Abstract
Human astroviruses (HAstVs) are a major cause of gastroenteritis. HAstV assembles from the structural protein VP90 and undergoes a cascade of proteolytic cleavages. Cleavage to VP70 is required for release of immature particles from cells, and subsequent cleavage by trypsin confers infectivity. We used electron cryomicroscopy and icosahedral image analysis to determine the first experimentally derived, three-dimensional structures of an immature VP70 virion and a fully proteolyzed, infectious virion. Both particles display T = 3 icosahedral symmetry and nearly identical solid capsid shells with diameters of ~ 350 Å. Globular spikes emanate from the capsid surface, yielding an overall diameter of ~ 440 Å. While the immature particles display 90 dimeric spikes, the mature capsid only displays 30 spikes, located on the icosahedral 2-fold axes. Loss of the 60 peripentonal spikes likely plays an important role in viral infectivity. In addition, immature HAstV bears a striking resemblance to the structure of hepatitis E virus (HEV)-like particles, as previously predicted from structural similarity of the crystal structure of the astrovirus spike domain with the HEV P-domain [Dong, J., Dong, L., Méndez, E. & Tao, Y. (2011). Crystal structure of the human astrovirus capsid spike. Proc. Natl. Acad. Sci. USA108, 12681–12686]. Similarities between their capsid shells and dimeric spikes and between the sequences of their capsid proteins suggest that these viral families are phylogenetically related and may share common assembly and activation mechanisms.
Abbreviations: HAstV, human astrovirus; HEV, hepatitis E virus; cryoEM, electron cryomicroscopy; ORF, open reading frame; EM, electron microscopy; 3D, three‐dimensional; HEV‐lp, HEV‐like particle; PDB, Protein Data Bank; NIH, National Institutes of Health
Keywords: virus structure, electron microscopy, cryomicroscopy, image analysis
Graphical Abstract
Highlights
► Three‐dimensional electron cryomicroscopy density maps of immature and mature HAstV. ► Mature virions only display 30 of 90 spikes after proteolytic cleavage. ► Immature virions are remarkably similar to HEV.
Introduction
Human astroviruses (HAstVs) are an important cause of gastroenteritis in children, the elderly, and immunocompromised adults. The virus was first identified in 1975 in infants who developed diarrhea in hospital nurseries in the United Kingdom.1., 2. HAstV is a small, single-stranded, positive-sense RNA virus with an ~ 6.8‐kb genome. The 5′-terminal two-thirds of the genome [open reading frame (ORF)-1a and ORF-1b] encodes two major nonstructural proteins, a serine protease whose structure has been solved by X-ray crystallography3 and an RNA-dependent RNA polymerase. The 3′-terminal one-third of the genome (ORF-2) encodes the structural protein VP90, which is translated as an 87‐ to 90‐kDa capsid precursor protein. VP90 has a highly conserved N-terminal domain and a variable C-terminal domain among astrovirus serotypes.
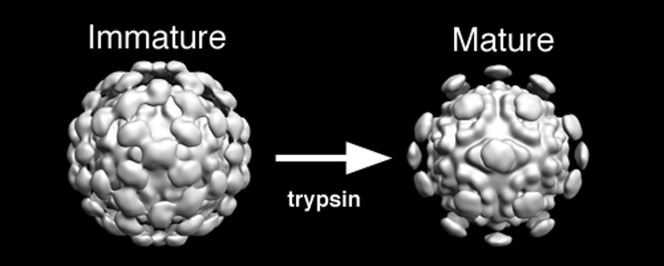
To date, eight serotypes have been identified of which type 1 is the most prevalent, and type 8 has been studied extensively. While their genomes are fairly well conserved,4., 5., 6. the mature virions of different serotypes vary somewhat in the exact size and number of structural proteins that form the capsid (reviewed by Krishna7). The current consensus is that morphogenesis of infectious particles relies on a series of proteolytic cleavages of the capsid precursor protein (Fig. 1 ). VP90 assembles in infected cells and undergoes C-terminal cleavages by caspases to generate VP70 (a 70‐ to 79‐kDa protein) for release from the cells as immature virus particles.8 Generation of infectious particles is dependent upon further trypsin cleavages usually resulting in three structural proteins, VP34, VP27, and VP25, each of which range in actual size and name depending on the serotype.9., 10., 11. VP34 contains the conserved N-terminal domain and is ascribed to the capsid, while the overlapping VP27 and VP25 subunits contain the variable C-domain and are ascribed to the viral spikes.7., 12. VP25 is generated by additional trimming at the N-terminus of VP27.11
Fig. 1.
