosis read-out is shown in left panel. Cytotoxicity results are expressed as the number of cells ± SD and experiments were performed in triplicate. Apoptosis results are expressed as the mean of the percentages of early apoptotic (Q4 region), late apoptotic (Q2 region) or necrotic cells (Q1 region) ± SD (P < 0.05). (C) DC from Balb/c or C57BL/6 mice were differentiated in vitro with 30 ng/ml of GM-CSF for 5 days, detached and positively selected using anti-mouse CD11c-coated magnetic beads and MACS separation columns. CD11c+ DC were loaded ex vivo with different peptides in the presence of 50 μg/ml of Advax: LLO91–99, LLO189–200, LLO190–201, LLO189–201, LLO296–304, GAPDH1–15 or GAPDH1–22 peptides (50 μg/ml) for 24 h or left unloaded (DC-CONT). Cells were washed and analysed using a FACSCanto flow cytometer. The percentages of CD11c+MHC-IIhighCD40+CD86+ positive cells are shown. Results are expressed as the mean ± SD of triplicate samples (P < 0.05). (D) C57BL/6 (black bars) or Balb/c mice (white bars) were immunised i.p with 5 × 103 CFU of Listeria/mouse for 7 days and then left hind footpads were inoculated with 1 × 106 cells of different DC-LLO and DC-GAPDH vaccines or saline (NT) for 24 h in the presence of Advax (250 μg), while right hind footpads were not inoculated and served as controls. Footpad swelling was measured with a calliper and expressed as the differences in mm between left and right hind footpads. Results are expressed as the mean ± SD of three different experiments (P < 0.05).
Analysis of LLO for epitopes indicated that LLO91–99 was predicted to be a strong binder to H-2Kd (0.7 values) and also a weak binder to H-2Kb MHC-I (67 values); LLO296–304 peptide was predicted to be a strong binder to H-2Kb (score 0.2), a weak binder to H-2Kd (score 32) and an intermediate binder to H-2Ld (score 12.5) and LLO189–201 was predicted to be a strong binder to IAb (score 51.3) and low binder to IAd (score 5326). LLO189–200 and LLO190–201 peptides were not predicted to bind to IAd (score > 10000) and to be weak binders to IAb (scores 85 and 75, respectively) (Calderon-Gonzalez et al., 2015). All LLO epitope predictions were in agreement with the previously MHC-I and MHC-II eluted LLO epitopes (Geginat et al., 2001, Skoberne and Geginat, 2002). However, the bioinformatic analysis also predicted new MHC binding abilities for these LLO epitopes. Hence, the LLO91–99 epitope previously eluted from H-2Kd was predicted to be a weak binder to H-2Kb, LLO296–304 epitope eluted from H-2Kb was predicted to be a weak binder to H-2Kd, LLO189–201 and LLO190–201 epitopes eluted from IAb were predicted to be weak binders to IAd, and LLO189–200 epitope eluted from IAd was predicted to be a weak binder to IAb. Therefore, the bioinformatics analysis identified a higher number of potential binding epitopes than the elution methods. We next studied where the predicted LLO epitopes were situated within the recently reported LLO crystal structure (Köster et al., 2014) PBDI: 4CDB (left image, Fig. 1A). LLO showed 4 domains (D1, D2, D3 and D4) that contained several β-sheets and α-helices with D4 being the binding domain to the phagosomal membranes. Since no crystal structure from Listeria GAPDH was available, we used the Automated Comparative Protein Modelling Server SWISS-MODEL available at: (http://www.expasy-ch/swissmod/SWISS-MODEL.html) to produce a theoretical 3D model (right image, Fig. 1A). LLO epitopes that bind to MHC-I were tight or wide loops (pink and green LLO sequences, respectively, Fig. 1A), while epitopes that bind to MHC-II contained α-helices (purple LLO sequence, Fig. 1A). A similar pattern was observed for GAPDH, with GAPDH1–22 and GAPDH1–15 epitopes that bind to MHC-I containing tight loops and epitopes that bind to MHC-II molecules containing two α-helices. According to this rule, we predicted that GAPDH1–22 and GAPDH1–15 epitopes containing tight loops and two α-helices that are able to simultaneous bind to H-2Kb, H-2Kd, H-2Ld, IAb and IAd might be the best candidates for a T-cell based listeriosis vaccine. Next, the LLO91–99 epitope containing tight loops and LLO296–304 epitope with wide loops that both bind to MHC-I molecules and the LLO189–201 epitope containing two complete α-helices that bind to MHC-II molecules, were identified as good vaccine candidates as they were able to bind to MHC from both mice strains. LLO189–200 and LLO190–201 epitopes that contained one α-helix and bind to MHC-II molecules of one mice strain exclusively were classified using this algorithm as poor vaccine candidates.
3.2. Evaluation of the quality of DC vaccine formulations
Having identified candidate vaccine epitopes we next wished to exclude any negative effects of the relevant peptides on the viability of the proposed DC vaccine vectors. DC vectors need to remain viable to fully activate T cells in the draining lymph nodes, a process that takes at least 24 h. Therefore, we examined the effect of the relevant LLO and GAPDH peptides on DC cytotoxicity and activation after 24 h of culture, the time required for DC loading with peptides (Pion et al., 2010, Kono et al., 2012, Vacas-Cordoba et al., 2013, Calderon-Gonzalez et al., 2014). Cytotoxicity was examined with Trypan-blue staining and apoptosis with 7-ADD and annexin-V fluorescence (Fig. 1B and Table 1 ) using different concentrations of LLO91–99, LLO189–201, LLO189–200, LLO190–201, LLO296–304, GAPDH1–15 or GAPDH1–22 peptides (0–500 μg/ml). None of the LLO or GAPDH peptides at doses of 50 μg/ml and below caused DC cytotoxicity, early (E-apo of Q4 region or E-apoptosis in black bars of Fig. 1B) or late apoptosis (l-apop of Q2 region or l-apoptosis in white bars of Fig. 1B). As the highest concentrations of peptides (500 μg/ml) were associated with slightly lower DC viability (Table 1), we selected 50 μg/ml as the on-going loading concentration for our DC vaccines.
