Abstract
Different viruses can be responsible for similar clinical manifestations of respiratory infections. Thus, the etiological diagnosis of respiratory viral diseases requires the detection of a large number of viruses. In this study, 6 duplex real-time PCR assays, using EvaGreen intercalating dye, were developed to detect 12 major viruses responsible for respiratory diseases: influenza A and B viruses, enteroviruses (including enterovirus spp, and rhinovirus spp), respiratory syncytial virus, human metapneumovirus, coronaviruses group I (of which CoV 229E and CoV NL63 are part) and II (including CoV OC43 and CoV HKU1), parainfluenza viruses type 1, 2, 3 and 4, human adenoviruses and human bocaviruses. The 2 target viruses of each duplex reaction were distinguishable by the melting temperatures of their amplicons. The 6 duplex real time PCR assays were applied for diagnostic purpose on 202 respiratory samples from 157 patients. One hundred fifty-seven samples were throat swabs and 45 were bronchoalveolar lavages. The results of the duplex PCR assays were confirmed by comparison with a commercial, validated, assay; in addition, the positive results were confirmed by sequencing. The analytical sensitivity of the duplex PCR assays varied from 103 copies/ml to 104 copies/ml. For parainfluenza virus 2 only it was 105 copies/ml. Seventy clinical samples (35%) from 55 patients (30 children and 25 adults) were positive for 1 or more viruses. In adult patients, influenza A virus was the most frequently detected respiratory virus followed by rhinoviruses. In contrast, respiratory syncytial virus was the most common virus in children, followed by enteroviruses, influenza A virus and coronavirus NL63. The small number of samples/patients does not allow us to draw any epidemiological conclusion. Altogether, the results of this study indicate that the 6 duplex PCR assays described in this study are sensitive, specific and cost-effective. Thus, this assay could be particularly useful to identify the main respiratory viruses directly from clinical samples, after nucleic acid extraction, and, also, to screen a large number of patients for epidemiological studies.
Keywords: Duplex real time PCR, Respiratory viruses detection
Highlights
-
•
We developed 6 real time PCRs to detect 12 respiratory viruses.
-
•
The amplicons of each duplex were distinguishable by the melting temperatures.
-
•
Altogether, the assay can be performed in about 2 h.
-
•
The cost of each duplex-PCR is about 3 euros.
-
•
202 clinical samples have been analyzed for the detection of 12 respiratory viruses.
1. Introduction
Viral respiratory acute infections are common and contribute significantly to morbidity and mortality worldwide [1]. Many different viruses can determine respiratory tract infections and most of them belong to the Orthomyxoviridae, Coronaviridae, Picornaviridae, Paramyxoviridae, Adenoviridae and Parvoviridae families. Viral respiratory infections may be either asymptomatic, or they may occur with mild symptoms or even cause severe diseases.
In patients with predisposing conditions, the outcome of these infections can be more severe and require hospitalization, sometimes even in intensive care units (ICUs), following the development of pneumonia and acute respiratory distress syndrome (ARDS). Respiratory viruses account for about 30% of pneumonia cases in adult patients, hospitalized in ICU, with mortality rates comparable to those of bacterial pneumonia [2]. In children, especially those younger than 2 years, they frequently cause pneumonia [3], [4].
A rapid and accurate etiological diagnosis is essential for prompt patient management, ruling out non-viral infection, limiting the spread of infections and, when available, initiating timely therapeutic treatments. For many years the diagnosis of viral respiratory tract infections has been made by non-molecular approaches such as antigen detection by direct immunofluorescence and viral culture. These methods, although effective and often complementary, are time-consuming, labour-intensive and, often, lack of sensitivity or specificity [5], [6], [7]. Compared to classical methods, molecular methods have significantly improved the diagnosis of acute respiratory tract infections as they offer high sensitivity and provide specific results within a shorter period of time and for a larger number of pathogens [8], [9], [10], [11], [12], [13], [14]. Consequently, a number of different molecular systems have become commercially available for fast and accurate detection of respiratory viruses [7], [8], [9], [10], [15], [16], [17].
Different viruses can be responsible for similar clinical manifestations of respiratory infections. Thus, the etiological diagnosis of respiratory viral diseases requires the detection of a large number of viruses. Both commercial kits and “in-house” methods, which can simultaneously detect many respiratory viruses, have been described [11], [12], [13], [14], [15], [16], [17], [18], [19] but most of them are expensive and require specific detection instruments.