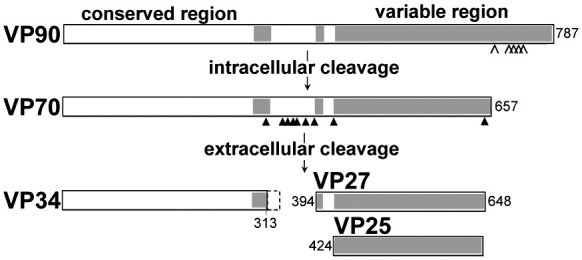
Protein organization and proteolysis. Schematic diagram showing proteolytic processing of the HAstV coat protein. Sites for caspase and trypsin cleavage are indicated with open and filled arrowheads, respectively. Trypsin cleavage of the coat protein between the conserved (white boxes) and variable (shaded boxes) domains is required for viral maturation. The specific pattern of peptide products varies between serotypes. In these experiments, we examined uncleaved particles of HAstV-8 formed by VP70 and fully cleaved particles of HAstV-1.
Astroviruses were initially named for the distinctive star-like appearance of their viral surface observed in ~ 10% of the fecally shed viral particles evaluated by negative-stain electron microscopy (EM). EM ultrastructural analysis of infectious HAstV generated in cell culture showed spherical particles with a surface that was studded with spikes and an external diameter of ~ 410 Å.13 A recent milestone was determination of a high-resolution X-ray structure of the P-domain of the astrovirus spike VP25 (ordered residues 430–645).12 Similarities in sequence and domain organization of hepatitis E virus (HEV) and HAstV enabled the building of a homology model of HAstV.
In this study, we sought to gain direct insight into the structure and assembly of astrovirus by electron cryomicroscopy (cryoEM) of immature and proteolytically activated particles. We examined (1) immature HAstV-8 particles composed of the 70‐kDa intermediate protein, which has undergone caspase but not trypsin cleavage and is therefore not infectious, and (2) fully cleaved, mature infectious particles of HAstV-1. The sequences of the two strains are extremely similar, with 83% identity in the conserved domain and 60% identity in the variable domain. Therefore, we expect that our three-dimensional (3D) reconstructions are representative of the Astroviridae family. Although the capsid shells of both immature and mature HAstVs are nearly identical, there is a striking difference in the apparent stoichiometry of the surface spikes. Immature, uncleaved particles, which are strikingly similar in appearance to HEV-like particles (HEV-lp),14 have 90 spikes, whereas mature, cleaved particles only display 30 spikes, all located at the icosahedral 2-fold axes. Loss of the 60 peripentonal spikes likely plays an important role in viral infectivity.
Results and Discussion
Astrovirus particles have T = 3 icosahedral symmetry
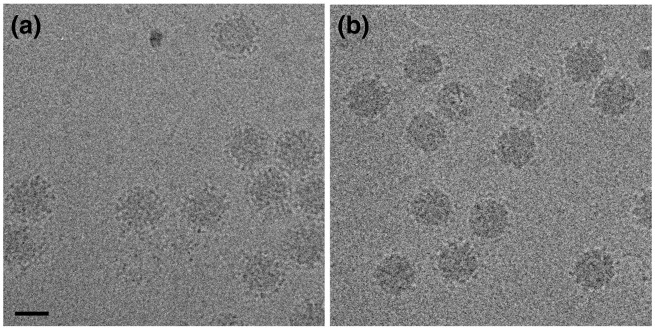
Purification of HAstV has been very challenging, and sufficient material was only available to perform cryoEM once for immature HAstV-8 and mature HAstV-1. Icosahedral image analysis of electron cryomicrographs (Fig. 2 ) yielded 3D maps at ~ 25 Å resolution (Fig. 3a and b). Both particles display two concentric layers of density that conform to T = 3 icosahedral lattice symmetry. A nearly identical inner layer forms a solid capsid shell ~ 45 Å thick with an outer diameter of ~ 350 Å. This capsid shell has a chiseled appearance assembled from 180 protein subunits in which trimeric facets form a plateau (Fig. 3a and b, bottom). The trimeric facets are in close contact across the icosahedral 2-fold (2F) symmetry axes, with depressions at the 5-fold (5F) and 3-fold (3F) symmetry axes and a groove across the local 2F axes. Distal from the solid capsid shell is an outer layer of globular spike-like densities. The spikes are ~ 40 Å in diameter, with a somewhat elliptical shape, yielding an outer particle diameter of ~ 440 Å. The stoichiometry suggests that the spikes are dimers with contributions from adjacent trimer facets (Fig. 3a, color code and labels). Thin linker densities connect the spikes to the capsid shells and are visible in 3D reconstructions of negative-stained particles (Fig. S1) or when the isosurface is lowered significantly in the cryoEM maps. The lack of well-defined linker density may be the result of the narrowness of the linker, minor heterogeneity in maturation cleavage, or wobbling of the spike head, which would obscure the connections during icosahedral symmetry averaging.
Fig. 2.
Electron cryomicrographs of frozen-hydrated samples. Immature HAstV-8 (a) and mature HAstV-1 (b) have identical capsid diameters, but the immature particles have substantially more surface density. The scale bar represents 50 nm.
Fig. 3.
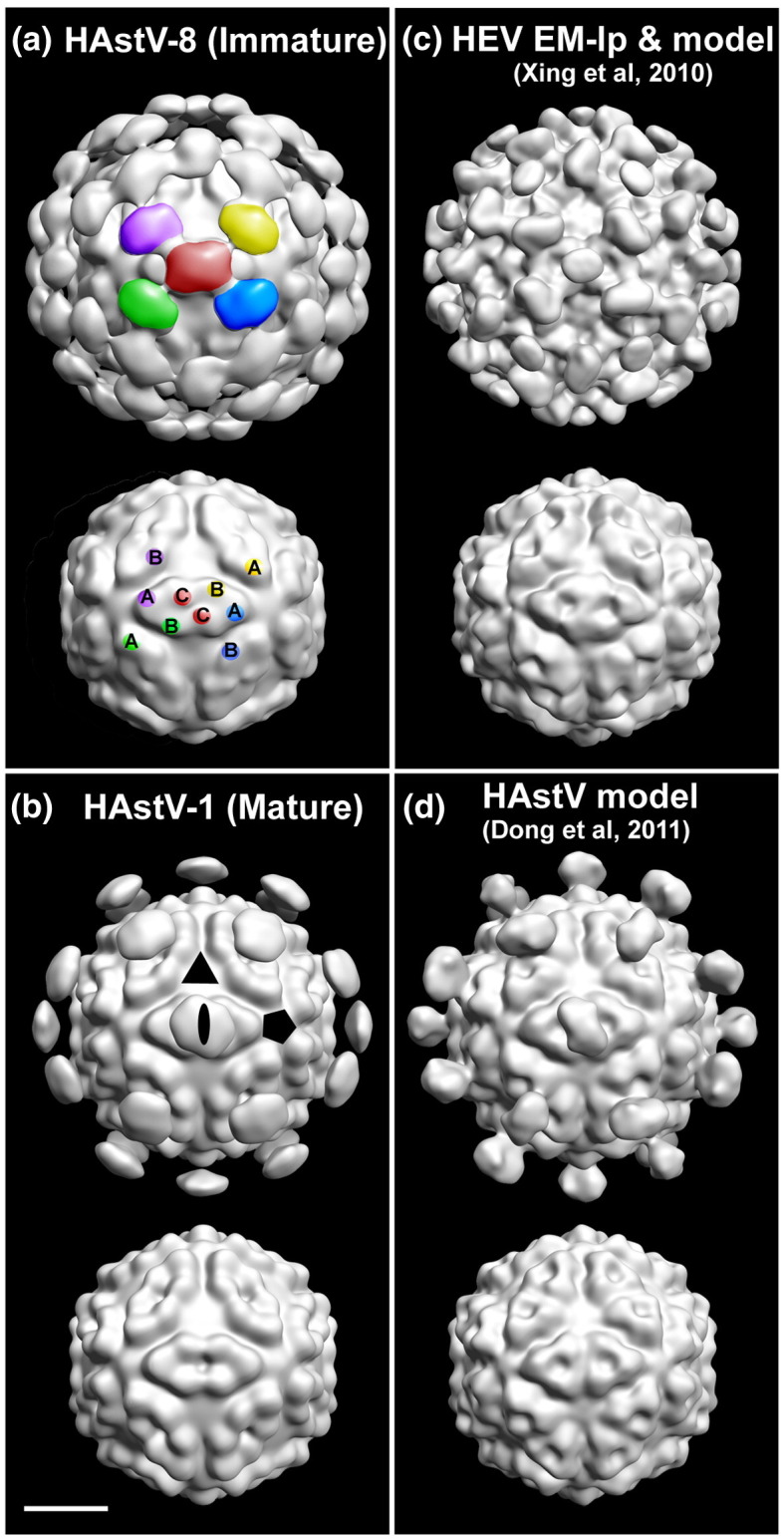
Reconstruction of HAstV-8 and HAstV-1 by cryoEM and icosahedral image analysis, and comparison to model structures. Surface-shaded 3D reconstructions of (a) immature HAstV-8 and (b) mature HAstV-1 display the same diameters (440 Å) and T = 3 icosahedral symmetry. The uncleaved HAstV-8 particles [(a) top panel] display 90 disconnected spikes compared to 30 on the cleaved HAstV-1 particles [(b) top panel]. Maps truncated at a radius of 180 Å (lower panels) display a very similar chiseled morphology and diameter of the capsid shells, which are most likely assembled from the conserved N-terminal domain of VP34. The surface spikes are attributed to dimers of the VP25/VP27 variable domain. A possible scheme for the attachment of the surface spikes to the capsid shell is indicated on the HAstV-8 map (a) by the paired colors of the globular spikes (top panel) and presumed attachment sites (bottom panel), with A, B, and C subunits labeled. Symmetry axes are indicated by symbols on the HAstV-1 map [(b) top panel)]. (c) T = 3 HEV EM structure (emd ID: 5173) low-pass filtered at 25 Å resolution looks remarkably similar to the architecture of immature HAstV [(a) top panel]. The modeled HEV backbone (PDB ID: 3IYO) was used to more clearly visualize the capsid by generating a 25-Å‐resolution map without the P‐domains. (d) A model of HAstV calculated by homology with the HEV T = 1 crystal structure12 (kindly provided by the authors). This structure was similarly truncated at 25 Å resolution for comparison, and generated with (top panel) and without (bottom panel) the spikes. The scale bar represents 100 Å.
Dramatic surface spike changes upon trypsin cleavage
Although the capsid shells of both immature and mature HAstVs are nearly identical, there is a striking difference in the apparent stoichiometry of the surface spikes. Immature, uncleaved particles have 90 spikes (Fig. 3a, top). Thirty are located at the icosahedral 2F symmetry axes, and sixty surround the 5F vertices, centered at the local 2F axes. Mature, cleaved particles only display 30 spikes, all located at the icosahedral 2F axes (Fig. 3b, top). The packing of the viral proteins displays two arrangements for the dimer interaction of the spikes—one that places the dimer densities in close proximity (CC dimers) and one that spans nearly twice the distance between the contributing subunits (AB dimers) (Fig. 3a, bottom labels). This difference in separation may expose different cleavage sites during maturation.
Proteolytic cleavage as a mechanism for activation
Proteolytic cleavage is a common mechanism for activation of both enveloped and nonenveloped viruses, including influenza virus,15., 16. coronavirus,17 rotavirus,18 reovirus,19 and alphaviruses.20 A recurring theme is that cleavage triggers a conformational change in a surface spike or other cell-attachment domain. For astrovirus, the uncleaved, immature T = 3 particle contains 180 copies of VP70, in which the viral capsid is attributed to the N-terminal domains and the 90 spikes are attributed to dimers of the variable C-terminal domain. Similarly, trypsin cleavage of the immature particles produces three products, of which we correspondingly attribute VP34 to the capsid shell and the two different C-terminal products, VP27 and VP25, to the spikes.
SDS-PAGE suggests that trypsin cleaves virtually all of the VP70 molecules (Fig. S2). In addition, the 3D reconstruction in Fig. 3b suggests that trypsin cleavage results in release of two-thirds of the spikes in the mature particles. However, we note that the bands of VP27 and VP25 on SDS-PAGE gels display intensities similar to that of VP34,10 suggesting that trypsin cleavage might lead rather to conformational flexibility and that the products of proteolysis, or a significant percentage of them, could remain noncovalently attached to the capsid shell. Limitations in material have precluded careful quantitative analysis to resolve this question. However, we note that the trypsin-treated particles in Fig. 2b have noticably less surface density than the particles in Fig. 2a. If the cleaved proteins remain noncovalently associated with the capsid shell, then one would still expect to see surface density in the images of individual particles, even if the polypeptides do not conform to icosahedral symmetry. Consequently, our working hypothesis is that trypsin cleavage leads to loss of 60 peripentonal spikes, which confers infectivity, possibly by exposing additional receptor binding sites on the capsid surface.
The structures of immature HAstV-8 and HEV‐lp are remarkably similar
The size, shape, and architecture of immature HAstV-8 are very similar to those reported for a T = 3 HEV-lp14 (Fig. 3c). In the case of HEV-lp, a T = 1 X-ray crystal structure allowed detailed modeling of the protein subunits, which led to the proposal that the capsid is assembled from dimers around the 5F axes and across the icosahedral 2F axes. The HEV capsid protein is divided into three domains, defined as the shell (S-domains), intermediate (M-domains), and protruding densities (P-domains). Dong et al. identified the HEV P-domain as structurally similar to their crystal structure of the astrovirus dimer (construct P2412–646), which represents most of VP25 by DALI alignment, and from which they proposed a phylogenetic relationship.12 They also used the HEV T = 1 X-ray structure to create a homology model of the astrovirus core sequences (residues 82–406) and docked their crystal structure of residues 430–645 in place of the HEV P-domain.
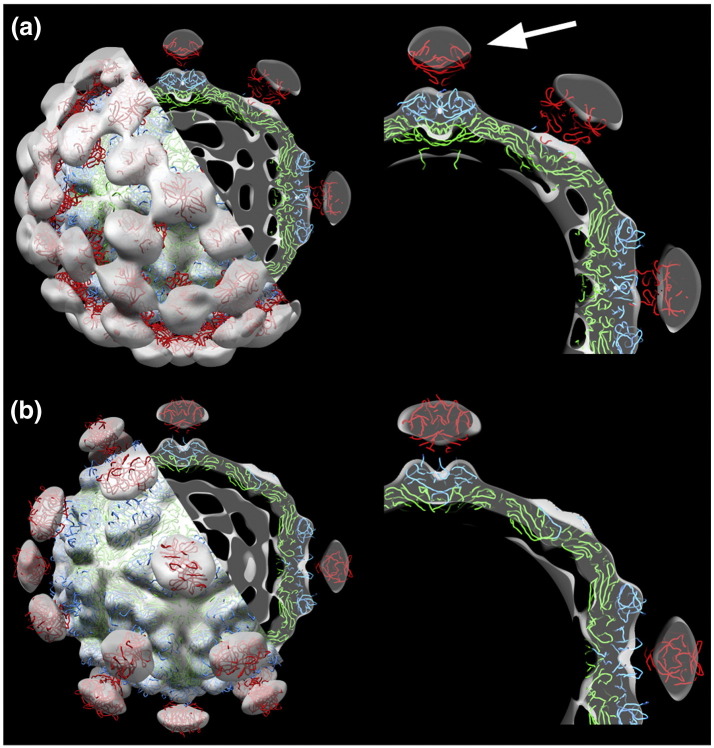
Our experimental results support the proposed relationship between HAstV and HEV. Both in silico atomic models match exceedingly well with the HAstV density maps and dock without any scaling: the HEV-lp model into our VP70 immature structure (Figs. 3c and 4a) and the HAstV model into our mature structure (Figs. 3d and 4b). The similarity reinforces the supposition that VP34 forms the capsid shell and that VP27/VP25 complexes comprise the spikes. The one noticeable difference between the structures is that the astrovirus spikes extend to a radius greater than that of HEV-lp (Fig. 4a, arrow). In comparison, structures from the Caliciviridae family, many of which are also T = 3 viruses that assemble as dimers, do not dock nearly as well, with differences in capsid features and orientation of their P-domains (data not shown).
Fig. 4.
Superposition of the HEV and HAstV X-ray models with native HAstV maps. The T = 3 HEV model (PDB ID: 3IYO) backbone trace was docked into the 3D cryoEM density map of HAstV-8 (a), and the HAstV model from Dong et al.12 was docked into the 3D cryoEM density map of HAstV-1 (b). The close-up view at the right shows a 20-Å‐thick central slab of the density maps. With no scaling or translation, the maps align very well, except that the HAstV spikes extend to a higher radius (arrow) than those of HEV. Density maps are in gray scale, and for clarity in the enlarged views, the interior density was set to zero at a radius of 120 Å. The backbone chains are color coded by domain as defined in (a) for HEV; S-domain (green), M-domain (blue), and P-domain (red).
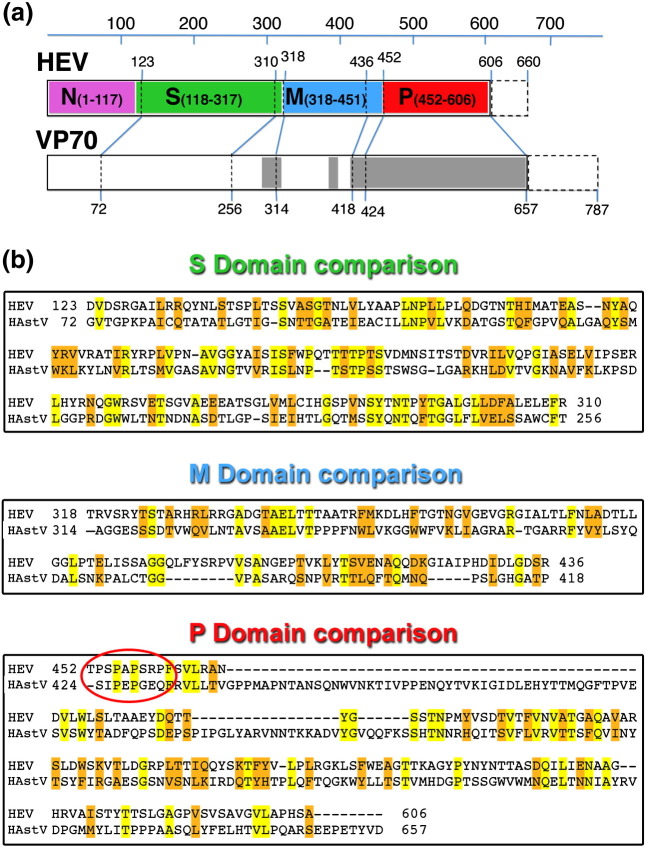
Furthermore, although the conservation is low, the top score for viral proteins from a naïve National Center for Biotechnology Information BLAST (Basic Local Alignment Search Tool) search of the Protein Database for the HAstV capsid protein identified similarity with the HEV capsid protein (E = 2e− 3) (Fig. 5 ). The sequence in the HAstV conserved domain, amino acids 72–256, has ~ 21% identity with the capsid S-domain of HEV. Regions of the HAstV VP90 sequence can also be aligned to the HEV M- and P-domains with moderate identity (~ 13%) (Fig. 5b). When the HEV capsid protein is used as the reference sequence, the search identifies the conserved domain from multiple astrovirus species, with the strongest scores for bat and bovine astroviruses (E < 1e− 8). It is also interesting to note that, while most astroviruses cause gastroenteritis in their host, duck astrovirus causes fatal hepatitis.21
Fig. 5.
HAstV protein alignment with HEV. (a) Schematic diagram of the HEV and HAstV (VP70) capsid protein sequences. Separations for alignment were based on the domains defined for HEV. The dashed boxes represent the C-terminal portion that is missing from the structures—by caspase cleavage for HAstV and by in vitro deletion for HEV. (b) From the similarity identified from a National Center for Biotechnology Information BLAST search, HAstV sequence (GenBank ID: CAA80132) was aligned to each of the HEV domains (GenBank ID: AAA45736). A pair of proline residues in HAstV were aligned with the HEV hinge (red ellipse), and a long spacer inserted for HAstV to account for the greater distance from the capsid observed for its spike. Yellow highlights identical residues, and orange highlights similar residues.
Possible similarities in assembly and activation of HAstV and HEV
Although the astrovirus polyprotein is larger by ~ 120 residues, there may also be biological similarities between HAstV and HEV. For instance, in vitro assembly of expressed HEV capsid protein requires deletion of the 52 C-terminal residues, which may be analogous to caspase cleavage of VP90 and removal of ~ 125 amino acids to generate VP70 (Fig. 1). As there is no cell culture system currently capable of propagating HEV, little is known about its infectivity. Nevertheless, it is reasonable to speculate that a cleavage cascade similar to HAstV may be required for both assembly and activation. Furthermore, by analogy with the HEV-lp model, we may be able to infer that the differences between the AB and CC dimers of immature astrovirus is relevant for assembly, in addition to activation. In the case of HEV-lp, there is a measureable difference in the hinge angles between the AB and CC dimers, which may explain the similar differences observed in HAstV. It was proposed for HEV that the flatter angle of the CC dimers is required to make T = 3 rather than T = 1 particles and only happens in the presence of RNA.14., 22. The need for RNA as a structural component may account for the inability, to date, to assemble stable astrovirus-like particles from expressed proteins. Both systems may provide insights for the other.
Materials and Methods
Production and purification of HAstV-8
The astrovirus strain Yuc8 was isolated from a natural infection and adapted to grow in Caco-2 cells.4 To propagate the virus, we washed the cells twice with minimal essential medium without fetal bovine serum and inoculated them with Yuc8, previously treated with 200 μg/ml of trypsin for 1 h, followed by a 1‐h treatment with soybean trypsin inhibitor at the same concentration. Infected cells were incubated at 37 °C and harvested 1 day post-infection. The cells were lysed by three freeze–thaw cycles and extracted once with an equal volume of Genetron 113 (trichlorotrifluoroethane). From this point, every step was carried out at 4 °C, and solutions were maintained in an ice-water bath. The aqueous phase obtained after centrifugation at 5000g for 30 min was pelleted via ultracentrifugation for 4 h at 85,000g (Beckman SW28 rotor). The pellet was resuspended in TNE buffer [50 mM Tris–HCl (pH 7.4), 0.1 M NaCl, and 10 mM ethylenediaminetetraacetic acid] and layered onto a cesium chloride solution (1.36 g/ml). A density gradient was formed during ultracentrifugation for 16–18 h at 110,000g (Beckman SW50.1 rotor), and the opalescent band corresponding to viral particles was collected and diluted in TNE buffer. The final pellet was isolated by ultracentrifugation of this suspension for 2.5 h at 150,000g (Beckman SW50.1 rotor), which was resuspended in TNE buffer and stored at 4 °C. Infectivity assays and protein analysis of trypsin-treated virus were performed to ensure the identity of the purified particles.11
Production and purification of HAstV-1
Monolayers of a continuous line of colon carcinoma cells (Caco-2) were infected with HAstV serotype 1 (Oxford strain) that was initially adapted to grow in a continuous line of monkey kidney epithelial cells (LLCMK2). Virus propagation required the addition of trypsin (10 μg/ml, type IX; Sigma) to the culture medium (RPMI 1640). At 48 h post-infection, cells were lysed by three freeze–thaw cycles, and the lysates were partially purified by fluorocarbon extraction and differential centrifugation. The viral pellets were solubilized in TNMC buffer [50 mM Tris–HCl (pH 8), 100 mM NaCl, 10 mM MgCl2, and 10 mM CaCl2]. The suspensions were then layered on a cesium chloride density gradient (1.37 g/ml). Following isopycnic centrifugation, the fraction containing peak enzyme immunoassay activity corresponded to a density of 1.37 g/ml. This fraction was diluted 10-fold with TNMC buffer, and viral particles were concentrated by ultracentrifugation for 2.5 h at 4 °C (150,000g).
Electron cryomicroscopy
Samples were prepared for cryoEM by standard methods. In brief, 3‐μl aliquots from each viral preparation were applied to freshly glow-discharged, holey carbon grids, blotted to near dryness, and flash frozen in a slurry of liquid ethane. Grids were maintained at liquid nitrogen temperatures in a Gatan 626 cryo-holder and examined using an FEI CM120 electron microscope (Eindhoven) under low electron dose conditions as previously described.23 Images were recorded on Kodak SO-163 low-density film at nominal magnifications of 35,000 or 45,000×. Micrographs with minimal astigmatism and drift were digitized on a Zeiss microdensitometer (Z/I Imaging) at a 7-μm sampling interval corresponding to 2.0 or 1.56 Å resolution at the specimen.
Image processing
Particles were digitally extracted from the micrographs using the program X3D.24 A linear background gradient was subtracted from the masked particle images, and the contrast was normalized. Underfocus values were calculated using Robem‡ and ranged from − 1.45 to − 1.85 μm. Correlation methods (PPFT) were used to derive particle orientations, and the 3D reconstructions were generated by Fourier–Bessel inversion.25 Initial models were determined from 10 cycles of the Random Model Calculation.26 Additional rounds of image processing restricted the data search to the outer radii (r = 150–230 Å), but definition of the surface spikes did not improve. The data sets were randomly divided into two groups of particle images, and the two resulting maps were compared by Fourier shell correlation analysis, from which the resolution for both HAstV-1 and HAstV-8 maps was estimated to be ~ 25 Å using a correlation coefficient cutoff value of 0.5. The final 3D HAstV-8 and HAstV-1 maps were calculated from 332 (of 451) and 397 (of 583) particles, respectively, with selection based on their correlation coefficients. The handedness of both maps was set by matching the capsid features to those of the HEV structure.
Alignment search
The amino acid sequences of HAstV-1 (GenBank ID: CAA80132), HAstV-8 (GenBank ID: CAA91443), and human HEV (GenBank ID: AAA45736) were analyzed using the programs BLAST§ and clustalw2∥ . BLAST searches were run using the default selections and with multiple database options [nonredundant protein sequences, Protein Data Bank (PDB) sequences, and Swiss Protein sequences].
Density maps calculated from PDB models
PDB files containing full T = 3 icosahedrally related copies of the HEV (PDB ID: 3IYO 14) or HAstV (kindly provided by Dr. Yizhi Tao12) asymmetric unit (or spike-truncated sequences) were generated using the ViperDB utility GenerateOligomers. 27 Density maps of the models were then calculated from the PDB files with the EMAN program28 pdb2mrc using a step‐size equivalent to the cryoEM maps and truncated at 25 Å resolution. The HEV cryoEM map (emd ID: 5173) was low-pass filtered with the EMAN program proc3d.
Graphics
Contour levels were set to encompass the expected volume of 180 copies of VP70 (HAstV-8), or 180 copies of VP34 plus 60 copies of VP27, based on a protein density of 1.35 g/cm3. Figures of the maps and docking of PDB coordinates were performed using Chimera.29
Acknowledgements
We gratefully acknowledge Dr. Yizhi Tao for providing her model of astrovirus VP25. We also thank Dr. Barbie Ganser-Pornillos for helpful discussions and Yunuen Acevedo for technical support. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco [supported by National Institutes of Health (NIH) P41 RR001081]. This work was supported by funding from the Austrian Science Fund (J1348) to N.N., DGAPA-UNAM (219910) and CONACyT (79574) to E.M., NIH R21 AI43513 to S.M.M., and NIH R01 GM066087 to M.Y.
Edited by E. Nogales
Footnotes
Electon density maps have been deposited in the Electron Microscopy Data Bank and will be released upon publication.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2012.06.029
Appendix A. Supplementary Data
Supplementary materials
References
- 1.Appleton H., Higgins P.G. Letter: viruses and gastroenteritis in infants. Lancet. 1975;1:1297. doi: 10.1016/s0140-6736(75)92581-7. [DOI] [PubMed] [Google Scholar]
- 2.Madeley C.R., Cosgrove B.P. Letter: 28 nm particles in faeces in infantile gastroenteritis. Lancet. 1975;1:451–452. doi: 10.1016/s0140-6736(75)90858-2. [DOI] [PubMed] [Google Scholar]
- 3.Speroni S., Rohayem J., Nenci S., Bonivento D., Robel I., Barthel J. Structural and biochemical analysis of human pathogenic astrovirus serine protease at 2.0 Å resolution. J. Mol. Biol. 2009;387:1137–1152. doi: 10.1016/j.jmb.2009.02.044. [DOI] [PubMed] [Google Scholar]
- 4.Méndez-Toss M., Romero-Guido P., Munguía M.E., Méndez E., Arias C.F. Molecular analysis of a serotype 8 human astrovirus genome. J. Gen. Virol. 2000;81:2891–2897. doi: 10.1099/0022-1317-81-12-2891. [DOI] [PubMed] [Google Scholar]
- 5.Monroe S.S., Jiang B., Stine S.E., Koopmans M., Glass R.I. Subgenomic RNA sequence of human astrovirus supports classification of Astroviridae as a new family of RNA viruses. J. Virol. 1993;67:3611–3614. doi: 10.1128/jvi.67.6.3611-3614.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willcocks M.M., Brown T.D.K., Madeley C.R., Carter M.J. The complete sequence of a human astrovirus. J. Gen. Virol. 1994;75:1785–1788. doi: 10.1099/0022-1317-75-7-1785. [DOI] [PubMed] [Google Scholar]
- 7.Krishna N.K. Identification of structural domains involved in astrovirus capsid biology. Viral Immunol. 2005;18:17–26. doi: 10.1089/vim.2005.18.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Méndez E., Salas-Ocampo E., Arias C.F. Caspases mediate processing of the capsid precursor and cell release of human astroviruses. J. Virol. 2004;78:8601–8608. doi: 10.1128/JVI.78.16.8601-8608.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez-Fauquier A., Carrascosa A.L., Carrascosa J.L., Otero A., Glass R.I., Lopez J.A. Characterization of a human astrovirus serotype 2 structural protein (VP26) that contains an epitope involved in virus neutralization. Virology. 1994;201:312–320. doi: 10.1006/viro.1994.1296. [DOI] [PubMed] [Google Scholar]
- 10.Bass D.M., Qiu S. Proteolytic processing of the astrovirus capsid. J. Virol. 2000;74:1810–1814. doi: 10.1128/jvi.74.4.1810-1814.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Méndez E., Fernández-Luna T., López S., Méndez-Toss M., Arias C.F. Proteolytic processing of a serotype 8 human astrovirus ORF2 polyprotein. J. Virol. 2002;76:7996–8002. doi: 10.1128/JVI.76.16.7996-8002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong J., Dong L., Méndez E., Tao Y. Crystal structure of the human astrovirus capsid spike. Proc. Natl Acad. Sci. USA. 2011;108:12681–12686. doi: 10.1073/pnas.1104834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Risco C., Carrascosa J.L., Pedregosa A.M., Humphrey C.D., Sánchez-Fauquier A. Ultrastructure of human astrovirus serotype 2. J. Gen. Virol. 1995;76:2075–2080. doi: 10.1099/0022-1317-76-8-2075. [DOI] [PubMed] [Google Scholar]
- 14.Xing L., Li T.-C., Mayazaki N., Simon M.N., Wall J.S., Moore M. Structure of hepatitis E virion-sized particle reveals an RNA-dependent viral assembly pathway. J. Biol. Chem. 2010;285:33175–33183. doi: 10.1074/jbc.M110.106336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klenk H.-D., Rott R., Orlich M., Blödorn J. Activation of influenza A viruses by trypsin treatment. Virology. 1975;68:426–439. doi: 10.1016/0042-6822(75)90284-6. [DOI] [PubMed] [Google Scholar]
- 16.Lazarowitz S.G., Choppin P.W. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology. 1975;68:440–454. doi: 10.1016/0042-6822(75)90285-8. [DOI] [PubMed] [Google Scholar]
- 17.Du L., Kao R.Y., Zhou Y., He Y., Zhao G., Wong C. Cleavage of spike protein of SARS coronavirus by protease factor Xa is associated with viral infectivity. Biochem. Biophys. Res. Commun. 2007;359:174–179. doi: 10.1016/j.bbrc.2007.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Estes M.K., Graham D.Y., Mason B.B. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J. Virol. 1981;39:879–888. doi: 10.1128/jvi.39.3.879-888.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bodkin D.K., Nibert M.L., Fields B.N. Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J. Virol. 1989;63:4676–4681. doi: 10.1128/jvi.63.11.4676-4681.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salminen A., Wahlberg J.M., Lobigs M., Liljeström P., Garoff H. Membrane fusion process of Semliki Forest virus II: cleavage-dependent reorganization of the spike protein complex controls virus entry. J. Cell Biol. 1992;116:349–357. doi: 10.1083/jcb.116.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gough R.E., Collins M.S., Borland E., Keymer L.F. Astrovirus-like particles associated with hepatitis in ducklings. Vet. