Abstract
Versatile, simple and efficient sample preparation is desirable for point-of-care testing of emerging diseases such as zoonoses, but current sample preparation assays are insensitive, labour-intensive and time-consuming and require multiple instruments. We developed a single-tube sample preparation approach involving direct pathogen enrichment and extraction from human specimens using diatomaceous earth (DE). Amine-modified DE was used to directly enrich a zoonotic pathogen, Brucella, in a large sample volume. Next, a complex of amine-modified DE and dimethyl suberimidate was used for nucleic acid extraction from the enriched pathogen. Using our single-tube approach, the pathogen can be enriched and extracted within 60 min at a level of 1 colony formation unit (CFU) from a 1 ml sample volume in the same tube. The performance of this approach is 10–100 times better than that of a commercial kit (102 to 103 CFU/ml) but does not require a large centrifuge. Finally, we combined the single-tube approach with a bio-optical sensor for rapid and accurate zoonotic pathogen detection in human urine samples. Using the combination system, Brucella in human urine can be efficiently enriched (~ 8-fold) and the detection limit is enhanced by up to 100 times (1 CFU/ml bacteria in urine) compared with the commercial kit. This combined system is fast and highly sensitive and thus represents a promising approach for disease diagnosis in the clinical setting.
Keywords: Bio-sensing, In vitro diagnostics, Sample enrichment and extraction, Single tube approach, Zoonosis
Highlights
-
•
A single-tube sample preparation approach involving direct pathogen enrichment and extraction.
-
•
Pathogen can be enriched and extracted within 60 min at a level of 1 colony.
-
•
Combination of the single-tube approach with a bio-optical sensor for rapid and accurate zoonotic pathogen detection.
-
•
This system can be efficiently enriched (~ 8-fold) and the detection limit is enhanced by up to 100 times.
1. Introduction
In the past decade, due to the increased emergence of zoonotic diseases such as the highly pathogenic viral infections H7N9 avian influenza, Zika, Ebola, middle east respiratory syndrome-coronavirus, and highly contagious infections Brucellosis, Bartonellosis in both human and animal, attention has turned to the rapid, reliable and straightforward enrichment and identification of pathogens (Ai et al., 2016, Baker and Gray, 2009, Carroll et al., 2015, Durai et al., 2015, Kang et al., 2012, Mäkelä et al., 1998, Nilsson and Mandenius, 1994, Xavier et al., 2010). In addition, pharmaceutical companies are constantly expanding to meet the needs of patients worldwide, with some research focussing on the detection of zoonotic pathogens (Mackey and Liang, 2013). Several detection methods have been developed, including the classic microbial culture and improved techniques such as immunological assays and biochemical kits (Boehme et al., 2010, Islam et al., 2004, Kinjo et al., 2005, Silk and Donnelly, 1997). However, most of these methods suffer from several drawbacks—they are time-consuming, labour-intensive, insensitive and expensive—that greatly limit their commercialization (Ivnitski et al., 1999). Because low-cost, mass-produced and adequately sensitive commercial products play a crucial role in global disease surveillance, especially in developing countries, new solutions that are simple, rapid, sensitive, reliable and cheap are vital to detect pathogens in the clinical setting (Daar et al., 2002).
To overcome these issues as they relate to the diagnosis of zoonotic diseases, a novel approach is desired that can efficiently enrich and extract pathogens on-site, particularly for point-of-care diagnostics, which are vital for disease surveillance (Chin et al., 2012). Traditional pathogen enrichment methods are based on density gradient centrifugation and cell culture, which are still used in clinical analysis (Dorn et al., 1976, Hanff et al., 1984, Lagier et al., 2015). Several promising methods have been developed to capture pathogens in a small sample volume, such as antibody-, lectin- and antimicrobial peptide-based biological methods (Bicart-See et al., 2016, Chairatana and Nolan, 2014, Pal et al., 2017, Russo et al., 2014). A small sample amount (a few hundred microliters) is used in conventional assays bcause of their intrinsic limitations. This decreases detection sensitivity by excluding the pathogens in the remaining sample. Therefore, a new assay that can use the entire sample and subsequently enhance sensitivity is desired. Regarding detection, nucleic acid extraction methods can be integrated with the enrichment step to remove various constraints, including contamination, pathogen loss and spread, and the need for large instruments during the reaction. One report has focused on pathogen enrichment and in situ DNA extraction in a tube (Won et al., 2010). However, the commercialization of this method is challenging due to its complex fabrication process and the high-cost of raw materials.
Here, we report a simple and low-cost single-tube method for the diagnosis of zoonotic disease that involves pathogen enrichment from a large sample volume and on-site nucleic acid extraction. Our approach is the first to use diatomaceous earth (DE) as a principal substance for diagnostic purposes. DE is a natural, low-cost and bio-comfortable silica used in diverse applications such as light harvesting, photonics, sensing and drug delivery (Ehrlich et al., 2010, Rosi et al., 2004). DE promises high-performance in many research areas because of its large surface/volume ratio and strong adsorption capacity when combined with other materials such as nano-silicon anode and grapheme (Campbell et al., 2016, Chen et al., 2016, Losic et al., 2009, Melzak et al., 1996, Qian et al., 2016). To enable DE to be used for diagnostic purposes, pure DE was amino-functionalized by 3-aminopropyltriethoxysilane (APTES), which directly contributes to pathogen enrichment via electrostatic absorption. Additionally, to obviate the need for a chaotropic agent for nucleic acid extraction, dimethyl suberimidate (DMS) was used as a novel non-chaotropic agent for DNA extraction due to the reversible crosslinking reaction between DNA fragments and the amine groups of APTES-functionalized DE (termed APTES-DE).
