Abstract
Poly(ADP-ribosyl)ation (PARylation), a protein post-translational modification that was originally connected to the DNA damage response, is now known to engage in a continuously increasing number of biological processes. Despite extensive research and ceaseless, important findings about its role and mode of action, poly(ADP-ribose) remains an enigma regarding its structural complexity and diversity. The recent identification and structural characterization of four different poly(ADP-ribose) binding motifs represents a quantum leap in the comprehension of how this molecule can be decoded. Moreover, the recent discovery of a direct connection between PARylation and poly-ubiquitylation in targeting proteins for degradation by the proteasome has paved the way for a new interpretation of this protein modification. These two novel aspects, poly(ADP-ribose) recognition and readout by the ubiquitylation/proteasome system are developed here.
Keywords: Poly(ADP-ribose), NAD+, ubiquitylation, post-translational modifications, macro domains, zinc fingers
The diversity in origin, nature and outcome of PARylation
PARylation is a post-translational modification of proteins that is catalyzed by poly(ADP-ribose) polymerases (PARPs) 1, 2, 3, 4. PARP hydrolyses nicotinamide adenine dinucleotide (NAD+) and transfers the ADP-ribose (ADPR) moiety to acceptor proteins, including itself. The transfer of one ADPR unit leads to mono-ADP-ribosylation, whereas subsequent additions of ADPR units through 2′,1′′-O-glycosidic ribose-ribose bonds can lead to long and linear poly(ADP-ribose) (PAR) polymers; for some PARPs, an even more complex branched structure can be obtained through 2′′,1′′′-glycosidic bonds (Figure 1 ). PARPs form a family of 17 members in humans, all having in common a sequence homologous to the catalytic domain of the founding and most described PARP, PARP-1. Some members lack critical residues for PAR synthesis, and therefore display only mono-ADP-ribosyl-transferase (MART) activity (transfer of a single ADPR unit to acceptor protein), or are completely inactive [5].
Figure 1.
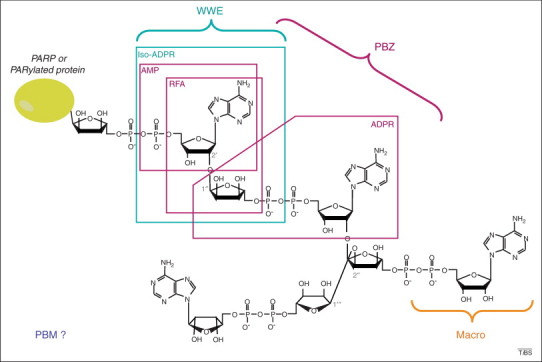
Structure of poly(ADP-ribose) (PAR) and recognition by PAR-binding motifs (PBMs). The chemical structure of branched PAR, covalently attached to an acceptor protein (which can be poly(ADP-ribose) polymerase itself), is shown. The characteristic linkages of linear (1→2′ O-glycosidic bound) and branched (1→2′′ O-glycosidic bound) are indicated. The brackets illustrate the region recognized by each PAR-binding domain. The ADP-ribose (ADPR) derivatives used in structural characterization of PAR-binding domains are framed. The RNF146 PAR-binding domain WWE was co-crystallized with iso-ADPR [46], supporting a mechanism of internal recognition of PAR. The solution structure of the two aprataxin and PNK-like factor (APLF) PAR-binding zinc fingers (PBZ motifs) bound to 2′-O-α-D-ribofuranosyl-adenosine (RFA) was solved by nuclear magnetic resonance (NMR) [38], whereas the CHFR PBZ domain was co-crystallized with AMP and ADPR [41]. This PBZ domain was also co-crystallized with P1P2-diadenosine 5′-pyrophosphate (AMP2), which is not a derivative of PAR and thus cannot be illustrated in this scheme, but which supports the hypothesis that the PBZ motif recognizes two successive ADPR units. Macrodomains bind ADPR units, but capping of the 2′OH from ADP prohibits internal recognition of PAR 14, 24. How the PBMs recognize PAR remains unknown, as illustrated by the open question mark.
PARylation is involved in many processes such as DNA repair, replication, chromatin structure, transcription, telomere homeostasis, chromosome segregation, cell differentiation, cell proliferation or cell death. From a physiological point of view, PARP activity has been directly linked to energy metabolism, spermatogenesis, innate and acquired immunity, biological clock or memory 1, 2, 3, 4. Like any other post-translational modification, PARylation can modify the biochemical and functional properties of the target protein and as such, regulates protein–protein or protein–nucleic acid (both DNA and RNA) interactions. PAR is also receiving increasing attention as a signaling molecule, because it can define the outcome of a cell whose DNA integrity has been injured 6, 7. Another well-established function of PAR is to direct the recruitment of proteins to a defined site. This is particularly well-documented in the DNA damage response, in which PARP-1 rapidly detects DNA breaks through its nick-sensing activity, is activated, and synthesizes PAR, leading to the immediate recruitment of repair factors displaying one of the four known PAR-binding modules to the site of DNA damage (Box 1 ). Finally, much attention has been given in the past decade to investigating PARP inhibitors as an anticancer strategy. PARP-1 inhibition potentiates the killing efficiency of anticancer genotoxic drugs, and is synthetically lethal in tumors that are defective in the homologous recombination repair pathway (e.g. breast cancers with BRCA1 or BRCA2 mutations) 4, 8, 9 or, as recently shown, in Ewing's sarcoma cells [10].
Box 1. Binding to PAR, a mode of recruitment to DNA damage sites.