In this study, 6 duplex real-time PCR assays, using EvaGreen intercalating dye, were developed to detect in a rapid and cost effective way the main viruses involved in respiratory diseases. In particular, the object of this study was the direct detection in respiratory samples, after nucleic acid extraction, of influenza viruses type A and type B, enteroviruses (EV) (including enterovirus spp, and rhinovirus spp, hRV), respiratory syncytial virus (hRSV), human metapneumovirus (hMPV), coronaviruses group I (CoVI, of which CoV 229E and CoV NL63 are part) and II (CoVII, including CoV OC43 and CoV HKU1), parainfluenza viruses type 1, 2, 3 and 4 (PIV1, 2, 3, 4), human adenoviruses (hAdV) and human bocaviruses (HBoV).
2. Materials and methods
2.1. Respiratory viruses and primers design
The viruses object of this study weretype A and type B influenza viruses, EV (including hRV spp and EV spp), hRSV, hMPV, CoVI and CoVII, PIV1 and 3 (Respirovirus genus), PIV2 and PIV4, hAdV, HBoV.
Twelve primer pairs (Table 1 ) were designed targeting a conserved and specific region in the genome of each virus included in this study. For this purpose, from 100 to 656 sequences of each virus have been downloaded from GenBank (NCBI) and aligned using ClustalW v1.4 included in BioEdit v7.0.0. The primer sequences were chosen in order to use the same assay for the amplification reactions For some viruses, published primer sequences could be used [20], [21], [22], [23], [24], [25], [26], as shown in Table 1. In addition, Table 1 shows the gene target for each virus, the sequence of the chosen primers, the size of the amplicons and annealing temperature used.
Table 1.
Primers used to perform duplex real time PCR assays.
| Viruses | Gene | Primer sequences | Annealing | Amplicon size | Ref. |
|---|---|---|---|---|---|
| Influenza A | M | PF:TCAGGCCCCCTCAAAGCC PR:GGGCACGGTGAGCGTRAA |
55 °C | 158 | This study |
| Influenza B | M | PF:TAGCAGAAGGCCATGAAAGCTC PR:CGTTCCTAGTTTTACTTGCATTGAAT |
55 °C | 94 | [20] |
| PIV1 and 3 | L | PF:GAGACTCTGAGCTGTTTTTTAAC PR:GCTGTACTTTCAAATCTCCA |
55 °C | 71 | This study |
| PIV2 | L | PF:TGCATGTTTTATAACTACTGATCTTGCTAA PR:GTTCGAGCAAAATGGATTATGGT |
55 °C | 77 | [21] |
| PIV4 | L | PF:CGCTAAATGAGCCAAGAAGG PR:GACCCATGAAATCGAGTGCT |
55 °C | 182 | This study |
| CoVI | Polymerase | PF:CAACGTATGTGTGCTATAGGC PR:GTATTAACTATTTCAGCAGGAC |
55 °C | 74 | This study |
| CoVII | Polymerase | PF:TGGTGGCTGGGATGATATGT PR:GGCATAGCACGATCACACTTCAA |
55 °C | 96 | [22] |
| hRV/EV | 5′UTR | PF:AGTCCTCCGGCCCCTGAA PR:GAAACACGGCACCCCAAAGT |
55 °C | 120 | [23] |
| hRSV | L | PF:AATACAGCCAAATCTAACCAACTTTA PR:GCCAAGGAAGCATGCAATAAA |
55 °C | 94 | [24] |
| hMPV | F | PF:GAGAGCTGAAAGAATTTGTGAGC PR:GGTCCAATGATATTGCTGGTGTTA |
55 °C | 174 | This study |
| hAdV | Exon 6 | PF:CCCITTYAACCACCG PR:ACATCCTTBCKGAAGTTCCA |
55 °C | 167 | [25] |
| HBoV | NS | PF:CTTGGGCGGGACAGAATGC PR:AACAGAATTGCCACCAACAACC |
55 °C | 120 | [26] |
2.2. Clinical samples
A total of 202 respiratory samples from 157 patients were analysed to detect respiratory viruses. From all patients (93 children and 64 adults) a throat swab was obtained. Of 64 adult patients 45 were hospitalized in intensive care unit (ICU) and a bronchoalveolar lavage (BAL) was also made available. The clinical samples were collected between September 2011 and February 2015 from patients with suspected influenza virus infection.