Brucella, one of the major global zoonotic pathogens that causes both huge financial losses and high human morbidity (Boschiroli et al., 2001), is used to assay the performance of this approach. Incidence rate of Brucella in specific occupations and endemic regions was much higher than in the general population. The annual incidence rates per million populations. Incidence rates in Saudi Arabia (214.4), Turkey (262.2), and Syria (1603.4), had been reported (Pappas et al., 2006; WHO, 1997). There were occupations with high incidence rate of Brucella; butchers and abattoir workers (12.7), laboratory personnel (3.1) and veterinarians (53.2) (Lytras et al., 2016). Using our single-tube approach, the pathogen can be enriched and extracted within 60 min at a level of 1 colony formation unit (CFU) from a 1 ml sample volume in the same tube. The performance of this approach is 10–100 times better than that of a commercial kit, but does not require a large centrifuge. Additionally, for the rapid diagnosis of zoonotic pathogens, we combined this method with a label-free, real-time, fast and highly sensitive isothermal solid-phase amplification/detection (ISAD) system that has been developed in our report (Shin et al., 2013). Thus, the target DNA obtained from the single-tube approach was detected within 20 min by label-free and real-time monitoring of the optical wavelength shift due to the changing reflection index of a silicon microring resonator. Using the combination system, Brucella in human urine can be efficiently enriched (~ 8-fold) and the detection limit is enhanced by up to 100 times compared with the commercial kit. Therefore, the combined system can detect zoonotic pathogens without any sample loss in a total sample volume of 1–2 ml within 80 min.
2. Methods
2.1. Preparation of clean DE and the APTES functionalization process
Commercial DE was washed with distilled water (DW) for 30 min with vigorous stirring. Sediments with impurities were removed after short gravity settling of the mixture. After being washed with 70% and 99.9% ethanol, clean DE was collected by centrifugation. DW was used to prepare 80 ml of 95% ethanol. To form reactive silanol, 2.4 ml of APTES was added into the solution dropwise with stirring for 3 min at room temperature. Then, 500 mg clean DE was dispersed in this solution under vigorous stirring for 4 h. After centrifugation, the precipitates were washed twice with ethanol to remove free silanol. The APTES-functionalized DE was collected via centrifugation and subsequently dried under vacuum overnight at ambient temperature. Finally, the APTES-DE was redispersed into DW at a concentration of 50 mg/ml for subsequent use.
2.2. Enrichment process
The procedure for pathogen enrichment is described in Fig. 2a. First, 50 mg/ml APTES-DE was added into 1 ml DW. Uniformly dispersed suspensions were subsequently formed by vortex. Then, 40 μl of the APTES-DE suspension was pipetted immediately into a 1 ml sample solution. Pathogens were absorbed on the surface of APTES-DE after 40 min incubation in an 850 rpm shaker at room temperature. After being washed with PBS buffer, pathogen-attached APTES-DE was collected via a spin-down device. Then, 200 μl of elution buffer was used to detach pathogens. Thus, pathogens contained in a large volume sample were enriched in 200 μl of elution buffer. DNA was extracted by the Qiagen kit and subjected to fluorescent qRT-PCR analysis. Detailed primer sequences are attached in Supplementary Table 1.
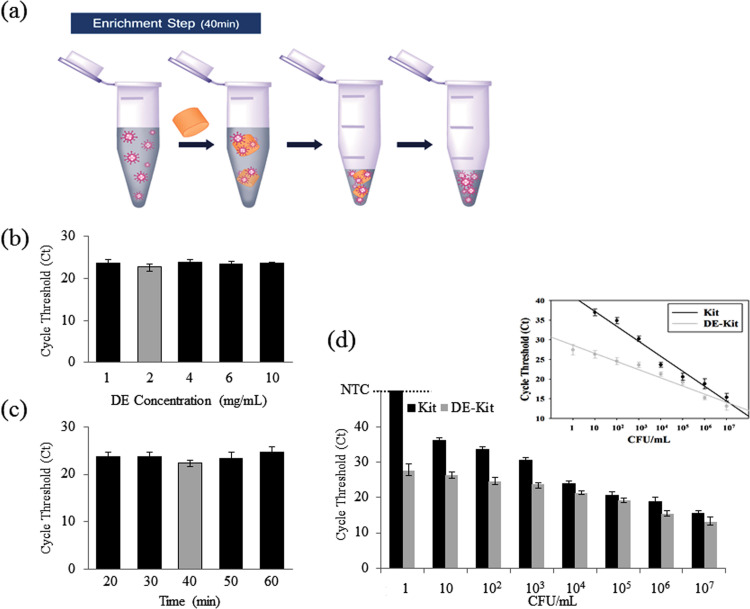
Fig. 2.
Characterization of the pathogen enrichment process using APTES-DE. (a) Experimental pathogen enrichment procedure. (b–c) Optimized protocols for enrichment were demonstrated using qRT-PCR with serially diluted concentrations of DE (b) and at varying incubation times (c). (d) Parallel experiments comparing the performances of the APTES-DE assay and a commercial kit with a series of cell concentrations; inset, linear relationship of both approaches between cycle threshold (Ct) and concentrations of pathogen. Bars extended to a negative control (NTC) line here means no amplification signals.
2.3. DNA extraction process
For DNA extraction from HCT116 cancer cells, 200 μl of sample solution was added into a tube with 20 μl of protease K and 100 μl lysis buffer, mixed gently with a pipette and incubated at room temperature for 1 min. Then, 4 mg/ml DE was added, followed by 100 μl of 100 mg/ml DMS solution. The solution was mixed well, incubated at 56 °C in a thermal shaker at 850 rpm for 30 min and centrifuged with a spin-down device. The supernatant was then removed. The precipitate was washed twice with 200 μl of PBS buffer and all of the solution was mixed by pipetting up and down. Finally, 100 μl of elution buffer was added and the solution was incubated for 1 min at room temperature. It was then centrifuged with a spin-down device and the supernatant was moved to a 1.5 ml tube. The extracted DNA was stored at – 20 °C until use it for downstream analysis.
2.4. Single-tube approach
In this step, 2 mg/ml APTES-DE solution was added into 1 ml of a 105 CFU/ml Brucella sample. The mixture was incubated at ambient temperature for 40 min with gentle shaking to avoid precipitation. It was centrifuged with a spin-down device and the supernatant was removed. The precipitate was washed with 1 ml DW twice and all the solution was mixed by pipetting up and down. Then, 100 μl of elution buffer was added, the solution was incubated for 1 min, and 150 μl of lysis buffer was added with 20 μl of protease K and the mixture was gently pipetted up and down ten times. Another 4 mg/ml APTES-DE solution was again added, followed by 100 μl of 100 mg/ml DMS. After being gently pipetted up and down ten times, the mixture was incubated for 20 min at 56 °C on a thermal shaker at 850 rpm. It was centrifuged with a spin-down device and the supernatant was removed. It was washed twice with 200 μl of PBS buffer and the solution was mixed by pipetting. Finally, 100 μl of elution buffer was added and the solution was incubated for 1 min at room temperature. After spin-down, the supernatant was moved to a 1.5 ml tube. The extracted DNA was stored at – 20 °C until use.
2.5. ISAD system assay
The ISAD system assay process was simply modified from the previously reported method (Shin et al., 2013; Shin et al., 2015a, Shin et al., 2015b). The silicon microring chip was treated with oxygen plasma for 10 min and subsequently incubated in a fresh solution of 2% APTES in ethanol (95%) for 2 h. After being washed with ethanol (99.9%) and deionized (DI) water in turn, the chip was dried in an oven at 120 °C for 15 min. Next, the chip was incubated again in fresh solution containing 2.5% glutaraldehyde and 5 mM sodium cyanoborohydride for another 1 h, followed by a DI water wash. Next, we immobilized the ISAD forward primer of Brucella on the chip by incubating it in PBS (pH 7.4, 1 ×) containing 1 μM forward primer and 5 mM sodium cyanoborohydride at room temperature overnight. Detailed primer sequences are attached in Supplementary Table 1. After being washed with PBS and dried by nitrogen, an acrylic well (6 mm in length, 1.5 mm in width and 1 mm in height) was pasted as a reaction chamber onto the silicon microring area of the chip. The fabricated chip then underwent optical measurement. To determine the optimal conditions, a mixture of 29.5 μl of rehydration buffer, 15 μl of nuclease-free water and 2.5 μl of reverse primers were injected immediately into a vial containing dried enzyme. Then, 2.5 μl of magnesium acetate solution was injected with 15 s vortexing steps. Next, 5 μl of the sample (target DNA or negative control) and 10 μl of the mixture were added into the acrylic well and covered with 10 μl mineral oil. All of the above operations were performed at room temperature. Finally, the chip was placed on a thermoelectric cooler and maintained at 38 °C under an incident laser. The resonance signal from the refracted light was immediately recorded and used as a reference to obtain a baseline. Thereafter, the amplification of target DNA was monitored in real time by recording the wavelength shift every 3 min for up to 20 min.
3. Results
3.1. Diatom earth as a novel substance for diagnostic purposes
Purified DE mainly presents a single morphology, a micro-scale hollow in the centre with numerous nano-scale pores in the wall, approximately 3 µm and 200 nm in size, respectively (Supplementary Fig. 1). This regularly ordered nanoporous structure offers a strong physical absorption property and substantial reaction area (Yang et al., 2011). In addition, there are abundant hydroxyl groups on its surface, which allow its chemical modification and subsequent use in biological applications, such as biotechnology and biomimetics (Rosi et al., 2004, Townley et al., 2008). Here, APTES is selected not only for its robust coating of saline due to covalent bond formation, but also for its chemical stability compared with alkane thiols, which can be oxidized. This means that APTES-DE allows long-term storage under standard conditions. Washed and dried APTES-DE is stored in a tube at room temperature and redispersed in distilled water at 50 mg/ml for experimental use. This process is critical because it is the foundation of all subsequent experiments.
Fourier transform infrared spectroscopy (FTIR) is used to confirm the APTES functionalization process. The samples are washed three more times to adequately remove impurities and any free APTES. The FTIR spectra of plain DE and APTES-DE are presented in Supplementary Fig. S2. The absorption peak at 1072 cm−1 is ascribed to the asymmetric stretching vibrations of Si-O-Si, whereas the absorption peaks at 3414 and 794 cm−1 are attributed to Si-OH on the surface of DE. On the other hand, after the APTES modification, new absorption peaks appear at 3663, 2987, 2901, 1649, 1406, 1251, 1228 and 893 cm−1. The peaks at 3663, 1649 and 893 cm−1 are attributed to N-H stretching, in-plane bending and out-of-plane bending vibrations, respectively. These results confirm the modification of amine groups on the surface of DE. Meanwhile, the peaks observed at 2987 and 2901 cm−1 can be assigned to C-H stretching vibrations and those at 1251 and 1228 cm−1 to C-N stretching vibrations due to amine groups directly bonded to the DE. In addition, the surface morphology of DE is changed after APTES treatment, as shown in Fig. S1 d. The pore sizes of APTES-DE are significantly reduced due to the self-polymerization of APTES. These characteristics provide direct evidence for the efficient amine functionalization of DE by APTES, which means that APTES-DE can be used in biological applications.
3.2. Principle of the single-tube approach for pathogen enrichment and extraction
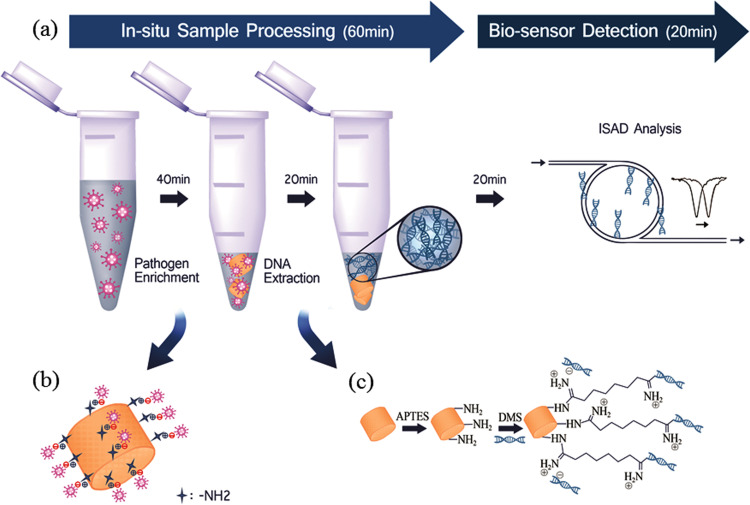
The principle of the single-tube approach involving DE for the diagnosis of zoonotic pathogens is presented in Fig. 1a. We report a single-tube approach for the enrichment and extraction of pathogens using APTES-DE. APTES-DE efficiently concentrates pathogens on its surface via both electrostatic and physical absorption (Fig. 1b). SEM images clearly show Brucella enriched on the surface of APTES-DE within 40 min (Fig. S1e,f). Next, DMS is combined with APTES-DE for the extraction of high-quality nucleic acids. This acts as a novel capture reagent that recognizes the amine groups on the sticky ends of nucleic acid fragments and enables positively charged amidine linkage that further enhances the isolation of negatively charged DNA via the electrostatic binding of anionic DNA and cationic amidine (Fig. 1c). Thus, DNA is extracted from the enriched pathogen via crosslinking reactions between the amines of the DMS/DNA complex and APTES-DE within 20 min. For diagnostic purposes, we added the ISAD system for the rapid detection of the pathogen DNA obtained from the single-tube approach. In this system, the specific target primer is directly immobilized on the surface of a silicon microring sensor device, and the DNA amplification is guided on-site, which changes the reflective index. When a laser light passes through the sensor, a shifted wavelength triggered by the amplified DNA is observed. This resonance optical signal is specifically related to the DNA concentration of the detected sample (Shin et al., 2013; Shin et al., 2015a, Shin et al., 2015b). Therefore, using this combination single-tube approach and ISAD system, zoonotic pathogens can be rapidly and simply detected within 80 min with high sensitivity Fig. 1).
Fig. 1.
Schematic diagram of rapid pathogen diagnostic system using a single-tube approach. (a) In situ sample processing involves pathogen enrichment by APTES-modified DE (diagram) within 40 min and DNA extraction by DMS (dimethyl suberimidate)-assisted APTES-DE (diagram) within 20 min. The ISAD biosensor is then used to detect pathogen obtained from the in situ sample processing within 20 min (b) Pathogen enriched via electrostatic interaction. (c) DNA extraction by DMS (dimethyl suberimidate)-assisted APTES-DE. This novel system can be used for the rapid, simple and sensitive diagnosis of pathogens.
3.3. Characterization of pathogen enrichment using APTES-DE
Efficient pathogen enrichment in a large sample volume (1 ml or 2 ml) is always challenging, particularly without expensive biological agents such as antibodies or biotin (Jing et al., 2013). The gold standard method for the diagnosis and successful isolation of pathogens was determined to be a solution-based pathogen culture method, especially for the diagnosis of brucellosis, an emerging zoonotic pathogen, because its clinical diagnosis is extremely difficult due to its broad spectrum of nonspecific clinical manifestations (Bosilkovski et al., 2010b). Blood culture is the classic method for brucellosis diagnosis but it has limited sensitivity and requires a long cultivation period (Bosilkovski et al., 2010a, Rich et al., 2000). In addition, only a small amount of sample (200 μl out of 1 or 2 ml) is used in conventional assays because of their intrinsic limitations, or an ultra-centrifugation is used for sample pre-concentration in large volume of samples. To overcome the limitations of these assays, APTES-DE was used for pathogen enrichment in larger sample volumes. Due to the low isoelectric point of Brucella (Geresu and Kassa, 2016, Saha et al., 1990), a non-covalent approach, namely, electrostatic binding, can be used for enrichment of the pathogen to the surface of DE. Because APTES-DE generates a positive charge due to functionalized amino groups in the reaction buffer, the pathogen can be electrically driven onto the surface of APTES-DE. The strong physical absorption property of DE also boosts the pathogen enrichment process. Thus, APTES-DE can efficiently enrich the pathogenic bacteria Fig. 2 and Fig. S1 e,f). For optimal enrichment, we assessed the optimal DE concentration and incubation time (Fig. 2b,c). Based on our quantitative real-time PCR (qRT-PCR) results, with a lower cycle threshold (Ct) from qRT-PCR reflecting higher amounts of targeted DNA (Heid et al., 1996), 2 mg/ml DE and a 40-min incubation time were the optimal conditions. To test the usefulness of APTES-DE as a tube-based enrichment assay, we used serially diluted pathogen amounts in 1 ml solutions. Using the optimal conditions, we first enriched the pathogen from a 1 ml solution using APTES-DE, and then the enriched pathogen was eluted in a 200 μl solution for DNA extraction using a Qiagen kit. Finally, qRT-PCR was performed to confirm the amount of DNA in the samples. Fig. 2d shows that the APTES-DE assay in a 1 ml sample can useful for the enrichment of pathogen from 1 CFU. In addition, to compare the utility of APTES-DE and the Qiagen kit, 200 μl out of the 1 ml sample was used for direct DNA extraction without any enrichment step using the Qiagen kit. The detection limit of the Qiagen kit was 10 CFU (Fig. 2d). The Ct from the qRT-PCR in the enriched samples using APTES-DE was much lower than in the initial samples. The inset shows the linear relationship of Ct as a function of the diverse pathogen concentrations of both assays. We further assessed the performance of this enrichment procedure at low pathogen concentrations in 2 ml samples (Fig. S3). After APTES-DE enrichment, the Ct values from the qRT-PCR were still much lower than those in the kit. The sensitivity of the APTES-DE assay was found to be 10-times higher than that of the kit method. In addition, our proposed assay does not require any ultra centrifugation for pathogen enrichment in a large sample volume. Therefore, APTES-DE in a single tube can be a useful tool for the effective enrichment of low-concentration pathogens in large volume samples within 40 min.
3.4. Characterization of DNA extraction using a complex of APTES-DE and DMS
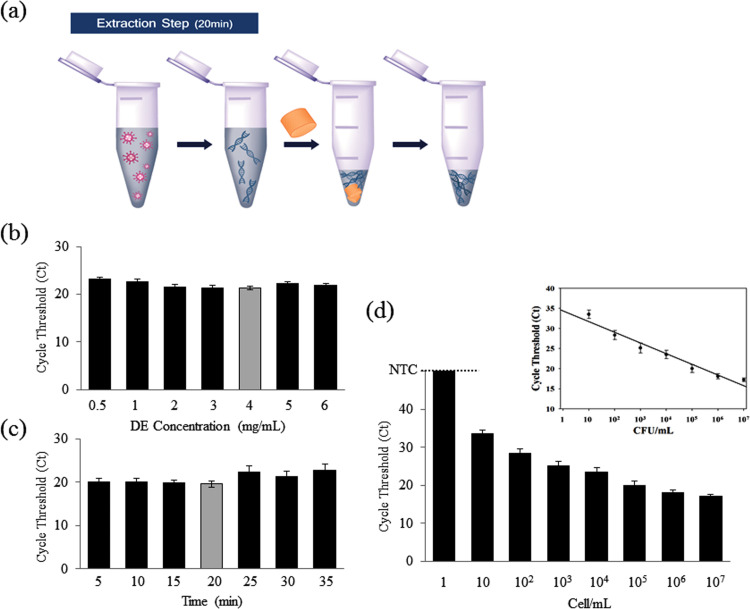
Next, to simplify the reaction step, we developed a DNA extraction assay to be performed in the same tube after the enrichment reaction ( Fig. 3a). First, 200 μl of sample solution was mixed with 100 μl of lysis buffer and 20 μl of protease K in a 1.5 ml tube. Then, 40 μl of APTES-DE and 100 μl of DMS were added in that order. The mixture was subsequently incubated for 10 min in a shaker at 850 rpm and 56 °C. During the reaction, DNA and DMS complexes bound to APTES-DE and were collected via a spin-down device. After being washed with PBS buffer, DNA was eluted via the addition of 100 μl of elution buffer and pipette mixing and spin-down (Fig. 3a). We report the DNA extraction using APTES-DE and the non-chaotropic agent DMS. DMS is selected here not only for the specific binding process, but also because the binding is reversible at high pH (Shin et al., 2015b). To determine the optimal conditions for DNA extraction, we assessed the optimal DE concentration and incubation time (Fig. 3b,c). Based on the qRT-PCR results, 4 mg/ml of APTES-DE and a 20 min incubation time were the optimal conditions. Using these conditions, the performance of APTES-DE as a DNA extraction method was assessed with serially diluted samples containing from 1 to 107 cells/ml of pathogen. A linear relationship between the Ct and cell concentration was visible (Fig. 3, inset). This extraction process detects up to 10 cells/ml with a high linear correlation coefficient (Fig. 3d and R 2 = 0.9898). What's more, we evaluated the performance of APTES-DE by comparing with various silica matrixes (i.e., silica gel, silica sand). The qRT-PCR results indicated that APTES-DE was better candidate to compare with others (Fig. S4). Therefore, this combination of APTES-DE and DMS can act as a novel DNA extraction tool via the assistance of a non-chaotropic agent.
Fig. 3.
Characterization of the DNA extraction process using APTES-DE. (a) Experimental DNA extraction procedure. (b–c) Optimized protocols for DNA extraction were demonstrated using qRT-PCR with serially diluted concentrations of DE (b) and at varying incubation times (c). (d) Performance of the DNA extraction assay using APTES-DE with a series of cell concentrations 1–107 cells/ml); inset, linear relationship of the cycle threshold (Ct) as a function of the serial cell concentration. Bars extended to a negative control (NTC) line here means no amplification signals.
3.5. Single-tube approach for pathogen enrichment and extraction
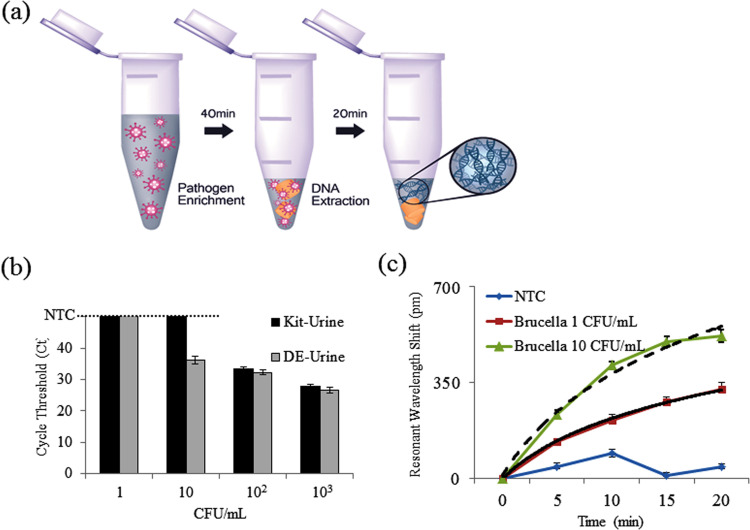
Because APTES-DE is used as the principal substance for both pathogen enrichment and DNA extraction, as detailed above, we combined the two processes in a single tube ( Fig. 4). Brucella in 1 ml of PBS buffer was first enriched by APTES-DE. Then, lysis buffer was directly added to the tube, followed by DMS. Next, we tested the DNA obtained from the single-tube approach using qRT-PCR (Fig. S5). We confirmed the ability of the single-tube approach to detect pathogen in samples containing as little as 10 CFU/ml. We then examined whether this approach can be useful for the diagnosis of zoonotic pathogens in human specimens. As a systemic infection, human brucellosis appears a wide clinical spectrum while Infections of the genitourinary system are the second most common complication. It was reported that for Brucella ovis diagnosis, preputial wash samples and urine were suitable specimens (Xavier et al., 2010). Considering of the convenience, we selected urine samples as main matrix in this study. We used spiked Brucella in human urine samples at 102 CFU/ml. Brucella was detected two cycles sooner with qRT-PCR analysis via the single-tube approach than with the Qiagen kit (Fig. S6a), which means an ~8-fold concentration increase (He et al., 2002, Heid et al., 1996). In addition, conventional PCR analysis also indicated that high-quality DNA was extracted via the single-tube approach (due to the presence of a clear target band) (Fig. S6b). We also evaluated our system using spiked Brucella in animal urines (ram, dog). Similar performance was shown in Fig. S7a. Since this single-tube approach is based on electrostatic interaction for pathogen enrichment, the selectivity of this system is not based on the enrichment step but the amplification step with the specific primer. In order to test this capability of the system, we used the samples of mixed pathogens including Brucella, Salmonella, E.coli (Fig. S7 b). When PCR was used to amplify Salmonella, the target primers could be amplified the target pathogen only. Also, when PCR was used to amplify E.coli, the primers could be amplified the target only. Fig. S7 b presented that target specific primers could only amplify the target pathogen's DNA. We further investigated this single-tube approach using low-concentration samples ranging from 1 to 103 CFU/ml of Brucella in urine. The Ct value of extracted DNA by qRT-PCT in this single-tube approach was much lower than that of the Qiagen kit (Fig. 4b). In addition, the performance limit was 10 times higher at 10 CFU/ml than that of the Qiagen kit (102 CFU/ml). Thus, we confirmed that high levels of high-quality DNA improved the detection limit of the diagnostic system. Finally, due to its highly sensitive nucleic acid detection, we used ISAD to analyze the pathogen obtained from the single-tube approach in urine. Signals from shifted wavelengths can be measured using the ISAD system because the gradually amplified DNA leads to changes in the reflective index. Regular changes are shown in Fig. 4c and are specific to DNA from Brucella due to the selected primers. Even at 1 CFU/ml, we can still obtain a specific response signal via the ISAD system. Because the detection limit of the Qiagen kit is only 102 CFU/ml in urine (Fig. 4b), the limit is thus increased 100 times with the single-tube approach and ISAD system. In addition, the detection time for the ISAD system is only 20 min, whereas both qRT-PCR and conventional PCR need 2–3 h (Fig. 4c).
Fig. 4.
Characterization of the single-tube approach and the combination system with optical sensor. (a) Experimental single-tube approach. (b) Parallel experiments comparing the performance of the commercial kit with that of the single-tube approach for analysis of Brucella in urine using qRT-PCR at low concentrations. (c) Combination system with ISAD sensor. Analysis of pathogenic DNA obtained from the single-tube approach at low concentrations within 80 min (R2 = 0.98515 for 10 CFU/ml and R2 = 0.99872 for 1 CFU/ml). Bars extended to a negative control (NTC) line here means no amplification signals.
4. Conclusion
Despite numerous technological advances in the field of disease diagnostics in recent decades, many scientists, engineers and clinicians desire more appropriate technology for clinical settings. Importantly, detection sensitivity in clinical applications can be improved when high levels of high-quality nucleic acids are obtained from large sample volumes. Thus, the new technology would need to encompass the following features: (I) efficient enrichment of pathogens in a large sample volume without the need for ultracentrifugation, (II) nucleic acid extraction from pathogens without a chaotropic agent, (III) easy integration with the detection system for point-of-care testing, and (IV) rapid and efficient pathogen diagnosis. To achieve these characteristics, we used a single-tube approach that enables efficient pathogen enrichment and extraction for enhanced diagnostic performance. The relevant issues have been innovatively addressed. First, Brucella was directly enriched in a 1–2 ml sample without ultra-centrifugation. Second, DNA was extracted from the enriched pathogen in the same tube without the need for large instruments. High levels of high-quality DNA were captured through DMS-assisted crosslinking reactions. Third, using the single-tube approach, pathogenic Brucella in human urine was efficiently enriched and concentrated by approximately 8-fold. Lastly, a combination of the single-tube approach and a bio-optical sensor enabled rapid (80 min), low-cost and simple analysis of complex samples, as well as ultra-sensitivity (1 CFU/ml). Our method involves the use of a single tube and a battery-driven spin-down device and is thus a promising candidate for point-of-care diagnosis. We envision that this combination system will be useful for other emerging diseases in both humans and animals.
Acknowledgements
This work was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health&Welfare, Republic of Korea (HI16C-0272–010016).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bios.2017.08.027.
Appendix A. Supplementary material
Supplementary material
References
- Ai J.-W., Zhang Y., Zhang W. Emerg. Microbes Infect. 2016;5(3):e21. doi: 10.1038/emi.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker W.S., Gray G.C. J. Am. Vet. Med. Assoc. 2009;234(10):1271–1278. doi: 10.2460/javma.234.10.1271. [DOI] [PubMed] [Google Scholar]
- Bicart-See A., Rottman M., Cartwright M., Seiler B., Gamini N., Rodas M., Penary M., Giordano G., Oswald E., Super M. PloS One. 2016;11(6):e0156287. doi: 10.1371/journal.pone.0156287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehme C.C., Nabeta P., Hillemann D., Nicol M.P., Shenai S., Krapp F., Allen J., Tahirli R., Blakemore R., Rustomjee R. N. Engl. J. Med. 2010;363(11):1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschiroli M.-L., Foulongne V., O'Callaghan D. Curr. Opin. Microb. 2001;4(1):58–64. doi: 10.1016/s1369-5274(00)00165-x. [DOI] [PubMed] [Google Scholar]
- Bosilkovski M., Katerina S., Zaklina S., Ivan V. Comp. Immunol. Microb. Infect. Dis. 2010;33(5):435–442. doi: 10.1016/j.cimid.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Bosilkovski M., Krteva L., Dimzova M., Vidinic I., Sopova Z., Spasovska K. Croat. Med. J. 2010;51(4):327–336. doi: 10.3325/cmj.2010.51.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell B., Ionescu R., Tolchin M., Ahmed K., Favors Z., Bozhilov K.N., Ozkan C.S., Ozkan M. Sci. Rep. 2016;6:33050. doi: 10.1038/srep33050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll M.W., Matthews D.A., Hiscox J.A., Elmore M.J., Pollakis G., Rambaut A., Hewson R., García-Dorival I., Bore J.A., Koundouno R. Nature. 2015;524:97–101. doi: 10.1038/nature14594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chairatana P., Nolan E.M. J. Am. Chem. Soc. 2014;136(38):13267–13276. doi: 10.1021/ja5057906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Li C., Shi L., Gao T., Song X., Bachmatiuk A., Zou Z., Deng B., Ji Q., Ma D. Nat. Comm. 2016;7:13440. doi: 10.1038/ncomms13440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C.D., Linder V., Sia S.K. Lab Chip. 2012;12(12):2118–2134. doi: 10.1039/c2lc21204h. [DOI] [PubMed] [Google Scholar]
- Daar A.S., Thorsteinsdóttir H., Martin D.K., Smith A.C., Nast S., Singer P.A. Nat. Genet. 2002;32(2):229–232. doi: 10.1038/ng1002-229. [DOI] [PubMed] [Google Scholar]
- Dorn G.L., Burson G.G., Haynes J.R. J. Clin. Microb. 1976;3(3):258–263. doi: 10.1128/jcm.3.3.258-263.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durai P., Batool M., Shah M., Choi S. Exp. Mol. Med. 2015;47(8):e181. doi: 10.1038/emm.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich H., Demadis K.D., Pokrovsky O.S., Koutsoukos P.G. Chem. Rev. 2010;110(8):4656–4689. doi: 10.1021/cr900334y. [DOI] [PubMed] [Google Scholar]
- Geresu M.A., Kassa G.M. J. Vet. Sci. Technol. 2016;7(3):1000323. [Google Scholar]
- Hanff P.A., Norris S.J., Lovett M.A., Miller J.N. Sex. Transm. Dis. 1984;11(4):275–286. doi: 10.1097/00007435-198410000-00003. [DOI] [PubMed] [Google Scholar]
- He L., Chinnery P.F., Durham S.E., Blakely E.L., Wardell T.M., Borthwick G.M., Taylor R.W., Turnbull D.M. Nucleic Acids Res. 2002;30(14) doi: 10.1093/nar/gnf067. (e68-e68) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid C.A., Stevens J., Livak K.J., Williams P.M. Genome Res. 1996;6(10):986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Islam F.S., Gault A.G., Boothman C., Polya D.A. Nature. 2004;430(6995):68–71. doi: 10.1038/nature02638. [DOI] [PubMed] [Google Scholar]
- Ivnitski D., Abdel-Hamid I., Atanasov P., Wilkins E. Biosens. Bioelectron. 1999;14(7):599–624. doi: 10.1016/s0956-5663(99)00004-4. [DOI] [PubMed] [Google Scholar]
- Jing W., Zhao W., Liu S., Li L., Tsai C.-T., Fan X., Wu W., Li J., Yang X., Sui G. Anal. Chem. 2013;85(10):5255–5262. doi: 10.1021/ac400590c. [DOI] [PubMed] [Google Scholar]
- Kang D.H., Kim Y.J., Kim S.H., Sun B.J., Kim D.H., Yun S.C., Song J.M., Choo S.J., Chung C.H., Song J.K., Lee J.W., Sohn D.W. N. Engl. J. Med. 2012;366:2466–2473. doi: 10.1056/NEJMoa1112843. [DOI] [PubMed] [Google Scholar]
- Kinjo Y., Wu D., Kim G., Guo-Wen X. Nature. 2005;434(7032):520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- Lagier J.-C., Edouard S., Pagnier I., Mediannikov O., Drancourt M., Raoult D. Clin. Microb. Rev. 2015;28(1):208–236. doi: 10.1128/CMR.00110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losic D., Mitchell J.G., Voelcker N.H. Adv. Mat. 2009;21(29):2947–2958. [Google Scholar]
- Lytras T., Danis K., Dounias G. Int J. Occup. Environ. Med. 2016;7:221–226. doi: 10.15171/ijoem.2016.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey T.K., Liang B.A. Glob. Health. 2013;9(1):45. doi: 10.3402/gha.v6i0.19923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä M.J., Puhakka T., Ruuskanen O., Leinonen M., Saikku P., Kimpimäki M., Blomqvist S., Hyypiä T., Arstila P. J. Clin. Microbiol. 1998;36(2):539–542. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzak K.A., Sherwood C.S., Turner R.F., Haynes C.A. J. Colloid Interface Sci. 1996;181(2):635–644. [Google Scholar]
- Nilsson K., Mandenius C.-F. Nat. Biotech. 1994;12(13):1376–1378. doi: 10.1038/nbt1294-1376. [DOI] [PubMed] [Google Scholar]
- Pal M., Lee S., Kwon D., Hwang J., Lee H., Hwang S., Jeon S. Anal. Chim. Acta. 2017;952:81–87. doi: 10.1016/j.aca.2016.11.041. [DOI] [PubMed] [Google Scholar]
- Qian T., Li J., Deng Y. Sci. Rep. 2016;6:32392. doi: 10.1038/srep32392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas G., Papadimitriou P., Akritidis N., Chistou L., Tsianos E.V. Lancet Infect. Dis. 2006;6:91–99. doi: 10.1016/S1473-3099(06)70382-6. [DOI] [PubMed] [Google Scholar]
- Rich M., Bannatyne R.M., Memish Z.A. J. Clin. Microbiol. 2000;38(4) doi: 10.1128/jcm.38.4.1706-1706.2000. (1706-1706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosi N.L., Thaxton C.S., Mirkin C.A. Angew. Chem. Int. Ed. Engl. 2004;116(41):5616–5619. doi: 10.1002/anie.200460905. [DOI] [PubMed] [Google Scholar]
- Russo L., Taraballi F., Lupo C., Poveda A., Jiménez-Barbero J., Sandri M., Tampieri A., Nicotra F., Cipolla L. Interface Focus. 2014;4(1):20130040. doi: 10.1098/rsfs.2013.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A., Mukhopadhyay N., Dowling J., Ficht T., Adams L., Glew R. Infect. Immune. 1990;58(5):1153–1158. doi: 10.1128/iai.58.5.1153-1158.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y., Lim S.Y., Lee T.Y., Park M.K. Sci. Rep. 2015;5:14127. doi: 10.1038/srep14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y., Perera A.P., Kim K.W., Park M.K. Lab Chip. 2013;13(11):2106–2114. doi: 10.1039/c3lc50129a. [DOI] [PubMed] [Google Scholar]
- Shin Y., Perera A.P., Tang W.Y., Fu D.L., Liu Q., Sheng J.K., Gu Z., Lee T.Y., Barkham T., Park M.K. Biosens. Bioelectron. 2015;68:390–396. doi: 10.1016/j.bios.2015.01.030. [DOI] [PubMed] [Google Scholar]
- Silk T.M., Donnelly C.W. J. Food Prot. 1997;60(12):1483–1486. doi: 10.4315/0362-028X-60.12.1483. [DOI] [PubMed] [Google Scholar]
- Townley H.E., Parker A.R., White-Cooper H. Adv. Funct. Mater. 2008;18(2):369–374. [Google Scholar]
- Won J.Y., Min J., Park J.-H. Biosens. Bioelectron. 2010;26(4):1763–1767. doi: 10.1016/j.bios.2010.08.037. [DOI] [PubMed] [Google Scholar]
- World Health Organization Fact. Sheet. 1997;N173 [Google Scholar]
- Xavier M.N., Silva T.M., Costa É.A., Paixão T.A., Moustacas V.S., Júnior C.A.C., Sant’Anna F.M., Robles C.A., Gouveia A.M., Lage A.P. Vet. Microbiol. 2010;145(1):158–164. doi: 10.1016/j.vetmic.2010.02.037. [DOI] [PubMed] [Google Scholar]
- Yang W., Lopez P.J., Rosengarten G. Analyst. 2011;136(1):42–53. doi: 10.1039/c0an00602e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material