One of the best characterized functions of PARP-1 is the efficient detection of DNA breaks, triggering the immediate synthesis of PAR on itself (auto-PARylation) and on chromatin-associated proteins such as histones. The role of these PARylations is to promote DNA repair by fostering an appropriate chromatin status and by quickly recruiting factors to restore DNA and chromatin integrity. The base excision and single strand break repair scaffold protein X-ray repair cross-complementing gene 1 XRCC1 is the first of an expanding list of proteins that have been found to be recruited to the DNA damage site through binding to PAR that has been generated by PARP-1. Recognition of PAR can involve any of the four PAR-binding modules. These proteins can be involved in the DNA repair process itself [e.g. XRCC1, DNA ligase III, polynucleotide kinase (PNK), meiotic recombination 11 (Mre11), APLF]; in the structure and remodeling of chromatin to control accessibility of the lesion and surrounding chromatin compaction (e.g. ALC1 and macroH2A1.1); in the repression of transcription [e.g. the nucleosome remodeling and deacetylase (NuRD), the repressor proteins chromodomain 4 (CHD4), metastasis tumor antigen-1 (MTA1) and polycomb proteins Enhancer of Zeste homolog 2 (EZH2), B lymphoma Mo-MLV insertion region 1 (BMI1), chromobox 4 CBX4]; and in the decision process for whether a damaged cell will engage either a cell survival or a cell death program (e.g. RNF146 and AIF). Several recent reviews and articles cover this topic in depth 1, 4, 66, 67, 68, 69.
The first described PAR-binding motif (PBM)
Felix Althaus's group made the seminal discovery of the existence of a dedicated PBM, which is present in a still-growing list of proteins [11] and was recently refined to 8 amino acids with the following pattern: [HKR]1-X2-X3-[AIQVY]4-[KR]5-[KR]6-[AILV]7-[FILPV]8 [12]. In silico prediction of PAR binding proteins using this pattern has established a catalog of proteins with potential PAR-binding capacity; many of which have been confirmed [12]. The function of these PAR-binding proteins covers many biological processes, but some pathways are over-represented, such as DNA repair, DNA metabolism, chromosome organization, RNA metabolism and cell cycle regulation. One prominent example of a DNA repair factor with a PBM is the base excision/single strand break repair factor X-ray repair cross-complementing gene 1 (XRCC1), which is rapidly recruited to sites of DNA damage where PAR has been produced by PARP-1 (Box 1). Even though the PBM was the first PAR-binding domain to be described, and some 3D structures of PBM-containing polypeptides have been solved [i.e. the BRCT1 domain of XRCC1 (PDB 2d8m) or the apoptosis inducing factor AIF] [6], no precise structural information about how PAR is recognized by a PBM is currently available. This is because, in contrast to the three other PAR-binding modules described later, PBMs are not able to bind ADP-ribose monomers or derivatives besides PAR, thus preventing co-crystallization studies with such small PAR-derived molecules. Therefore, a characteristic of PBMs is their apparent lack of global structural conservation, because they can be interspersed within many types of functional domains, such as protein–protein or protein–nucleic acid interaction domains and nuclear localization or export signals, thus providing a regulatory role for PAR. The PBM basic amino acids, by forming an electropositive surface, probably favor interaction with the PAR molecule that is highly acidic due to the two phosphates per ADP-ribose unit.
The ADPR and PAR-binding macrodomain
The 130–190 amino acids, conserved macrodomain that was initially noted in the histone variant macroH2A [13], is found in all kingdoms of life. The first biochemical and structural evidence that macrodomains display ADPR-, and for some, PAR-binding activity, was reported for the macroprotein Af1521 from the thermophilic organism Archaeoglobus fulgidus [14]. That a bacterial protein could bind PAR, a molecule that at the time was not supposed to be produced in lower organisms, was somewhat unexpected. The recent demonstration of a structural link between the macrodomain and the catalytic site of the enzyme responsible for degrading PAR, the PAR glycohydrolase (PARG), and the discovery of bacterial PARG activities has now shed light on this new finding, and challenges the conventional paradigm [15].
In humans, 10 genes encode 11 macrodomain-containing proteins, which display one to three iterations of the motif (Figure 2a) 16, 17, 18. Of note, PARP-14 and PARP-15 combine a (mono)ADP-ribosylation activity (PARP-9 is likely inactive) together with an ADPR or even PAR binding activity 19, 20. However, if and how these two functional domains cooperate in the protein function is unknown. For example, both PARP-14 macrodomains and catalytic activity are involved in the regulation of interleukin (IL)-4-stimulated signal transducer and activator of transcription (Stat)6-dependent transcription 20, 21, but whether they act in a cooperative manner remains to be determined.
Figure 2.

Schematic domain architecture of macro-, poly(ADP-ribose)-binding zinc finger (PBZ) and WWE domain containing human proteins. Protein domains illustrated by colored boxes are defined according to the Pfam 26.0 database. (a) Schematic representation of human macrodomain (blue box) containing proteins. Their ability to bind poly(ADP-ribose) (PAR), ADP-ribose (ADPR), or O-acetyl-ADP-ribose (OAADPR), and their hydrolyzing activity toward ADPR-1′′P or OAADPR is indicated, if known. Structural information, when available in the protein data bank (PDB), is indicated. (b) Schematic representation of human PBZ domain containing proteins. Their ability to bind PAR (if known), and the availability of their 3D structures, are indicated. (c) Schematic representation of human WWE domain containing proteins. These WWE-containing proteins are classified according to whether their associated domains are either endowed with poly(ADP-ribose) polymerase (PARP) activity or with ubiquitin E3 ligase activity. Also indicated is whether these WWE domains possess the ability to bind PAR or isoADPR (if known), and whether 3D structural information is available. DDHD, Asp- and His-containing motif involved in phospholipase activity (light gray box); H2A, domain homologous to histone H2A (pale violet box); helicase-C, helicase superfamily C-terminal domain associated with DEXDc-, DEAD- and DEAH-box proteins (lavender box); FHA, forkhead associated (light pink box); HECT, homologous to E6-AP carboxyl terminus domain, displaying E3-ligase activity (light orange box); lactamase B, domain homologous to β-lactamase endowed with nuclease activity (gray box); macro, homologous to the nonhistone part of macroH2A, displaying PAR-binding activity (see text; blue box); PARP, catalytic domain, homologous to the poly(ADP-ribose) synthesis domain of PARP-1, endowed with mono- or poly(ADP-ribosyl)ation activity (green box); PBZ, PAR-binding zinc finger (see text; orange box); RING, really interesting new gene: zinc binding domain with ubiquitin E3 ligase activity (sienna box); SAM-L, sterile α motif-like (purple box); SNF2-N, SNF2 family N-terminal domain (light steel blue box); UBA, ubiquitin-binding domain (silver gray box); WWE, named after its three conserved residues Trp, Trp and Glu, displaying PAR-binding activity (see text; dark yellow box); Zf-CCCH, zinc finger motif (pink box).