2.3. Viral nucleic acids extraction
Extraction of viral RNAs and DNAs from clinical samples was carried out using a commercially available kit (QIAamp MinElute Virus, Valencia, CA, USA) for the simultaneous purification of viral RNA and DNA from body fluids. Following the kit instructions, viral nucleic acids were extracted from 200 μl of body fluids. The elution was carried out in a final volume of 150 μl.
2.4. Duplex real time PCR assays
For the detection of respiratory RNA viruses, after retrotranscription (RT) with random examer, 5 duplex real-time PCR, using EVAGreen fluorescent dye, were developed, a first one to detect influenza A and influenza B viruses; a second one to detect PIVs belonging to Respirovirus genus (PIV1 and PIV3) and hRSV; the viruses target of the third one were PIV type 2 and hMPV; a fourth duplex was performed to detect CoV I and EV/hRV; and a fifth duplex was performed to detect CoVII and PIV4. A sixth duplex was performed to detect hAdV and HBoV. In particular, after RT of 10 μl of the extracted sample with random examers (QuantiTect Reverse Transcription Kit, Qiagen, Valencia, CA, USA) in a final volume of 20 μl, 2× HRM PCR master mix (Qiagen, Valencia, CA, USA) was used for the amplification reaction. The reaction volume was 25 μl (12.5 μl of master mix, 1.75 μl of each primer [10 μM], 5 μl of cDNA or of extracted DNA and H2O to reach the final volume). After the initial activation step, 30 cycles of amplification (95 °C for 10 s, 55 °C for 30 s, 72 °C for 10 s (acquiring Green)) were performed. For the melting analysis, a ramp from 65 °C to 95 °C was used, rising by 0.1 °C each step. The reaction was performed on Rotor Gene 6000 (Qiagen, Valencia, CA, USA). The primers specific for each virus are listed in Table 1.
All duplex real time PCR assays were developed using standards prepared with reference strains obtained from the National Institute for Biological Standards and Controls (NIBSC) (CoV229E, hMPV, hRSVA2, PIV1, PIV2, PIV3, PIV4, hRVA) or already available in our laboratory (Influenza A and B strains, isolated and cultured in MDCK cells, and Coxsackie virus A6, hAdV type 2, CoV NL63, CoV OC43, HBoV, sequences obtained and characterised from clinical samples). The standards were prepared by cloning the selected sequences according to the standard protocol of pGEM-T Easy Vector System (Promega, Madison, Wisconsin, USA). The plasmid DNA was purified by QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA, USA). To obtain the RNA standard for each virus, each product of the cloning was transcribed with T7 RNA polymerase (Promega, Madison, Wisconsin, USA). The results of the duplex PCR assays were confirmed by sequencing positive samples, using the primers reported in Table 2 and by comparison with a commercial, validated, assay (r-gene for respiratory viruses detection, BioMérieux, Marcy-l’Etoile, France).
Table 2.
Primers used to perform sequencing reaction of positive samples.
| Viruses | Gene | Primer sequences | Annealing | Amplicon size | Ref. |
|---|---|---|---|---|---|
| Influenza A | M | PF:GAGTCTTCTAACMGAGGTCGAAACGTA PR:GCTGCCTGTTCACTCGATCC |
55 °C | 597 | This study |
| Influenza B | M | PF:CACTGTTGGTTCGGTGGGAA PR:ACAAAGCACAGAGCGTTCCT |
55 °C | 367 | This study |
| PIV2 | L | PF:TCTCGCAAATCATGCAGGTACT PR:GCCTTCAATACCTCCCTTGGA |
55 °C | 496 | This study |
| CoVI | polymerase | PF:GCYCAYGCTGCTGTTGATTC PR:ACTRGARCCATTGTCWACCTG |
55 °C | 534 | This study |
| hRV/EV | 5′UTR | PF:GCACTTCTGTTTCCCCG PR:GAAACACGGCACCCCAAAGT |
55 °C | 390 | [23] |
| hRSV | L | PF:TAAGRATTGCTAATTCWGAATTAGA PR:TMCCWGCTCCTTCACCTATGA |
55 °C | 501 | This study |
| hMPV | F | PF:AACCATMCGRCTTGAGAGTG PR:GCTYCCGTAGACCCCTATCAG |
55 °C | 418 | This study |
| hAdV | Exon 6 | PF:CAACACCTAYGASTACATGAA PR:KATGGGGTARAGCATGTT |
55 °C | 474 | [25] |
2.5. Melting temperature of targets
The melting temperature for each target sequence, as could be calculated after in silico studies, are reported in Table 3 . Altogether, the range of melting temperature of EV/hRV target sequence was comprised between 81 °C and 87.5 °C, as resulted after the in silico study of 656 sequences downloaded from GenBank (NCBI). In particular, this analysis showed that the melting temperature of hRV can vary from 81 °C to 85.5 °C, while the melting temperature of EV is variable from 83.5 °C to 87.5 °C.