Table 1.
Lack of cytoxicity induction of LLO and GAPDH peptides.
| Reagent | DC viabilitya |
|---|---|
| LLO91–99 (μg/ml) | |
| 500 | 1.69 ± 0.132 × 106 |
| 50 | 1.79 ± 0.120 × 106 |
| 5 | 1.78 ± 0.113 × 106 |
| 0.5 | 1.79 ± 0.128 × 106 |
| LLO189–201 (μg/ml) | |
| 500 | 1.69 ± 0.132 × 106 |
| 50 | 1.77 ± 0.120 × 106 |
| 5 | 1.78 ± 0.113 × 106 |
| 0.5 | 1.79 ± 0.128 × 106 |
| LLO190–201 (μg/ml) | |
| 500 | 1.69 ± 0.132 × 106 |
| 50 | 1.76 ± 0.120 × 106 |
| 5 | 1.78 ± 0.113 × 106 |
| 0.5 | 1.79 ± 0.128 × 106 |
| LLO190–201 (μg/ml) | |
| 500 | 1.67 ± 0.132 × 106 |
| 50 | 1.78 ± 0.120 × 106 |
| 5 | 1.78 ± 0.113 × 106 |
| 0.5 | 1.79 ± 0.128 × 106 |
| LLO296–304 (μg/ml) | |
| 500 | 1.61 ± 0.132 × 106 |
| 50 | 1.77 ± 0.120 × 106 |
| 5 | 1.78 ± 0.113 × 106 |
| 0.5 | 1.79 ± 0.128 × 106 |
| GAPDH1–15 (μg/ml) | |
| 500 | 1.73 ± 0.132 × 106 |
| 50 | 1.78 ± 0.120 × 106 |
| 5 | 1.78 ± 0.113 × 106 |
| 0.5 | 1.79 ± 0.128 × 106 |
| GAPDH1–22 (μg/ml) | |
| 500 | 1.69 ± 0.132 × 106 |
| 50 | 1.78 ± 0.120 × 106 |
| 5 | 1.78 ± 0.113 × 106 |
| 0.5 | 1.79 ± 0.128 × 106 |
In vitro cytoxicity assayed using DC incubated with different doses of peptides (0–500 μg/ml) for 24 h and then stained with Trypan blue. Results are expressed as the number of viable cells ± SD and experiments were performed in triplicates (P < 0.05).
An important issue for DC vaccines is their activation state, as this improves the quality of the resulting T-cell responses. Activated DCs show a characteristic CD11c+MHC-II+CD40+CD86+ phenotype that corresponds with higher vaccine efficiency (Pion et al., 2010, Kono et al., 2012, Vacas-Cordoba et al., 2013). Peptides by themselves generally do not activate DC, although longer peptides may sometimes do so (Calderon-Gonzalez et al., 2014). Since LLO and GAPDH peptides differ significantly in their length, we standardised the experiment by mixing the peptides with 50 μg/ml of Advax adjuvant prior to DC loading for 24 h. Advax was included, as it has been previously shown to modulate DC activity and potently enhance CD4+ and CD8+ T cell responses to Listeria antigens (Rodriguez-Del Rio et al., 2015). We also included LPS (10 ng/ml) as a classical DC activator (DC-LPS) (Kono et al., 2012, Calderon-Gonzalez et al., 2014) as well as untreated control DC (DC-CONT). LPS induced the highest frequency of CD11c+MHC-IIhighCD40+CD86+ DC (CD11c+ DC) followed by GAPDH1–22 and GAPDH1–15 peptides. LLO91–99 induced intermediate numbers of CD11c+ DC. LLO296–304 and LLO189–201 peptides also induced significant numbers of CD11c+ DC while LLO189–200 and LLO190–201 induced no CD11c+ DC at all (Fig. 1C).
We applied these results to the bioinformatics predictions to identify whether there were any epitope characteristics, which predicted the highest DC activation in both mice strains. Notably, the GAPDH1–15 epitope that induced a high DC activation (Fig. 1C) had four MHC-I binding sequences in tight loops and one MHC-II binding sequence containing two α-helices (Fig. 1A). Intermediate DC activation occurred with the LLO91–99 epitope (DC-LLO91–99 bars in Fig. 1C) that corresponded with a tight loop (pink LLO loop in Fig. 1A) that binds to MHC-I molecules in both mice strains. Lower DC activation was observed with LLO296–304 epitopes (DC-LLO296–304 bars in Fig. 1C), which corresponded with a wide loop that binds to MHC-I molecules in both mice strains (green LLO epitopes in Fig. 1A). Very low DC activation was observed only with LLO189–201 epitope (DC-LLO189–201 bars in Fig. 1C) that corresponded with two complete α-helices that bind to MHC-II molecules in both mice strains (purple LLO epitopes in Fig. 1A). No DC activation was observed with LLO189–200 or LLO190–201 epitopes (Fig. 1C) that corresponded with one α-helix that binds exclusively to MHC-II of only one mice strain. This analysis suggests that epitopes that bind MHC-I are more likely to induce DC activation than MHC-II binders.