Structures of macrodomains in complex with ADP-ribose derivatives are available for several human macroproteins (Figure 2a). These macrodomains show high structural homology, folding into a globular mixed α-helix/β-sheet structure that forms a deep groove, which is the ligand-binding pocket. However, binding to PAR and even to monomeric ADPR is not a general property of macrodomains. MacroH2A1.1, which is involved in the DNA damage response and transcriptional regulation, is able to bind PAR, ADPR and the SirT1 metabolite O-acetyl-ADP-ribose (OAADPR), which is produced during the NAD+-dependent deacetylation of acetylated proteins [22]. By contrast, due to subtle but sufficient structural changes, its splicing variant macroH2A1.2, like macroH2A2, is unable to bind any ADPR derivatives 22, 23, 24.
How do macrodomains recognize PAR? The crystal structure of the macroH2A1.1 macrodomain in complex with ADPR has revealed that access to the 2′ and 3′ OH groups in the proximal ribose is blocked, precluding any interaction with the internal ADPR unit within PAR, and thus designating macrodomains as recognizers of the last residue of PAR chains (Figure 1) [24]. Mutation of an Asp residue prevents PAR binding of the structurally unresolved macrodomain of ALC1 (amplified in liver cancer 1); a protein involved in chromatin remodeling 25, 26. This Asp is highly conserved within macrodomains and is directly involved in ADPR binding, as shown for Af1521 [14] and macroH2A1.1 18, 23, 24 where it engages in a hydrophobic bond with the NH2 of the adenine.
Several macrodomains found in positive-strand RNA viruses (alphaviruses, hepatitis E virus and coronaviruses, including the severe acute respiratory syndrome coronavirus SARS-CoV) are able to bind PAR within infected animal cells 27, 28, 29. This is an efficient strategy for viruses to hamper or hijack cellular pathways that are normally regulated by the host macroproteins. Some macrodomains have also been converted into powerful biological tools: the bacterial Af1521 macrodomain has been used as a PAR-binder to validate in vivo or in vitro protein PARylation, notably when anti-PAR antibodies are not sufficiently sensitive (or efficient, if PAR is too short) to detect the modification 30, 31.
Among the PAR-binding modules identified so far, macro is the only one for which an enzymatic activity on ADPR derivatives has been reported. Macrodomains from bacterial and viral proteins, and the human MDO1, MDO2 and orphan macrodomain (C6orf130) proteins display limited hydrolyzing activity on ADP-ribose-1′′-phosphate, a metabolite generated during tRNA splicing, and/or on OAADPR 16, 28, 32. No such OAADPR hydrolyzing activity has been detected for macroH2A1.1 despite its efficient binding to OAADPR [22]. Unfortunately, the very appealing hypothesis that macrodomains could hydrolyze PAR has never been validated, except for the macro-like PARG catalytic site 15, 33. The crystallographic structure of a DUF2263 fold from Thermonospora curvata, a distant relative of the PARG fold, has revealed striking similarities to macrodomains [15]. This PARG macro-like catalytic domain shows the closest structural and evolutionary relation to a macrodomain that displays OAADPR-hydrolyzing activity, MacroD1 [33]. Similar to macrodomains, the bacterial PARG recognizes ADP-ribose, with an analogous steric restraint precluding internal recognition of PAR and thus supporting exo- but not endoglycohydrolase activity [15]. However, this raises the question of the conservation of this macro-like structure in mammalian PARGs, because mammalian PARGs are endowed with both exo- and endoglycohydrolase activities [34]. The first crystal structure of a mammalian PARG now gives some clarification [35]. In comparison to the bacterial PARG, the catalytic domain of rat PARG shows: an extended catalytic groove that can accommodate the (n+1) ADP-ribose; the lack of the ribose cap that blocks the 2′-OH of the adenosine ribose in T. curvata PARG and thus excludes recognition of internal glycosidic linkages of PAR; and the presence of a unique flexible tyrosine clasp directly involved in substrate binding [35]. These structural differences explain how mammalian PARG, in contrast to bacterial PARG, is capable of endoglycohydrolase activity. Comparison of all the 3D structures of macrodomains now available sheds light on how these domains can differently act as ADP-ribose, PAR or OAADPR binders or hydrolyzers.
The PAR-binding zinc finger (PBZ)
The tumor suppressor CHFR [checkpoint with forkhead (FHA)-associated and really interesting new gene (RING) finger domains] is a mitotic checkpoint protein that prevents entry into mitosis upon mitotic stress elicited by microtubule poisons [36]. Analysis of its primary sequence has identified a C2H2 zing finger that has turned out to bind PAR efficiently [37], defining this motif as a new PAR binding module termed PBZ for PAR binding zinc finger. PBZ motifs display the consensus sequence [K/R]xxCx[F/Y]GxxCxbbxxxxHxxx[F/Y]xH, and have only been identified in two additional mammalian proteins so far, the histone chaperone and DNA repair protein aprataxin and PNK-like factor (APLF), and the interstrand crosslink repair protein SNM1 (Figure 2b). Interestingly, in other species, the PBZ domains are also found in proteins that are involved in the maintenance of genome integrity, and whose mammalian homologs are closely related to PAR metabolism. This is the case for the Dictyostelium discoideum tankyrase, chk2, and ku70; the Caenorhabditis elegans DNA ligase III; and the Drosophila melanogaster tdp1 [37]. Of note, some of the mammalian homologs (DNA ligase III, ku70) can bind PAR, but via a PBM instead of a PBZ motif [11]. Why different species have selected either a PBM or a PBZ for a protein homolog is an interesting issue, suggesting that there might be some redundant function between PBZ and PBM motifs.