Table 3.
Melting temperature of amplicons calculated in silico.
| Duplex PCR essays number | Target | Tm(°C) |
|---|---|---|
| 1 | Influenza A | 83 |
| Influenza B | 78 | |
| 2 | PIV2 | 74 |
| hMPV | 80 | |
| 3 | CoVI | 74.5–76 |
| hRV/EV | 81–87.5 | |
| 4 | PIV1 and 3 | 73 |
| hRSV | 77 | |
| 5 | CoVII | 77.5 |
| PIV4 | 80 | |
| 6 | hAdV | 81.5–88 |
| HBoV | 78 |
As hAdV is concerned, the study of the target region of about 500 sequences downloaded from GenBank (NCBI) showed that the melting temperature varies from 81.5 °C to 88 °C.
3. Results
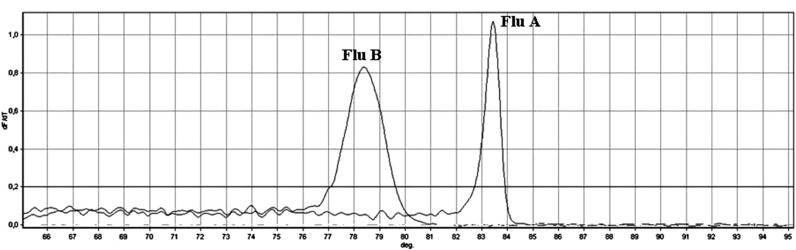
Six real-time duplex PCR assays, using EvaGreen fluorescent dye, were developed to identify 12 common respiratory viruses directly from clinical samples, after nucleic acid extraction. The amplicon size of the target region of each virus varied from 71 to 182 bp, with a melting temperature ranging from 73 °C to 88 °C. Each duplex reaction was designed so that the melting temperatures of the 2 target viruses were clearly distinguishable (Fig. 1 ).
Fig. 1.
Melting profile of influenza A and B amplicons obtained by amplification of the target sequence of the M gene, reported as an example.
The analytical sensitivity of all duplex PCR assays was determined using serial dilutions of the cloned target sequences, quantified by NanoDrop 1000 Spectrophotometer (ThermoScientific, Wilmington, DE, USA) (Table 4 ). In Table 4, the highest dilution which was always positive when repeated 10 times is shown; a further dilution was always negative, or positive in less than 50% of the times. The duplex real-time PCR 1 was able to detect 103 copies/ml of influenza A virus and 104 copies/ml of influenza B virus. The sensitivity of the duplex 2 was of 105 copies/ml for PIV2 and 104 copies/ml for hMPV. Reaction 3 detected 104 copies/ml of CoVI and 103 copies/ml of hRV/EV; 104 copies/ml and 103 copies/ml of PIV1,3 and hRSV, respectively, were detected by reaction 4. The sensitivity of reaction 5 was 104 copies/ml for both targets. The sensitivity of reaction 6 to detect hAdV and HBoV was 103 copies/ml for both targets.
Table 4.
Analytical sensitivity of each duplex PCR expressed in copies number/ml.
| Target | Copies number/ml |
|---|---|
| Influenza A | 103 |
| Influenza B | 104 |
| PIV2 | 105 |
| hMPV | 104 |
| CoVI | 104 |
| hRV/EV | 103 |
| PIV1 and 3 | 104 |
| hRSV | 103 |
| CoVII | 104 |
| PIV4 | 104 |
| hAdV | 103 |
| HBoV | 103 |
To evaluate the specificity of each duplex real-time PCR, each cloned target sequence (107 copies/μl) was tested individually by the 6 duplex reactions. In addition, a pool was prepared containing 107 copies/μl of each positive control. This pool was then tested with each duplex real-time PCR. The results confirmed that each duplex could identify only its specific targets.
Furthermore, in order to assess the intra-assay and inter-assay variability of each duplex real-time PCR, each clinical sample was assayed 3 times in the same run and all the positive samples were assayed in 3 different runs with reproducible results. Table 5 shows the results of this evaluation performed with the positive controls.
Table 5.
Intra-assay and inter-assay variability of the results of the 6 duplex PCR assays performed with the positive controls (mean of the melting temperature of each amplicon ± standard deviation).
| Target | Intra-assay variability | Inter-assay variability |
|---|---|---|
| Influenza A | 82.65 ± 0.04 | 82.78 ± 0.33 |
| Influenza B | 78.18 ± 0.07 | 78.08 ± 0.09 |
| PIV2 | 74.10 ± 0.04 | 74.22 ± 0.40 |
| hMPV | 80.50 ± 0.05 | 80.10 ± 0.50 |
| CoVI 229E | 76.40 ± 0.12 | 76.35 ± 0.37 |
| CoVI NL63 | 75.70 ± 0.07 | 75.83 ± 0.06 |
| EV | 85.88 ± 0.14 | 85.90 ± 0.28 |
| hRV | 84.35 ± 0.12 | 84.28 ± 0.25 |
| PIV1 and 3 | 73.05 ± 0.05 | 73.02 ± 0.20 |
| hRSV | 76.88 ± 0.12 | 76.93 ± 0.62 |
| CoVII OC43 | 77.5 ± 0.02 | 77.2 ± 0.2 |
| PIV4 | 80.05 ± 0.12 | 80.02 ± 0.50 |
| hAdV | 83.98 ± 0.02 | 84.07 ± 0.25 |
| HBoV | 77.7 ± 0.2 | 78.02 ± 0.33 |
Altogether, in this study 202 clinical samples from 157 patients, collected between September 2011 and February 2015, were analysed. A total of 70 clinical samples from 55 patients (30 children and 25 adults) were positive for 1 or more viruses. Influenza virus (23/157; 14.7%) was the most frequently detected respiratory virus, followed by EV/hRV (17/157; 10.8%). As regard influenza virus, in particular, 17 patients were positive for influenza A virus and 6 for influenza B virus. Seven patients positive for influenza A virus were hospitalized in ICU. In these patients the virus was detected in both throat swab and BAL. From 4 others adult patients, influenza A positives, only a throat swab was available; the other 6 influenza A positive patients were children. Influenza B virus was detected in 5 children and in 1 adult hospitalized in ICU, in both throat swab and BAL.
hRSV was detected in 8 (5%) patients, all children; CoVI was detected in 6 (3.8%) children while hAdV was detected in 5 (3.2%) patients, 4 children and 1 adult hospitalized in ICU. In this patient hAdV was detected only in BAL sample. PIV2 was detected only in the throat swab sample of 2 patients (1.3%), 1 adult and 1 child, and hMPV was detected only in the throat sample of 1 adult patient (0.64%). In 6 clinical samples 2 or more viruses were detected; in particular EV/hRV were the most common viruses detected in mixed infections (5/6) (Table 6 ). All patients with mixed infections were children.
Table 6.
Viruses detected in mixed infections.
| Sample | hRV | EV | hRSV A | Influenza A | CoV NL63 | PIV2 | hAdV |
|---|---|---|---|---|---|---|---|
| 1 | + | – | + | – | – | – | – |
| 2 | – | + | – | + | + | – | – |
| 3 | – | + | – | + | – | – | – |
| 4 | – | + | – | + | – | – | – |
| 5 | + | – | – | – | – | + | – |
| 6 | – | – | – | – | + | – | + |
The results of the duplex PCR assays, were confirmed by comparison with a commercial, validated, assay, (r-gene for respiratory viruses detection, Biomérieux, Marcy-l’Etoile, France) which gave similar results. In fact all the samples, positive with the duplex PCR assays, were positive also with the commercial assay. With reference to the 132 samples negative by the duplex PCR assays, 1 was positive for PIV with the commercial test. Thus, altogether, the sensitivity of the assay proposed was 98%. In addition, the positive results were confirmed by sequencing.
Concerning the 17 EV/hRV detected, their melting temperature varied from 82 °C to 85,90 °C. Four could be considered as hRV because of their melting temperature lower than 83.5°, 6 as EV, while 7 had a melting temperature ranging from 83.5 °C to 85 °C and could not be identified on this basis. The results of sequencing confirmed what had been inferred from the melting temperature data and allowed us to identify the 7 samples not previously determined as hRV. The sequencing of the hRSV showed that 7 were of type A and only 1 was of type B and all the CoV were NL63.