3.3. Quantification of T-cell responses elicited by DC-vaccine formulations
Next we sought to develop an assay to predict the DC epitopes that elicited the highest T cell responses, in vivo. For this purpose, we adapted a classical delayed-type-hypersensitivity (DTH) test (Mohamed et al., 2012), where DC vaccines loaded with LLO or GAPDH peptides plus Advax adjuvant (50 μg/ml) were inoculated into the foot pads of previously Listeria-challenged mice and the DTH response measured as the degree of footpad swelling 24 h post-inoculation (Fig. 1D). We included a positive control of DTH response, DC infected in vitro with wild type LM (DC-LMWT) that induced a high degree of footpad swelling as previously reported (Calderon-Gonzalez et al., 2015). In both mice strains, DC-GAPDH1–15 and DC-GAPDH1–22 vaccines generated the highest DTH responses, similar to the positive control DC-LMWT, followed by DC-LLO91–99, then DC-LLO296–304 and lastly DC-LLO189–201. DC-LLO190–201 induced a low DTH response only in susceptible Balb/c mice and DC-LLO189–200 induced no response in either strain (Fig. 1D). These results indicated that the size of the DTH responses elicited by the DC epitope vaccines formulated with Advax adjuvant, correlated well with the ability of the peptides to induce DC activation and with the initial bioinformatic predictions (Fig. 1B–C). The removal of Advax adjuvant from the DC vaccine formulations reduced all DTH results ~ 5-fold but the Advax adjuvant did not modify the controls (data not shown), confirming that its action was via enhancement of antigen-specific T-cell responses. This is consistent with enhancement of antigen-specific T-cell responses observed with Advax adjuvant in other contexts such as when combined with influenza, hepatitis B or SARS coronavirus vaccines (Honda-Okubo et al., 2012, Honda-Okubo et al., 2015, Saade et al., 2013).
Next, we tested whether the strong DTH results for specific DC vaccines correlated with a Th1 cytokine response, as reflected in the supernatants of DC loaded with the different LLO and GAPDH peptides in the presence of Advax adjuvant as in Fig. 1A. We observed that for both mouse strains, DC-GAPDH1–22, DC-GAPDH1–15 and DC-LLO91–99 vaccines induced the highest TNF-α levels followed by DC-LLO296–304. DC-LLO189–201, DC-LLO190–201 and DC-LLO189–200 vaccines induced no TNF-α (Table 2 ). Only DC-GAPDH1–22 and DC-GAPDH1–15 induced significant levels of IL-12 in both mice strains, with IL-12 having been shown to correlate with protective vaccine responses against listeriosis (Kono et al., 2012 and Calderon-Gonzalez et al., 2014). DC-LLO91–99 vaccines produced low but significant levels of IL-12 in both mice strains whereas DC-LLO296–304, DC-LLO189–201, DC-LLO190–201 and DC-LLO189–200 induced no detectable IL-12 levels (Table 2). Thus, only the DC epitopes originally predicted by bioinformatic analysis to be the best CD8 T-cell epitopes and which produced the highest DTH responses in vivo were associated with significant IL-12 production by DC.
Table 2.
Th1 cytokines production by DC vaccines loaded with peptides in the presence of Advax.
| Epitopea | Th1-cytokines (pg/ml)b |
||
|---|---|---|---|
| Cytokines | Balb/c | C57BL/6 | |
| LLO91–99 | TNF-α | 1495 ± 102 | 1475 ± 118 |
| IL-12 | 4.5 ± 0.1* | 0.45 ± 0.05 | |
| LLO296–304 | TNF-α | 315 ± 0.9 | 310 ± 0.9 |
| IL-12 | 0 ± 0 | 0 ± 0 | |
| LLO190–201 | TNF-α | 15 ± 0.7 | 10 ± 0.8 |
| IL-12 | 0.11 ± 0 | 0.12 ± 0.04 | |
| LLO189–200 | TNF-α | 10 ± 0.8 | 11 ± 0.7 |
| IL-12 | 0 ± 0 | 0 ± 0 | |
| LLO189–201 | TNF-α | 25 ± 0.9 | 20 ± 0.9 |
| IL-12 | 0 ± 0 | 0 ± 0 | |
| GAPDH1–22 | TNF-α | 1635 ± 100 | 1721 ± 113 |
| IL-12 | 25 ± 0.8* | 24 ± 0.8* | |
| GAPDH1–15 | TNF-α | 1515 ± 120 | 1510 ± 138 |
| IL-12 | 24 ± 0.8* | 22 ± 0.9* | |
Asterisks show the IL-12 significant levels.
Levels of pro-inflammatory TNF-α and IL-12 cytokines were analysed in sera of different DC vaccinated Balb/c or C57BL/6 mice in the presence of Advax adjuvant (50 μg/ml) by CBA. Results are expressed as cytokine concentration (pg/ml of mean ± SD, P < 0.05).
3.4. Validation of proposed methods of epitope prediction for listeriosis vaccines
Next, to verify the above predictions, susceptible Balb/c and resistant C57BL/6 mice were immunized with DC-LLO91–99, DC-LLO296–304, DC-LLO189–200, DC-LLO190–201, DC-LLO189–201, DC-GAPDH1–15, or DC-GAPDH1–22 together with Advax (50 μg/ml) or were left non-vaccinated (NV). Mice were immunized i.p with a single dose of vaccine and 7 days later were administered a Listeria challenge (see Methods section for procedures). Five days post-challenge, all mice were bled, sacrificed and spleens recovered to count Listeria CFU and analyse immune cell populations (Fig. 2A and Table 3 ). DC-GAPDH1–15 and DC-GAPDH1–22 provided the greatest protection in both Balb/c and C57BL/6 mice (~ 97 and 99%, respectively) (Fig. 2A). DC-LLO91–99 also conferred significant protection in both Balb/c and C57BL/6 mice (90 and 93%, respectively). DC-LLO296–304 vaccine conferred strong protection in C57BL/6 mice (~ 90%) (Fig. 2A) but only intermediate levels of protection in Balb/c mice (~ 60%) (white bars in Fig. 2A). DC-LLO189–200, DC-LLO190–201, and DC-LLO189–201 vaccines conferred no significant protection in either strain (2 and 5%, respectively). Vaccination efficiency was associated with normal spleen size (images of DC-GAPDH1–15 vaccine recipients shown in Fig. 2A) compared with enlarged spleens (images of NV mice in Fig. 2A) and granulomatous livers of non-vaccinated mice (data not shown). We also evaluated several immunological parameters induced by the most protective DC vaccines. DC-GAPDH1–15 and DC-GAPDH1–22 vaccines were found to induce the highest frequencies of IFN-γ-producing CD8+ T cells in both C57BL/6 and Balb/c mice (4.0–4.2%, respectively) followed by DC-LLO91–99 vaccine with 3.0% frequency as assessed by FACS using H2-Kb:Ig-peptide dimers or 2.16% frequency using H2-Ld:Ig-peptide dimers (right plot in Fig. 2B). DC-LLO296–304 vaccine induced very low 0.87% CD8+ T cell frequencies in C57BL/6 mice and undetectable frequencies in Balb/c mice (DC-LLO296–304 bars in Fig. 2B). Only DC-GAPDH1–15 and DC-GAPDH1–22 vaccines induced detectable numbers of IFN-γ producing peptide-specific CD4+ T cells (1.05 and 1.76%, respectively) (Fig. 2B show C57BL/6 data, values in Balb/c were 1.05 and 1.76%). In summary, our results with DC-GAPDH vaccines confirmed that CD8+ T cell responses were more relevant than CD4+ T cell responses for Listeria protection. GAPDH1–15 and GAPDH1–22 peptides presented high numbers of MHC-I binding epitopes in listeriosis resistant and sensible mice, explaining DC-GAPDH vaccines induced similar CD8+ T cell responses and Th1-cytokines in both mice strains.