The 3D structures of the APLF and CHFR PBZ motifs have been solved by nuclear magnetic resonance (NMR) spectroscopy (APLF, CHFR) and crystallography (CHFR) 38, 39, 40, 41. The solution structure of the tandem PBZ modules of APLF shows that the two motifs termed F1 and F2 are structurally independent and thus could independently recognize ADPR and PAR [39]. However, tandem PBZ domains are over 1000 times more efficient in PAR binding than isolated PBZ domain. It is proposed that F1 may serve as the primary high-affinity anchoring site for PAR, and that the close proximity between the two zinc fingers generates synergy in PAR binding [39]. Using 2′-O-α-d-ribofuranosyladenosine, which is the simplest adenosine derivative to contain the characteristic α(1->2) O-glycosidic bond between ribose rings, Eustermann and collaborators have showed by NMR that each PBZ module can recognize this internal fragment of PAR (Figure 1) [38]. The key residues designated by the solution structure to bind PAR have been confirmed by mutagenesis, because their substitution abolishes PAR binding in vitro and impairs recruitment of the PBZ mutants to laser-induced DNA damage sites [39] .
By solving the crystal structure of the CHFR PBZ domain, bound to ligands structurally similar to different regions of PAR (ADPR, AMP, Figure 1) or to AMP2 (to mimic two successive adenosines of PAR), Oberoi and collaborators have identified two distinct adenine binding sites that allow the PBZ domain to recognize simultaneously two ADP-ribose units of PAR. The APLF PBZ F1 could have the same capacity, whereas APLF F2, which lacks one key residue, would bind only one adenine [41]. Collectively, these structural data demonstrate that, because of the internal recognition of PAR, PBZ motifs are bona fide PAR binding modules. This internal recognition of PAR is a property shared by the fourth PAR-binding domain recently structurally characterized, the WWE domain.
The most recently reported PAR domain: WWE
The WWE domain, named after its most conserved residues, was initially described as a single or duplicate motif found in proteins either related to PARylation or to ubiquitylation (Figure 2c) [42]. The demonstration of a WWE domain binding to PAR was simultaneously reported for the RNF146 ubiquitin E3 ligase (also termed Iduna) in three different contexts: Wnt signaling, the DNA damage response, and neuroprotection 43, 44, 45. It was very exciting to discover that PARylation is a prerequisite for RNF146 activity in these cellular processes, leading to poly-ubiquitylation and subsequent protein degradation by the proteasome (discussed later, and Figure 3 ). The RNF146 WWE domain binds the smallest PAR structural unit that contains the ribose–ribose glycosidic bond that is unique to PAR, named iso-ADPR [46] (Figure 1). The crystal structure of the RNF146 WWE domain in complex with iso-ADPR [46] has revealed, however, significant differences compared to the NMR structure of the RNF146 WWE domain alone (PDB 1UJR), suggesting that conformational changes occur upon binding. Such changes probably accommodate PAR binding through the recognition of the internal iso-ADPR [46], which is supported by the copurification of PAR polymers of varying length with RNF146 [43]. RNF146 can bind PAR, but not ADPR, supporting the idea that WWE domain cannot recognize mono-ADP-ribosylated proteins [46]. Therefore, in contrast to the macrodomain, which could play a role in the biology of both MARTs and PARPs, the WWE domain, like the PBZ domain, is probably involved only in biological processes relying on PARP activities. The next section highlights how both the WWE and PBZ PAR reader domains interpret this modification and convert it to a signal for protein elimination by the proteasome.
Figure 3.
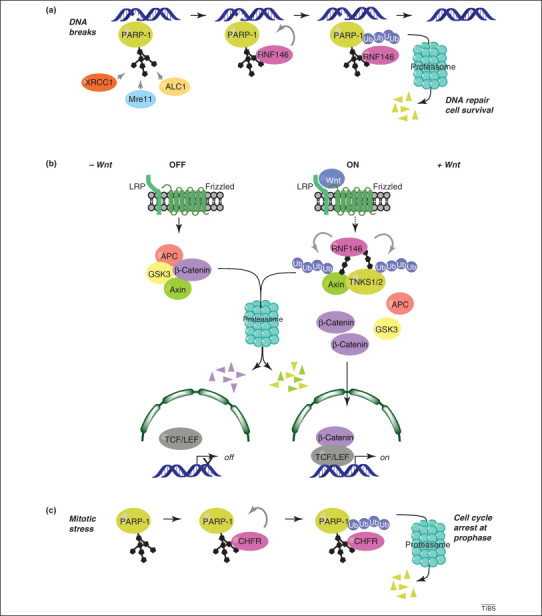
poly(ADP-ribose) (PAR)-dependent poly-ubiquitylation of proteins to promote their degradation by the proteasome. (a) PAR-dependent activation of RNF146 by poly(ADP-ribose) polymerase (PARP)-1 in the DNA damage response. PARP-1 detects DNA breaks, is activated, and auto- or heteromodifies acceptor proteins. PAR produced at DNA damage site has a signaling role and directly recruits factors involved in regulation of chromatin structure and DNA repair [for example amplified in liver cancer 1 (ALC1), X-ray repair cross-complementing gene 1 (XRCC1) or meiotic recombination 11 (Mre11), see Box 1]. The ubiquitin E3 ligase RNF146 is activated upon binding to poly(ADP-ribosyl)ated (PARylated) PARP-1 and poly-ubiquitylates PARP-1. Ubiquitylated PARP-1 is subsequently targeted to the proteasome for degradation. The timely and orchestrated poly(ADP-ribosyl)ation and ubiquitylation of PARP-1 regulates DNA repair and favors cell survival. (b) PAR-dependent activation of RNF146 by tankyrases during Wnt signaling. In the absence of Wnt, the multiprotein β-catenin destruction complex, which contains adenomatous polyposis coli (APC), glycogen synthase kinase 3 (GSK3) and axin triggers the proteasome-dependent degradation of β-catenin. The Wnt signaling cascade is activated by binding of Wnt to its receptor, the low-density lipoprotein receptor-related protein LRP/Frizzled. Tankyrase 1 and 2 (TNKS1/2) PARylate themselves and axin. How TNKS1/2 are activated during Wnt signaling is unknown, and it remains to be determined by which mechanism the activation signal is transmitted from Wnt receptor to TNKS1/2 (illustrated by a dashed arrow between the Wnt receptor and TNKS1/2/RNF146 complex). The PAR activates the ubiquitin E3 ligase RNF146, leading to TNKS1/2 and axin poly-ubiquitylation and subsequent degradation by the proteasome. The multiprotein β-catenin destruction complex is destabilized by the absence of axin, leading to β-catenin accumulation and translocation into the nucleus, where it coactivates the T-cell factor/lymphoid enhancer (TCF/LEF) transcription factor, to promote transcription of Wnt-dependent genes. (c) PAR-dependent activation of CHFR [checkpoint with forkhead (FHA)-associated and really interesting new gene (RING) finger domains] by PARP-1 during mitotic stress. In response to mitotic stress caused by drugs that affect microtubules, such as nocodazole or docetaxel, PARP-1 is activated by an as-yet-unknown process, leading to its auto-PARylation. This results in the PAR-dependent activation of the ubiquitin E3 ligase CHFR and subsequent poly-ubiquitylation of PARP-1 and degradation by the proteasome. Removal of PARylated PARP-1 is necessary to promote cell cycle arrest at prophase.
A new role for PAR: a label for subsequent ubiquitylation-mediated proteosomal degradation?
PARylation is a newcomer in the class of post-translational modifications that are known to mark proteins for elimination by the proteasome through the activation of the ubiquitylation process (Box 2 ). Several recently published concomitant studies have revealed the PAR-dependence of certain ubiquitin E3 ligases for targeting proteins for degradation. These E3 ligases bind PAR via either a WWE (RNF146/Iduna) 44, 45, 47 or a PBZ (CHFR) domain [48]. RNF146 poly-ubiquitylates automodified but not unmodified PARP-1 to target it for proteasomal degradation [44] (Figure 3a). RNF146 also ubiquitylates itself and poly-ubiquitylates many repair factors in a PAR-dependent manner, such as PARP-2, XRCC1, DNA ligase III, and ku70 [44]. Free PAR is sufficient to stimulate RNF146 activity, raising the possibility that the protein targeted for proteasomal degradation does not necessarily need to be PARylated itself [44]. RNF146 is rapidly recruited to laser-induced DNA-damaged sites; its PAR-dependent E3 ligase activity promotes DNA repair and prevents cell death induced by γ-irradiation, alkylating agents or hydrogen peroxide, but only at doses known to trigger cell death in a PARP-1/PAR dependent manner [44]. Eviction of PARylated proteins from the damaged site for their subsequent degradation probably contributes to the spatiotemporal regulation of the repair machinery. In addition, removing PARylated PARP-1 from the site of DNA damage could prevent sustained PARP activity and thus protect against PAR-dependent cell death, which is called parthanatos [7]. In this type of caspase-independent programmed cell death, PAR promotes the release of AIF from the mitochondria and its translocation to the nucleus, to launch genomic DNA fragmentation; a process called chromatinolysis [7]. By limiting PAR accumulation and preventing parthanatos, RNF146 is thought to have a pro-survival role in the DNA damage response [44]. Similarly, RNF146 protects neurons after an excitotoxic stimulus [43]. In this study, transgenic mice overexpressing RNF146, or mice that have had lentiviruses encoding wild-type RNF146, but not a mutant RNF146 that cannot bind PAR, injected in the brain, showed protection toward N-methyl-d-aspartate (NMDA) excitotoxicity and brain infarction, confirming the PAR-dependent neuroprotective effect of RNF146 in vivo [43].
Box 2. Ubiquitylation-mediated protein degradation.
Ubiquitin is a polypeptide of 76 amino acids that can be covalently transferred to a lysine residue of an acceptor protein in a post-translational process termed ubiquitylation [70]. Mono-ubiquitylation is involved in the regulation of protein function, whereas poly-ubiquitylation, in which ubiquitin molecules are sequentially transferred to form ubiquitin chains, is generally aimed to target proteins to degradation by the proteasome in a regulated manner. Ubiquitylation involves three classes of enzymes: an E1 enzyme activates ubiquitin and transfers it to an E2-conjugating enzyme; the E3-ubiquitin ligase enzyme then executes the transfer of ubiquitin to the protein substrate. It is the E3-enzyme that confers the specificity of the reaction by selecting the appropriate substrate [70].
RNF146 activity can be regulated by other PARPs such as the tankyrases TNKS1 and TNKS2, during Wnt/β-catenin signaling 45, 47, 49. By promoting the RNF146-dependent degradation of the negative regulator axin, tankyrases can activate the Wnt/β-catenin signaling pathway (Figure 3b). RNF146 WWE mutants that could not bind PAR also could not trigger axin degradation 45, 47. As observed for PARP-1, tankyrases themselves are subject to ubiquitylation and degradation during this process 45, 47. Tankyrase inhibitors thus appear as promising therapeutic drugs to antagonize constitutive activation of the Wnt cascade in Wnt-dependent cancers 49, 50. RNF146 is activated by PAR in different structures, such as the long and complex branched PAR generated by PARP-1 or the short and linear PAR generated by tankyrases, suggesting that many other PARPs could eventually modulate RNF146 to regulate the turnover of their substrate.
Besides axin, other targets of RNF146 that are regulated by tankyrase activity have been reported, such as the basic leucine zipper nuclear factor 1 (BLZ1), which is involved in the maintenance of Golgi structure; and the cancer susceptibility candidate 3 (CASC3), which is implicated in post-splicing events [45]. Similar to axin, which binds the ankyrin-repeat domain of tankyrase, [51], BLZ1 and CASC3 possess a tankyrase-interacting domain, which allows tankyrase to identify them for PARylation and subsequent ubiquitylation by RNF146 [45]. Tankyrase and RNF146 use a similar mechanism to regulate the stability of c-Abl Src homology 3 domain-binding protein-2, also referred to as SH3BP2 or 3BP2, in osteoclasts [52]. Mutation in 3BP2 causes the autosomal-dominant syndrome cherubism, which is characterized by destructive inflammatory bone lesions resulting in facial deformities. Cherubism mutations prevent the TNKS/RNF146-mediated proteasomal destruction of 3BP2, leading to its stabilization and to increased osteoclastogenesis [52]. Tnks−/−;Tnks2−/− mice die in utero [53], but lineage specific Tnks-depletion by shRNA in Tnks2−/− bone marrow cells phenocopied the knock-in cherubism mutation in 3BP2 gene, leading to bone loss and activation of osteoclasts [52]. The likely redundancy between TNKS1 and TNKS2, at least for this PAR-dependent ubiquitylation process, probably explains why knockout of a single tankyrase did not reveal a cherubism-like phenotype. In light of these new findings, the use of tankyrase inhibitors as a therapeutic strategy to target tumors with activated Wnt pathway as mentioned above should be cautiously examined, because sustained pharmacological inhibition of tankyrases might have unanticipated side effects such as osteoporosis, osteoclastogenesis and inflammation [52].
A functional link between PARP-1 auto-PARylation, subsequent poly-ubiquitylation, and proteasomal degradation has also been observed for a PBZ-containing ubiquitin E3 ligase, the CHFR mitotic checkpoint protein (Figure 3c) [48]. Mitotic stress induced by microtubule poisons such as nocodazole or docetaxel leads to PARP-1 automodification, poly-ubiquitylation by CHFR and degradation [48]. That the reduction in PARP-1 protein levels is required for cell cycle arrest at prophase is strengthened by the prolonged G2/M arrest observed in PARP-1-deficient cells treated with genotoxic drugs [54]. Conversely, PARP-1 activity might be required for proper downstream mitotic processes, as suggested by the presence of PARP-1 in mitotic centromeres and its interaction with and poly(ADP-ribosyl)ation of centromeric factors [55].
The close relation between PARylation and ubiquitylation to target proteins for degradation does not necessarily require the ubiquitin ligase to bind directly and/or be activated by PAR. For instance, the first target of TNKS to be discovered, the telomeric factor TRF1, is also degraded by ubiquitin-mediated proteolysis after release from telomeres upon PARylation by TNKS [56]. PARylation is required for TRF1 release from telomeres but is apparently dispensable for TRF1 proteasomal degradation [56], which is mediated by the SCFFbx4 E3 ligase [57]. However, SCFFbx4 has not been tested for direct PAR-binding or PAR-dependency for its catalytic activity. Another example is that in response to heat shock, PARP-1 is cleared from heat-shock-induced genes by sequential autoPARylation, sumoylation by the protein inhibitor of activated STAT-1 (signal transducer and activator of transcription-1) (PIAS1), and ubiquitination by the RING finger protein 4 (RNF4), leading to the subsequent degradation of PARP-1 [58]. Here again, there is currently no evidence that RNF4 catalytic activity depends on PAR.
It should be also emphasized that the link between PARylation and ubiquitylation-dependent protein degradation is not a general process: not every PARylated protein is led to degradation, and not every poly-ubiquitylated protein targeted to degradation requires prior PARylation. How this is sorted remains a major question. In addition, it is also possible that examples where PAR-dependent mono-ubiquitylation or poly-ubiquitylation does not result in protein degradation, but rather regulation of protein activities or localization, will arise in the future.
Concluding remarks
The recent structural characterization of new PAR-binding motifs, which brings the number of PAR readers that have their own specificity to four, has shed new light on the biological role of PAR and on how this molecule can be mechanistically decoded. Moreover, the discovery of a direct connection between PARylation and poly-ubiquitylation to target some proteins for degradation by the proteasome has opened the way for a new interpretation of the signaling function of PAR in several cellular processes: the degradation of proteins in a timely and orchestrated manner. However, in light of what has been discussed throughout this review, the major open question is: why is there a need for (at least) four different PAR-binding modules? It appears so far that PBM is probably a broad-spectrum PAR-binding domain that can regulate the many types of functional domains within which it can be inserted, whereas macro-, PBZ and WWE domains adopt specific structures that are probably associated with specific functions. This hypothesis is strengthened by the observation that macro-, PBZ and WWE domains do not recognize the same site within PAR (Figure 1), probably reflecting the different outcomes. However, both WWE and PBZ are involved in PAR-dependent ubiquitylation activity, thus defining a new family of PAR-dependent ubiquitin E3 ligases. As discussed in Box 1, all four types of PAR-binding domains could be involved in protein targeting to DNA-damage sites. This also raises the question of how can so many proteins endowed with PAR-binding domains and capable of binding free PAR, at least in vitro, discriminate between their PARylated protein targets (or even free PAR) in vivo? In light of this, a key point that needs further investigation is the structure of PAR itself. Although initial circular dichroism data have suggested that long PAR polymers adopt secondary structure [59], more recent NMR studies have indicated that free PAR is devoid of inherent regular structure [60]. It is tempting to speculate that, in physiological conditions, PAR might adopt the structural conformation that is favored by the physical and chemical interactions with its bound protein target. Different conformations could be recognized by specific, conformation-dependent PAR-binding motifs that have not yet been discovered. Some PAR/protein interactions have already been shown to depend on PAR chain length, such as the nonhistone chromosomal oncoprotein DEK, which efficiently binds 54mer but not 18mer PAR [61], supporting the idea of a PAR-structure dependent binding [62]. Therefore, we anticipate the discovery of additional types of PAR-binding modules, owing to the wide diversity of PAR types produced in terms of size and complexity (linear or branched). Many characterized PAR-binding proteins display none of the already described PAR-binding modules, supporting this hypothesis. Furthermore, having now identified domains that specifically recognize the 2′ → 1′′-O-glycosidic bond that defines linear PAR molecules, one can envision that dedicated domains might exist to specifically recognize the 2′′ → 1′′′-O-glycosidic bond that defines branched PAR molecules.