4. Discussion
The assay developed in this study can detect 12 different respiratory viruses, the most common viruses infecting the respiratory tract worldwide. The results indicate that the assay proposed gives sensitive, specific and reproducible results. In fact, the sensitivity of these duplex real-time PCR assays varies between 103 copies/ml and 105 copies/ml, which is comparable to that of other commercial and “in-house” assays. The thermal profile of each duplex allowed us to perform all the duplex real-time PCR assays simultaneously.
The assay takes about 2 h as most “home-made” assays described in the literature [8], [11], [13], [19], [27], whereas many commercial assays can be performed in 1 h or even less [28].
Regarding the cost of reagents (without the cost of the extraction) it can be calculated to be around 18 US dollars per sample to detect 12 different respiratory viruses while the cost of each duplex-PCR is about 3 US dollars. The costs of commercial assays may vary from about 10 US dollars to about 120 US dollars [15], [17], [29], [30]. However, the costs are strongly dependent on the number of target viruses.
The use of the melting temperature to distinguish the amplicons produced in each duplex allows to identify small size amplicons better than by the electrophoretic analysis.
In addition, the analysis of the melting temperature allows to partially distinguish between hRVs and EVs. In fact in 4 clinical samples the virus could be identified as hRV because of its melting temperature lower than 83.5 °C; in 6 clinical samples, instead, the virus showed a melting temperature slightly higher than 85.5 °C, suggesting a positivity for EVs. All these results were confirmed by sequence analysis. However the melting temperature of the viruses detected in 7 other clinical samples was between 83.5 °C and 85 °C, in a temperature range that does not allow to distinguish between the 2 species. In these cases the sequencing results showed that these samples were positive for hRVs.
A total of 70 out of 202 clinical samples from 55 of 157 patients (30 children and 25 adults) were positive for 1 or more viruses. Since the respiratory samples of adult patients were collected mainly during an influenza epidemic seasons, the influenza A viruses were the most frequently detected respiratory viruses followed by hRV. With reference to ICU patients, influenza viruses were demonstrable in both upper and lower respiratory tract samples, while AdV was detected only in BAL and PIV2 and hMPV only in throat swab. In contrast, hRSV was the most common virus detected in children, followed by EVs, influenza A virus and CoV NL63. However, the small number of samples/patients does not allow us to draw any epidemiological conclusion. Altogether, the number of positive samples for non-influenza respiratory viruses here observed was small as it could have been expected considering the relatively small number of examined samples.
The results reported in this study suggest that this assay could be particularly useful to identify the main respiratory viruses directly, after nucleic acid extraction, from clinical samples and also to screen a large number of patients for epidemiological studies.
In conclusion, the main positive aspect of this test is its cost effective, good technical outcome in terms of sensitivity, specificity, reproducibility and running time. In addition, its flexibility, allows the analysis of respiratory samples from 1 to 12 viral targets depending on the clinical needs.
However, the analysis of a larger number of samples should allow to have positive results for each target virus and strengthen the evidence that this assay performs equally well for all clinical samples.
Acknowledgements
This work was partially supported by a grant from the Foundation “Istituto di Ricerca Virologica Oretta Bartolomei Corsi”, Florence, Italy, and from the Tuscany Region (SETTORE SERVIZI DI PREVENZIONE IN SANITA' PUBBLICA E VETERINARIA).
Contributor Information
Rosaria Arvia, Email: rosaria.arvia@libero.it.
Fabiana Corcioli, Email: f.corcioli@virgilio.it.
Nunziata Ciccone, Email: cicconen@aou-careggi.toscana.it.
Nunzia Della Malva, Email: nunziadellamalva@yahoo.it.
Alberta Azzi, Email: alberta.azzi@unifi.it.
References
- 1.Williams B.G., Gouws E., Boschi-Pinto C., Bryce J., Dye C. Estimates of world wide distribution of child deaths from acute respiratory infections. Lancet Infect. Dis. 2002;2:25–32. doi: 10.1016/s1473-3099(01)00170-0. [DOI] [PubMed] [Google Scholar]
- 2.Choi S.H., Hong S.B., Ko G.B., Lee Y., Park H.J., Park S.Y., Moon S.M., Cho O.H., Park K.H., Chong Y.P., Kim S.H., Huh J.W., Sung H., Do K.H., Lee S.O., Kim M.N., Jeong J.Y., Lim C.M., Kim Y.S., Woo J.H., Koh Y. Viral infection in patients with severe pneumonia requiring intensive care unit admission. AJRCCM. 2012;186:325–332. doi: 10.1164/rccm.201112-2240OC. [DOI] [PubMed] [Google Scholar]
- 3.Sinaniotis C.A. Viral pneumonia in children: incidence and aetiology. Paediatr. Respir. Rev. 2004;5:197–200. doi: 10.1016/s1526-0542(04)90037-1. [DOI] [PubMed] [Google Scholar]
- 4.Perez C.M.B. Prevalence of viral pathogens among pediatric patients admitted for pneumonia in a local tertiary hospital. PIDSP J. 2012;13(no. 1) [Google Scholar]
- 5.Vallieres E., Renaud C. Clinical and economical impact of multiplex respiratory virus assays. Diagn Microbiol. Infect. Dis. 2013;76:255–261. doi: 10.1016/j.diagmicrobio.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng X., Todd K.M., Yen-Lieberman B., Kaul K., Mangold K., Shulman S.T. Unique finding of a 2009 H1N1 influenza virus-positive clinical sample suggests matrix gene sequence variation. J. Clin. Microbiol. 2010;48:665–666. doi: 10.1128/JCM.02318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho C.H., Lee C.K., Nam M.H., Yoon S.Y., Lim C.S., Cho Y., Kim Y.K. Evaluation of the AdvanSure™ real-time RT-PCR compared with culture and Seeplex RV15 for simultaneous detection of respiratory viruses. Diagnostic Microbiol. Infect. Dis. 2014;79:14–18. doi: 10.1016/j.diagmicrobio.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellau-Pujol S., Vabret A., Legrand L., Dina J., Gouarin S., Petitjean-Lecherbonnier J., Pozzetto B., Ginevra C., Freymuth F. Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J. Virol. Methods. 2005;126:53–63. doi: 10.1016/j.jviromet.2005.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahony J.B. Detection of respiratory viruses by molecular methods. Clin. Microbiol. Rev. 2008;21:716–747. doi: 10.1128/CMR.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caliendo A.M. Multiplex PCR and emerging technologies for the detection of respiratory pathogens. Clin. Infect. Dis. 2011;52(Suppl 4):S326–S330. doi: 10.1093/cid/cir047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bharaj P., Sullender W.M., Kabra S.K., Mani K., Cherian J., Tyagi V., Chahar H.S., Kaushik S., Dar L., Broor S. Respiratory viral infections detected by multiplex PCR among pediatric patients with lower respiratory tract infections seen at an urban hospital in Delhi from 2005 to 2007. Virol. J. 2009;6:89. doi: 10.1186/1743-422X-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brittain-Long R., Westin J., Olofsson S., Lindh M., Andersson L.M. Prospective evaluation of a novel multiplex real-time PCR assay for detection of fifteen respiratory pathogens-duration of symptoms significantly affects detection rate. J. Clin. Virol. 2010;47:263–267. doi: 10.1016/j.jcv.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahony J., Chong S., Merante F., Yaghoubian S., Sinha T., Lisle C., Janeczko R. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J. Clin. Microbiol. 2007;45:2965–2970. doi: 10.1128/JCM.02436-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pabbaraju K., Tokaryk K.L., Wong S., Fox J.D. Comparison of the Luminex xTAG respiratory viral panel with in-house nucleic acid amplification tests for diagnosis of respiratory virus infections. J. Clin. Microbiol. 2008;46:3056–3062. doi: 10.1128/JCM.00878-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bierbaum S., Forster J., Berner R., Rücker G., Rohde G., Neumann-Haefelin D., Panning M. CAPNETZ study group. Detection of respiratory viruses using a multiplex real-time PCR assay in Germany, 2009/10. Arch. Virol. 2013;159:669–676. doi: 10.1007/s00705-013-1876-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim H., Hur M., Moon H.W., Yun Y.M., Cho H.C. Comparison of two multiplex PCR assays for the detection of respiratory viral infections. Clin. Respir. J. 2013 doi: 10.1111/crj.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pretorius M.A., Madhi S.A., Cohen C., Naidoo D., Groome M., Moyes J., Buys A., Walaza S., Dawood H., Chhagan M., Haffjee S., Kahn K., Puren A., Venter M. Respiratory viral coinfections identified by a 10-Plex real-time reverse-transcription polymerase chain reaction assay in patients hospitalized with severe acute respiratory illness South Africa, 2009–2010. J. Infect. Dis. 2012;206:159–165. doi: 10.1093/infdis/jis538. [DOI] [PubMed] [Google Scholar]
- 18.Coiras M.T., Aguilar J.C., García M.L., Casas I., Pérez-Breña P. Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested-PCR assays. J. Med. Virol. 2004;72:484–495. doi: 10.1002/jmv.20008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choudhary M.L., Anand S.P., Heydari M., Rane G., Potdar V.A., Chadha M.S., Mishra A.C. Development of a multiplex one step RT-PCR that detects eighteen respiratory viruses in clinical specimens and comparison with real time RT-PCR. J. Virol. Methods. 2013;189:15–19. doi: 10.1016/j.jviromet.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.CDC . CDC; Atlanta, GA: 2008. 510(k) Summary for Centers for Disease Control and Prevention Human Influenza Virus Real-time RT-PCR Detection and Characterization Panel. [Google Scholar]
- 21.Kuypers J., Wright N., Ferrenberg J., Huang M.L., Cent A., Corey L., Morrow R. Comparison of real-time PCR assays with fluorescent-antibody assays for diagnosis of respiratory virus infections in children. J. Clin. Microbiol. 2006;44:2382–2388. doi: 10.1128/JCM.00216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuypers J., Martin E.T., Heugel J., Wright N., Morrow R., Englund J.A. Clinical disease in children associated with newly described coronavirus subtypes. Pediatrics. 2007;119:70–76. doi: 10.1542/peds.2006-1406. [DOI] [PubMed] [Google Scholar]
- 23.Deffernez C., Wunderli W., Thomas Y., Yerly S., Perrin L., Kaiser L. Amplicon sequencing and improved detection of human rhinovirus in respiratory samples. J. Clin. Microbiol. 2004;42:3212–3218. doi: 10.1128/JCM.42.7.3212-3218.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuypers J., Wright N., Morrow R. Evaluation of quantitative and type-specific real-time RT-PCR assays for detection of respiratory syncytial virus in respiratory specimens from children. J. Clin. Virol. 2004;31:123–129. doi: 10.1016/j.jcv.2004.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casas I., Avellon A., Mosquera M., Jabado O., Echevarria J.E., Campos R.H., Rewers M., Perez-Breña P., Lipkin W.I., Palacios G. Molecular identification of adenoviruses in clinical samples by analyzing a partial hexon genomic region. J. Clin. Microbiol. 2005;43:6176–6182. doi: 10.1128/JCM.43.12.6176-6182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bezerra P.G., Britto M.C., Correia J.B., Duarte Mdo C., Fonceca A.M., Rose K., Hopkins M.J., Cuevas L.E., McNamara P.S. Viral and atypical bacterial detection in acute respiratory infection in children under five years. PLoS One. 2011, 18;6(4):e18928. doi: 10.1371/journal.pone.0018928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chi H.C., Bayarjavkhlan C., Chang K.L., Myung H.N., Soo Y.Y., Chae S.L., Yunjung C., Young K.K. Evaluation of a novel real-time RT-PCR using TOCE technology compared with culture and Seeplex RV15 for simultaneous detection of respiratory viruses. J. Clin. Virol. 2013;57:338–342. doi: 10.1016/j.jcv.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piralla A., Lunghi G., Percivalle E., Vigano C., Nasta T., Pugni L., Mosca F., Stronati M., Torresani E., Baldanti F. FilmArray® respiratory panel performance in respiratory samples from neonatal care units. Diagn Microbiol. Infect. Dis. 2014;79:183–186. doi: 10.1016/j.diagmicrobio.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rand K.H., Rampersaud H., Houck H.J. Comparison of two multiplex methods for detection of respiratory viruses: filmarray RP and xTAG RVP. J. Clin. Microbiol. 2011;49:2449–2453. doi: 10.1128/JCM.02582-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin L., Shunxiang Q., Chen Z., Xiumei H., Hongwei S., Mengjie Y., Ji W., Miao W., Wenbo X., Xuejun M. A Two-Tube Multiplex Reverse Transcription PCR Assay for Simultaneous Detection of Sixteen Human Respiratory Virus Types/Subtypes. Biomed. Res. Int. 2013;2013:327620. doi: 10.1155/2013/327620. [DOI] [PMC free article] [PubMed] [Google Scholar]