Fig. 2.
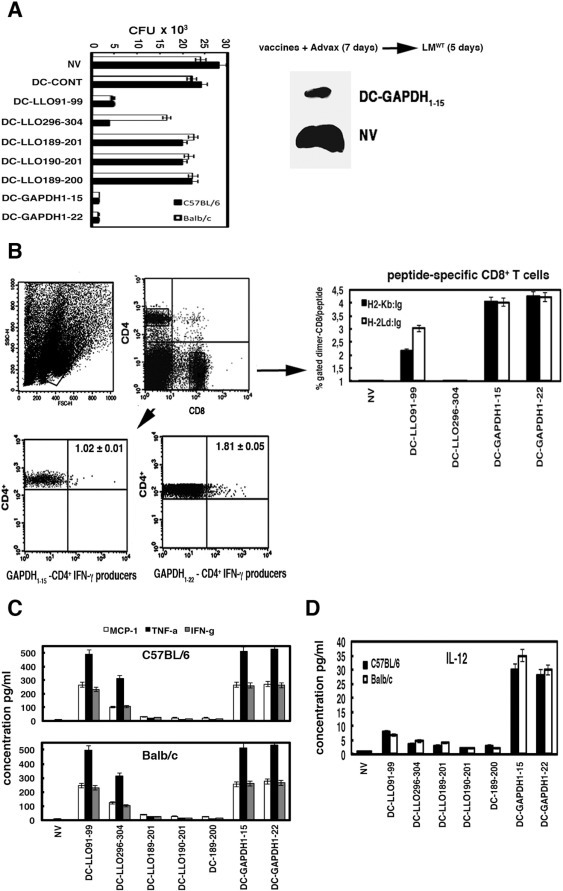
Experimental validation of epitope predictions with vaccination assays. DC vaccines were tested for their immunogenicity and ability to protect against listeriosis. (A) C57BL/6 (black bars) or Balb/c (white bars) mice were vaccinated i.p with different DC vaccine vectors (1 × 106 cells). DC-CONT, DC-GAPDH1–15, DC-GAPDH1–22, DC-LLO91–99, DC-LLO296–304, DC-LLO189–200, DC-LLO189–201 or DC-LLO190–201 in the presence of Advax (50 μg/ml) or left not vaccinated (NV) (n = 5 mice/group) and 7 days later were challenged i.p with 103 CFU LM (for Balb/c mice) or 104 CFU LM (for C57BL/6 mice). Results of spleen homogenates are expressed as CFU (mean ± SD) and obtained from triplicate samples of three independent experiments (P < 0.01). Images correspond to spleens of non-vaccinated mice (NV) showing splenomegaly versus DC-GAPDH1–15 vaccinated C57BL/6 mice. (B) C57BL/6 or Balb/c mice were vaccinated and then challenged with LM as in A and examined for specific GAPDH T-cell responses by intracellular cytokine staining (lower plots). Homogenates were stimulated 5 h with GAPDH1–15 or GAPDH1–22 peptides then intracellular cytokine staining performed. Histograms show the frequency of GAPDH1–15 (left lower plot) and GAPDH1–22-specific CD4+ T cells (right lower plot) and IFN-γ producers in C57BL/6 spleen homogenates. Experiments were performed in triplicate and results are expressed as mean ± SD (P < 0.05). To measure the frequencies of CD8+ GAPDH-specific immune responses (right plots), splenocytes from vaccinated mice were incubated with recombinant dimeric H-2Kb:Ig (black bars) or H-2Ld:Ig (white bars) fusion proteins loaded with LLO91–99, LLO296–304, GAPDH1–15 or GAPDH1–22 peptides, respectively. The staining cocktail contained the dimeric fusion protein loaded with the peptides, CD8 and IFN-γ antibodies. CD8+ cells were gated for anti-IFN-γ staining (% Gated dimer-CD8) to calculate the frequencies of CD8+-GAPDH1–15 or CD8+-GAPDH1–22 restricted cells and IFN-γ producers. Results were expressed as the mean ± SD of triplicates (P < 0.05). (C) Levels of pro-inflammatory cytokines MCP-1, TNF-α or IFN-γ were analysed in mouse sera from DC-vaccinated and LM challenged mice as in A using the CBA kit (BD Biosciences) by flow cytometry (see Material and Methods). Upper plot correspond to cytokine measurements of C57BL/6 mice and lower plot to Balb/c mice. Results expressed as cytokine concentration (pg/ml) of mean ± SD, P < 0.05. (D), IL-12 levels of sera from A measured with the CBA kit and expressed as cytokine concentration (pg/ml) of mean ± SD, P < 0.05.
Table 3.
B cell responses elicited by DC-vaccines in the presence of Advax.
| Vaccinea | Anti-peptide IgM antibodiesb |
|
|---|---|---|
| LLO91–99 | GAPDH1–22 | |
| Control | 0.156 ± 0.05 | 0.163 ± 0.04 |
| NV | 0.517 ± 0.02 | 1.021 ± 0.02 |
| DC-GAPDH1–22 | 0.712 ± 0.02 | 2.011 ± 0.05 |
| DC-LLO91–99 | 0.583 ± 0.03 | 1.276 ± 0.02 |
| Vaccinea | Spleen markersc |
|||
|---|---|---|---|---|
| CD19+ | CD4+ | CD8+ | CD11c+-CD86+ | |
| Control | 19 ± 0.5 | 7 ± 0.3 | 8 ± 0.2 | 0.5 ± 0.02 |
| NV | 15 ± 0.5 | 18 ± 0.6 | 17 ± 0.5 | 26 ± 0.8 |
| DC-GAPDH1–22 | 15 ± 0.3 | 26 ± 0.4 | 28 ± 0.5 | 70 ± 0.7 |
| DC-LLO91–99 | 15 ± 0.2 | 7 ± 0.2 | 27 ± 0.3 | 70 ± 0.6 |
C57BL/6 (Table) or Balb/c (data not shown) mice were vaccinated in the presence of Advax (50 μg/ml) with DC-GAPDH1–22 or DC-LLO91–99 vaccines or non-vaccinated (NV). 7 days post-vaccination, vaccinated and NV mice were infected with 5 × 103 CFU of LMWT for 5 days. Control mice were non-vaccinated and non-infected. Next, mice were sacrificed and sera collected and stored at − 80 °C. Spleens were homogenized for further analysis.
Sera from vaccinated, non-vaccinated or control mice were thawed and examined peptide-ELISA as described in Methods section. Reactions were developed with goat anti-mouse IgM and absorbances analysed at 450 nm. Results are the mean of triplicates ± SD.
Cell surface markers were examined in spleens of vaccinated, non-vaccinated or control mice after incubation with different antibodies conjugated with fluorochromes and analyse by FACS. Results corresponded to the mean of the percentages of positive cells ± SD.
Antibody production and B cell responses seemed to play no role in listeriosis (Pamer, 2004). However, antibodies against LLO seemed to participate in neutralization of Listeria growth (Edelson and Unanue, 2001). Here, we confirmed no variation in B cell numbers among non-vaccinated (NV), vaccinated mice (DC vaccines) or non-infected control mice (control) with similar percentages of CD19+ positive cells (Spleen markers rows in Table 3). Nevertheless, DC-GAPDH1–22 (DC-GAPDH1–22 rows in Table 3) or DC-GAPDH1–15 vaccines (data not shown) produced high levels of anti-GAPDH1–22 IgM antibodies compared to non-vaccinated mice (NV) that correlated with an increase in the percentages of CD4+ T cells. DC-LLO91–99 vaccinated mice produced no significant levels of anti-LLO91–99 IgM antibodies compared to NV and presented low percentages of CD4+ T cells (DC-LLO91–99 rows in Table 3). These data strongly suggested that vaccines conferring the highest protection in listeriosis induced prominent antigen-specific T cell responses and promoted also antigen-specific B cell responses, suggesting a collaborative role of B and T cell responses in listeriosis protection (Edelson and Unanue, 2001, Alvarez-Dominguez et al., 2008, Calderon-Gonzalez et al., 2014, Calderon-Gonzalez et al., 2015).
Finally, vaccination with DC-GAPDH1–15, DC-GAPDH1–22 or DC-LLO91–99 vaccine induced high levels of MCP-1, TNF-α and IFN-γ in both mice strains whereas DC-LLO296–304 vaccine induced only intermediate levels of TNF-α in C57BL/6 mice (DC-LLO296–304 bars in Fig. 2C). Only DC-GAPDH1–15 and DC-GAPDH1–22 vaccines induced high IL-12 levels, post-challenge, in both mice strains (right plot in Fig. 2C). DC-LLO91–99 vaccine induced low but significant levels of IL12 in both mice strains (Kono et al., 2012, Calderon-Gonzalez et al., 2014). Thus the Listeria T-cell epitope prediction algorithm described in the methods was validated in vivo and shown to predict vaccine effectiveness after Listeria vaccination of low and high susceptibility mice. Advax adjuvant was confirmed to be important to induction of a potent Listeria antigen specific T-cell response, consistent with its ability to enhance vaccine protection in other models including influenza, where it similarly enhanced CD4+ and CD8+ T-cell responses and Th1 cytokine production (Honda-Okubo et al., 2012). The present results confirm that Advax adjuvant has the ability to induce MHC I cross-presentation of Listeria antigens thereby resulting in activation of Listeria-specific CD8 T cells, although the mechanism by which it might induce cross-presentation is currently unknown. One possibility currently under investigation is that Advax induces cross-presentation via activation of CD4+ T helper cells that signal to upregulate cross-presentation pathways in antigen presenting cells. When considering design of Listeria vaccines, a major advantage of Advax over other potential T-cell adjuvants is its lack of toxicity as evidenced by data from preclinical studies (Lobgis et al., 2010, Cristillo et al., 2011, Eckersley et al., 2011, Layton et al., 2011, Larena et al., 2013, Petrovsky et al., 2013, Saade et al., 2013, Bielefeldt-Ohmann et al., 2014, Feinen et al., 2014, Honda-Okubo et al., 2014, Honda-Okubo et al., 2015) as well as human clinical trials (Gordon et al., 2012, Gordon et al., 2014).