Another intriguing question is how multiple PAR-binding motifs that are present within a single protein function together. Specifically, two macro-, PBZ or WWE domains have often been found within a single protein. PARP-14 even contains three macrodomains and one WWE domain, but the functionality of this WWE motif is unclear because some crucial residues for PAR recognition are not conserved [46]. The two PBZ domains of APLF, despite being in close proximity, are structurally independent [38]; however, the tandem APLF PBZ motifs also show synergy in PAR binding, suggesting a functional link between the two zinc fingers [39]. Structural modeling based on the structure of the unliganded Drosophila Deltex WWE domain suggests that two WWE domains could recognize two neighboring ADP-ribose units, supporting the idea of synergistic recognition of PAR by multiple PAR-binding domains present within a protein [46]. The next challenging issue is to determine whether a protein with multiple PAR-binding motifs could simultaneously bind different PARylated partners. This would multiply the possible mechanisms of tight regulation by functional interactions within protein complexes.
One final question is, what is the role of PARG in PARylation-coupled, ubiquitylation-mediated protein degradation, such as in the DNA damage response? The model described here, in which automodified PARP-1 is further ubiquitylated and rapidly degraded, does not take into account the efficient and quick PAR-degrading activity of PARG. Future experiments need to establish whether PARG, which is as essential as PARP-1 for prompt DNA breaks repair 63, 64, 65, is also involved in the PAR- and ubiquitin-dependent protein degradation process, or plays another PAR-dependent regulatory roles.
Acknowledgments
We apologize to investigators whose valuable contributions could not be cited in this review owing to space limitations. We thank the referees for their constructive suggestions. We acknowledge support from Centre National de la Recherche Scientifique, University of Strasbourg, Ligue contre le Cancer (comités du Bas-Rhin et du Haut-Rhin), Association pour la Recherche sur le Cancer, Agence Nationale de la Recherche, Electricité de France and Laboratory of Excellence Medalis. Our team is Equipe labellisée Ligue contre le Cancer.
References
- 1.Luo X., Kraus W.L. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hakmé A. The expanding field of poly(ADP-ribosyl)ation reactions. ‘Protein Modifications: Beyond the Usual Suspects’ Review Series. EMBO Rep. 2008;9:1094–1100. doi: 10.1038/embor.2008.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hottiger M.O. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Rouleau M. PARP inhibition: PARP1 and beyond. Nat. Rev. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kleine H. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos) Sci. Signal. 2011;4:ra20. doi: 10.1126/scisignal.2000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y. Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Exp. Neurol. 2009;218:193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol. Oncol. 2011;5:387–393. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Javle M., Curtin N.J. The role of PARP in DNA repair and its therapeutic exploitation. Br. J. Cancer. 2011;105:1114–1122. doi: 10.1038/bjc.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garnett M.J. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:509–642. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pleschke J.M. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 12.Gagné J.P. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pehrson J.R., Fried V.A. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- 14.Karras G.I. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slade D. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011;477:616–620. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen D. Identification of macrodomain proteins as novel O-acetyl-ADP-ribose deacetylases. J. Biol. Chem. 2011;286:13261–13271. doi: 10.1074/jbc.M110.206771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleine H., Luscher B. Learning how to read ADP-ribosylation. Cell. 2009;139:17–19. doi: 10.1016/j.cell.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 18.Han W. The macro domain protein family: structure, functions, and their potential therapeutic implications. Mutat. Res. 2011;727:86–103. doi: 10.1016/j.mrrev.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aguiar R.C. B-aggressive lymphoma (BAL) family proteins have unique domains that modulate transcription and exhibit Poly(ADP-ribose) polymerase activity. J. Biol. Chem. 2005;280:33756–33765. doi: 10.1074/jbc.M505408200. [DOI] [PubMed] [Google Scholar]
- 20.Goenka S., Boothby M. Selective potentiation of Stat-dependent gene expression by collaborator of Stat6 (CoaSt6), a transcriptional cofactor. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4210–4215. doi: 10.1073/pnas.0506981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehrotra P. PARP-14 functions as a transcriptional switch for STAT6 dependent gene activation. J. Biol. Chem. 2011;286:1767–1776. doi: 10.1074/jbc.M110.157768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Till S., Ladurner A.G. Sensing NAD metabolites through macro domains. Front. Biosci. 2009;14:3246–3258. doi: 10.2741/3448. [DOI] [PubMed] [Google Scholar]
- 23.Kustatscher G. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- 24.Timinszky G. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 25.Gottschalk A.J. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahel D. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egloff M.P. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006;80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neuvonen M., Ahola T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J. Mol. Biol. 2009;385:212–225. doi: 10.1016/j.jmb.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malet H. The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J. Virol. 2009;83:6534–6545. doi: 10.1128/JVI.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dani N. Combining affinity purification by ADP-ribose-binding macro domains with mass spectrometry to define the mammalian ADP-ribosyl proteome. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4243–4248. doi: 10.1073/pnas.0900066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guetg C. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol. Cell. 2012;45:790–800. doi: 10.1016/j.molcel.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 32.Peterson F.C. Orphan macrodomain protein (human C6orf130) is an O-acyl-ADP-ribose deacylase: solution structure and catalytic properties. J. Biol. Chem. 2011;286:35955–35965. doi: 10.1074/jbc.M111.276238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hassler M. PARG: a macrodomain in disguise. Structure. 2011;19:1351–1353. doi: 10.1016/j.str.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 34.Brochu G. Mode of action of poly(ADP-ribose) glycohydrolase. Biochim. Biophys. Acta. 1994;1219:342–350. doi: 10.1016/0167-4781(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 35.Kim I.K. Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element. Nat. Struct. Mol. Biol. 2012;19:653–656. doi: 10.1038/nsmb.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scolnick D.M., Halazonetis T.D. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature. 2000;406:430–435. doi: 10.1038/35019108. [DOI] [PubMed] [Google Scholar]
- 37.Ahel I. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 38.Eustermann S. Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose) Nat. Struct. Mol. Biol. 2010;17:241–243. doi: 10.1038/nsmb.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li G.Y. Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9129–9134. doi: 10.1073/pnas.1000556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isogai S. Solution structure of a zinc-finger domain that binds to poly-ADP-ribose. Genes Cells. 2010;15:101–110. doi: 10.1111/j.1365-2443.2009.01369.x. [DOI] [PubMed] [Google Scholar]
- 41.Oberoi J. Structural basis of poly(ADP-ribose) recognition by the multizinc binding domain of checkpoint with forkhead-associated and RING domains (CHFR) J. Biol. Chem. 2010;285:39348–39358. doi: 10.1074/jbc.M110.159855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aravind L. The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 2001;26:273–275. doi: 10.1016/s0968-0004(01)01787-x. [DOI] [PubMed] [Google Scholar]
- 43.Andrabi S.A. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat. Med. 2011;17:692–699. doi: 10.1038/nm.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang H.C. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2011;108:14103–14108. doi: 10.1073/pnas.1108799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat. Cell Biol. 2011;13:623–629. doi: 10.1038/ncb2222. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 2012;26:235–240. doi: 10.1101/gad.182618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Callow M.G. Ubiquitin ligase RNF146 regulates tankyrase and axin to promote Wnt signaling. PLoS ONE. 2011;6:e22595. doi: 10.1371/journal.pone.0022595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kashima L. CHFR regulates the mitotic checkpoint by targeting PARP-1 for ubiquitination and degradation. J. Biol. Chem. 2012;287:12975–12984. doi: 10.1074/jbc.M111.321828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang S.M. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 50.Waaler J. A novel tankyrase inhibitor decreases canonical Wnt signaling in colon carcinoma cells and reduces tumor growth in conditional APC mutant mice. Cancer Res. 2012;72:2822–2832. doi: 10.1158/0008-5472.CAN-11-3336. [DOI] [PubMed] [Google Scholar]
- 51.Morrone S. Crystal structure of a Tankyrase–Axin complex and its implications for Axin turnover and Tankyrase substrate recruitment. Proc. Natl. Acad. Sci. U.S.A. 2012;109:1500–1505. doi: 10.1073/pnas.1116618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levaot N. Loss of Tankyrase-mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism. Cell. 2011;147:1324–1339. doi: 10.1016/j.cell.2011.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chiang Y.J. Tankyrase 1 and tankyrase 2 are essential but redundant for mouse embryonic development. PLoS ONE. 2008;3:e2639. doi: 10.1371/journal.pone.0002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Menissier de Murcia J. Requirement of poly(ADP-ribose)polymerase in recovery from DNA damage in mice and in cells. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saxena A. Centromere proteins Cenpa, Cenpb, and Bub3 interact with poly(ADP-ribose) polymerase-1 protein and are poly(ADP-ribosyl)ated. J. Biol. Chem. 2002;277:26921–26926. doi: 10.1074/jbc.M200620200. [DOI] [PubMed] [Google Scholar]
- 56.Chang W. TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev. 2003;17:1328–1333. doi: 10.1101/gad.1077103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng Z. Structural basis of selective ubiquitination of TRF1 by SCFFbx4. Dev. Cell. 2010;18:214–225. doi: 10.1016/j.devcel.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin N. PARP-1 transcriptional activity is regulated by sumoylation upon heat shock. EMBO J. 2009;28:3534–3548. doi: 10.1038/emboj.2009.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Minaga T., Kun E. Probable helical conformation of poly(ADP-ribose). The effect of cations on spectral properties. J. Biol. Chem. 1983;258:5726–5730. [PubMed] [Google Scholar]
- 60.Schultheisz H.L. Enzymatic synthesis and structural characterization of 13C, 15N-poly(ADP-ribose) J. Am. Chem. Soc. 2009;131:14571–14578. doi: 10.1021/ja903155s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fahrer J. High-affinity interaction of poly(ADP-ribose) and the human DEK oncoprotein depends upon chain length. Biochemistry. 2010;49:7119–7130. doi: 10.1021/bi1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fahrer J. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007;35:e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fisher A.E. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol. Cell. Biol. 2007;27:5597–5605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amé J.C. Radiation-induced mitotic catastrophe in PARG-deficient cells. J. Cell Sci. 2009;122:1990–2002. doi: 10.1242/jcs.039115. [DOI] [PubMed] [Google Scholar]
- 65.Erdelyi K. Dual role of poly(ADP-ribose) glycohydrolase in the regulation of cell death in oxidatively stressed A549 cells. FASEB J. 2009;23:3553–3563. doi: 10.1096/fj.09-133264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Vos M. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem. Pharmacol. 2012;84:137–146. doi: 10.1016/j.bcp.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 67.Krishnakumar R., Kraus W.L. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chou D.M. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ismail, I.H. et al. (2012) CBX4-mediated SUMO modification regulates BMI1 recruitment at sites of DNA damage. Nucleic Acids Res. 10.1093/nar/gks222 [DOI] [PMC free article] [PubMed]
- 70.Weissman A.M. The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat. Rev. Mol. Cell Biol. 2011;12:605–620. doi: 10.1038/nrm3173. [DOI] [PMC free article] [PubMed] [Google Scholar]