4. Conclusions
New methods of T-cell epitope screening would greatly assist design of DC vaccine formulations, including for protection against listeriosis. Here we have developed a method for DC vaccine design that combines bioinformatic prediction of MHC binding epitopes with 3D structural predictions and DC functional assays utilising Advax, a potent T-cell adjuvant, to amplify the signal. Using these methodologies, we successfully predicted Listeria epitopes that when incorporated in DC vaccines and administered to either Balb/c or C57BL/6 mice were able to confer protection against Listeria challenge. Peptides binding at least four MHC-I molecules with high affinity due to tight loops and also containing a weaker MHC-II binding epitope containing two α-helices, elicited the highest DC activation, the strongest DTH immune responses and the highest production of IL-12. These properties conferred the greatest protection in mouse strains at either high or low susceptibility to listeriosis. The GAPDH1–15 epitope possessed these minimal requirements and proved to have similar protective efficacy to the longer GAPDH1–22 epitope. This methodology might help to design new human and veterinary vaccines against listeriosis, an area of need given the increasing incidence of this disease in European countries (Peña-Sagredo et al., 2008, Valero and Rafart, 2014, Pérez-Trallero et al., 2014). The methodology used in this study to predict the best T-cell epitopes for vaccine design, might be useful in design of DC vaccines against other infectious diseases, and notably the protective GAPDH1–15 epitope showed 98% homology with S. pneumonia and Streptococcus pyogenes (Alvarez-Dominguez et al., 2008).
Disclosures
NP is a Director of Vaxine Pty Ltd, which holds rights over Advax™ adjuvant.
Acknowledgements
This study was supported in part by MINECO grants, SAF2009-08695 and SAF2012-34203 and Institutional grants API2012/03/SAF2009-08695 and AIP2014/SAF2012-34203 (to C. A-D). We acknowledge the laboratory technical assistance of L. V-R, the peptide synthesis by F. Roncal (CNB, CSIC, Madrid), the IDIVAL institutional assistance of C. Santacruz-Llata with FACS analysis and the Listeria strains provided by D. A. Portnoy (UCLA. USA). Development of Advax™ adjuvant was supported by contract HHSN272200800039C to Vaxine from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. This paper's contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
References
- Alvarez-Dominguez C., Madrazo-Toca F., Fernandez-Prieto L., Vandeckerhove J., Pareja E., Tobes R., Gomez-Lopez M.T., Del Cerro-Vadillo E., Fresno-Escudero M., Leyva-Cobián F., Carrasco-Marin E. Characterization of a Listeria monocytogenes protein interfering with Rab5a. Traffic. 2008;9:325. doi: 10.1111/j.1600-0854.2007.00683.x. [DOI] [PubMed] [Google Scholar]
- Angelakopoulos H., Loock K., Sisul D.M., Jensen E.R., Miller J.F., Hohmann E.L. Safety and shedding of an attenuated strain of Listeria monocytogenes with a deletion of actA/plcB in adult volunteers: a dose escalation study of oral inoculation. Infect. Immun. 2002;70(7):3592. doi: 10.1128/IAI.70.7.3592-3601.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielefeldt-Ohmann H., Prow N.A., Wang W., Tan C.S., Coyle M., Douma A., Hobson-Peters J., Kidd L., Hall R.A., Petrovsky N. Safety and immunogenicity of a delta inulin-adjuvanted inactivated Japanese encephalitis virus vaccine in pregnant mares and foals. Vet. Res. 2014;45:130. doi: 10.1186/s13567-014-0130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon-Gonzalez R., Frande-Cabanes E., Bronchalo-Vicente L., Lecea-Cuello M.J., Pareja E., Bosch-Martinez A., Fanarraga M.L., Yañez-Diaz S., Carrasco-Marin E., Alvarez-Dominguez C. Cellular vaccines in listeriosis: role of the Listeria antigen GAPDH. Front. Cell. Infect. Microbiol. 2014;4(22):1–11. doi: 10.3389/fcimb.2014.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon-Gonzalez R., Frande-Cabanes E., Tobes R., Pareja E., Alaez-Alvarez L., Alvarez-Dominguez C. A dendritic cell targetted vaccine loaded with a glyceraldehyde-3-phosphate-dehydrogenase peptide proposed for individuals at high risk of listeriosis. J. Vaccines Vaccin. 2015;6:266. [Google Scholar]
- Cohen N., Margalit R., Pevsner-Fischer M., Yona S., Jung S., Eisenbach L., Cohen I.R. Mouse dendritic cells pulsed with capsular polysaccharide induce resistance to lethal pneumococcal challenge: roles of T cells and B cells. PLoS One. 2012;7(6):e39193. doi: 10.1371/journal.pone.0039193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper P.D., Petrovsky N. Delta inulin: a novel, immunologically active, stable packing structure comprising β-d-[2- > 1] poly(fructo-furanosyl) α-d-glucose polymers. Glycobiology. 2011;21(5):595. doi: 10.1093/glycob/cwq201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper P.D., Barclay T.G., Ginic-Markovic M., Petrovsky N. The polysaccharide inulin is characterized by an extensive series of periodic isoforms with varying biological actions. Glycobiology. 2013;23(10):1164. doi: 10.1093/glycob/cwt053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper P.D., Barclay T.G., Ginic-Markovic M., Gerson A.R., Petrovsky N. Inulin isoforms differ by repeated additions of one crystal unit cell. Carbohydr. Polym. 2014;103:392. doi: 10.1016/j.carbpol.2013.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper P.D., Rajapaksha K.H., Barclay T.G., Ginic-Markovic M., Gerson A.R., Petrovsky N. Inulin cristal initiation via a glucose-fructose cross-link of adjacent polymer chains: atomic force microscopy and static molecular modelling. Carbohydr. Polym. 2015;117:964. doi: 10.1016/j.carbpol.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristillo A.D., Ferrari M.G., Hudacik L., Lewis B., Galmin L., Bowen B., Thompson D., Petrovsky N., Markham P., Pal R. Induction of mucosal and systemic antibody and T-cell responses following prime-boost immunization with novel adjuvanted human immunodeficiency virus-1-vaccine formulations. J. Gen. Virol. 2011;92(Pt 1):128. doi: 10.1099/vir.0.023242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersley A.M., Petrovsky N., Kinne J., Wernery R., Wernery U. Improving the dromedary antibody response: the hunt for the ideal camel adjuvant. J. Camel Pract. Res. 2011;18(1):p35. [Google Scholar]
- Edelson B., Unanue E.R. Intracellular antibody neutralizes Listeria growth. Immunity. 2001;14(5):503. doi: 10.1016/s1074-7613(01)00139-x. [DOI] [PubMed] [Google Scholar]
- Feinen B., Petrovsky N., Verma A., Merkel T.J. Advax-adjuvanted recombinant protective antigen provides protection against inhalational anthrax that is further enhanced by addition of murabutide adjuvant. Clin. Vaccine Immunol. 2014;21(4):580. doi: 10.1128/CVI.00019-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromen C.A., Robbins G.R., Shen T.W., Kai M.P., Ting J.P., DeSimone J.M. Controlled analysis of nanoparticle charge on mucosal and systemic antibody responses following pulmonary immunization. Proc. Natl. Acad. Sci. U. S. A. 2015;112(2):488. doi: 10.1073/pnas.1422923112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geginat G., Schenk S., Skoberne M., Goebel W., Hof H. A novel approach of direct ex vivo epitope mapping identifies dominant and subdominant CD4 and CD8 T cell epitopes from Listeria monocytogenes. J. Immunol. 2001;166:1877. doi: 10.4049/jimmunol.166.3.1877. [DOI] [PubMed] [Google Scholar]
- Gordon D.L., Sajkov D., Woodman R.J., Honda-Okubo Y., Cox M.J., Heinzel S., Petrovsky N. Randomized clinical trial of immunogenicity and safety of a recombinant H1N1/2009 pandemic influenza vaccine containing Advax™ polysaccharide adjuvant. Vaccine. 2012;30:5047. doi: 10.1016/j.vaccine.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D., Kelley P., Heinzel S., Cooper P., Petrovsky N. Immunogenicity and safety of Advax™, a novel polysaccharide adjuvant based on delta inulin, when formulated with hepatitis B surface antigen: a randomized controlled phase 1 study. Vaccine. 2014 doi: 10.1016/j.vaccine.2014.09.034. (Sep 26. pii: S0264-410X(14)01293-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda-Okubo Y., Saade F., Petrovsky N. Advax™, a polysaccharide adjuvant derived from delta inulin, provides improved influenza vaccine protection through broad-based enhancement of adaptive immune responses. Vaccine. 2012;30(36):5373–5381. doi: 10.1016/j.vaccine.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda-Okubo Y., Kolpe A., Li L., Petrovsky N. A single immunization with inactivated H1N1 influenza vaccine formulated with delta inulin adjuvant (Advax™) overcomes pregnancy-associated immune suppression and enhances passive neonatal protection. Vaccine. 2014;32(36):4651. doi: 10.1016/j.vaccine.2014.06.057. (2014 Aug 6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda-Okubo Y., Barnard D., Ong C.H., Peng B.H., Tseng C.T., Petrovsky N. Severe acute respiratory syndrome-associated coronavirus vaccines formulated with delta inulin adjuvants provide enhanced protection while ameliorating lung eosinophilic immunopathology. J. Virol. 2015;89(6):2995. doi: 10.1128/JVI.02980-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki N., Rillahan C.D., Cheng T.Y., Van Rhijn I., Macauley M.S., Moody D.B., Paulson J.C. Targeted delivery of mycobacterial antigens to human dendritic cells via Siglec-7 induces robust T cell activation. J. Immunol. 2014;193(4):1560. doi: 10.4049/jimmunol.1303278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Ponomarenko J., Zhu Z., Tamang D., Wang P., Greenbaum J., Lundegaard C., Sette A., Lund O., Bourne P.E., Nielsen M., Peters B. Immune epitope database analysis resource. Nucleic Acids Res. 2012;40(Web Server issue):W525. doi: 10.1093/nar/gks438. (Epub 2012 May 18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono M., Nakamura Y., Suda T., Uchijima M., Tsujimura K., Nagata T., Giermasz A.S., Kalinski P., Nakamura H., Chida K. Enhancement of protective immunity against intracellular bacteria using type-1 polarized dendritic cell (DC) vaccine. Vaccine. 2012;30:2633. doi: 10.1016/j.vaccine.2012.02.026. [DOI] [PubMed] [Google Scholar]
- Köster S., van Pee K., Hudel M., Leustik M., Rhinow D., Kühlbrandt W., Chakraborty T., Yildiz Ö. Crystal structure of listeriolysin O reveals molecular details of oligomerization and pore formation. Nat. Commun. 2014;5:3690. doi: 10.1038/ncomms4690. [DOI] [PubMed] [Google Scholar]
- Larena M., Prow N.A., Hall R.A., Petrovsky N., Lobigs M. JE-ADVAX vaccine protection against Japanese encephalitis virus mediated by memory B cells in the absence of CD8(+) T cells and pre-exposure neutralizing antibody. J. Virol. 2013;8:4395. doi: 10.1128/JVI.03144-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer P., Hanson B., Lemmens E.E., Liu W., Luckett W.S., Leong M.L., Allen H.E., Skoble J., Bahjat K.S., Freitag N.E., Brockstedt D.G., Dubensky T.W. Constitutive activation of the PrfA regulon enhances the potency of vaccines based on live-attenuated and killed but metabolically active Listeria monocytogenes strains. Infect. Immun. 2008;76(8):3742. doi: 10.1128/IAI.00390-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton R.C., Petrovsky N., Gigliotti A.P., Pollock Z., Knight J., Donart N., Pyles J., Harrod K.S., Gao P., Koster F. Delta inulin polysaccharide adjuvant enhances the ability of split-virion H5N1 vaccine to protect against lethal challenge in ferrets. Vaccine. 2011;29(37):6242. doi: 10.1016/j.vaccine.2011.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobgis M., Pavy M., Hall R.A., Lobgis P., Cooper P., Komiya T., Toriniwa H., Petrovsky N. An inactivated vero cell-grown Japanese encephalitis vaccine formulated with Advax, a novel inulin-based adjuvant, induces protective neutralizing antibody against homologous and heterologous flaviviruses. J. Gen. Virol. 2010;91:1407. doi: 10.1099/vir.0.019190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainou-Fowler T., MacGowan A.P., Postlethwaite R. Virulence of Listeria spp.: course of infection in resistant and susceptible mice. J. Med. Microbiol. 1988;27(2):131. doi: 10.1099/00222615-27-2-131. [DOI] [PubMed] [Google Scholar]
- Mohamed W., Sethi S., Tchatalbachev S., Darji A., Chakraborty T. Protective immunity to Listeria monocytogenes infection mediated by recombinant Listeria innocua harboring the VGC locus. PLoS One. 2012;7(4):e35503. doi: 10.1371/journal.pone.0035503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M., Lundegaard C., Worning P., Lauemøller S.L., Lamberth K., Buus S., Brunak S., Lund O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12(5):1007. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamer E.G. Immune responses to Listeria monocytogenes. Nat. Rev. Immunol. 2004;4:812. doi: 10.1038/nri1461. [DOI] [PubMed] [Google Scholar]
- Peña-Sagredo J.L., Hernández M.V., Fernandez-Llanio N., Giménez-Ubeda E., Muñoz-Fernandez S., Ortiz A., Gonzalez-Gay M.A., Fariñas M.C., Biobadaser group Listeria monocytogenes infection in patients with rheumatic diseases on TNF-alpha antagonist therapy: the Spanish Study Group experience. Clin. Exp. Rheumatol. 2008;26(5):854. [PubMed] [Google Scholar]
- Pérez-Trallero E., Zigorraga C., Artieda J., Alkorta M., Marimón J.M. Two outbreaks of Listeria monocytogenes infection, Northern Spain. Emerg. Infect. Dis. 2014;20(12):2155. doi: 10.3201/eid2012.140993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B., Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinformatics. 2005;31(6):132. doi: 10.1186/1471-2105-6-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovsky N., Larena M., Siddharthan V., Prow N.A., Hall R.A., Lobigs M., Morrey J. An inactivated cell culture Japanese encephalitis vaccine (JE-ADVAX) formulated with delta inulin adjuvant provides robust heterologous protection against West Nile encephalitis via cross-protective memory B cells and neutralizing antibody. J. Virol. 2013;18:b10324. doi: 10.1128/JVI.00480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pion M., Serramia M.J., Diaz L., Bryszewska M., Gallart T., Garcia F., Gomez R., de la Mata F.J., Muñoz-Fernandez M.A. Phenotype and functional analysis of human monocytes-derived dendritic cells loaded with a carbosilane dendrimer. Biomaterials. 2010;31:8749. doi: 10.1016/j.biomaterials.2010.07.093. [DOI] [PubMed] [Google Scholar]
- Poulsen K.P., Faith N.G., Steinberg H., Czuprinsky C.J. Pregnancy reduces the genetic resistance of C57BL/6 mice to Listeria monocytogenes intragastric inoculation. Microb. Pathog. 2011;50(6):360. doi: 10.1016/j.micpath.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey-Ladino J., Ross A.G.P., Cripps A.W. Immunity, immunopathology, and human vaccine development against sexually transmitted Chlamydia trachomatis. Human Vaccines Immunother. 2014;10(9):2664. doi: 10.4161/hv.29683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Del Rio E., Marradi M., Calderon-Gonzalez R., Frande-Cabanes E., Penades S., Petrovsky N., Alvarez-Dominguez C. A gold-glyconanoparticle carrying a listeriolysin o peptide and formulated with Advax™ delta inulin adjuvant induces robust T-cell protection against Listeria infection. Vaccine. 2015;33(12):1465. doi: 10.1016/j.vaccine.2015.01.062. [DOI] [PubMed] [Google Scholar]
- Saade F., Honda-Okubo Y., Trec S., Petrovsky N. A novel hepatitis B vaccine containing Advax™, a polysaccharide adjuvant derived from delta inulin, induces robust humoral and cellular immunity with minimal reactogenicity in preclinical testing. Vaccine. 2013;31(15):1999. doi: 10.1016/j.vaccine.2012.12.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J., Assarsson E., Moore C., Ngo S., Pinilla C., Sette A., Peters B. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 2008;4:2. doi: 10.1186/1745-7580-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoberne M., Geginat G. Efficient in vivo presentation of Listeria monocytogenes derived CD4 and CD8 cell epitopes in the absence of IFN-γ. J. Immunol. 2002;168:1854. doi: 10.4049/jimmunol.168.4.1854. [DOI] [PubMed] [Google Scholar]
- Stephenson K.E., Neubauer G.H., Reimer U., Pawlowski N., Knaute T., Zerweck J., Korber B.T., Barauch D.H. Quantification of epitope diversity of HIV-1-specific binding antibodies by peptide microarrays for global HIV-1 vaccine development. J. Immunol. Methods. 2015;416:105. doi: 10.1016/j.jim.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacas-Cordoba E., Pion M., Rasines B., Filippini D., Komber H., Ionov M., Bryszewska M., Appelhans D., Muñoz-Fernandez M.A. Glycodendrimers as new tools in the search for effective anti-HIV DC-based immunotherapies. Nanomedicine. 2013;9(7):972. doi: 10.1016/j.nano.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Valero F.P., Rafart J.V. Incidence study of listeriosis in Spain. Gac. Sanit. 2014;28(1):74. doi: 10.1016/j.gaceta.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Wood L.M., Paterson Y. Attenuated Listeria monocytogenes: a powerful and versatile vector for the future of tumor immunotherapy. Front. Cell. Infect. Microbiol. 2014;4:51. doi: 10.3389/fcimb.2014.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Petrovsky N., Schönbach C., Koh J.Y., Brusic V. Methods for prediction of peptide binding to MHC molecules: a comparative study. Mol. Med. 2002;8(3):137. [PMC free article] [PubMed] [Google Scholar]


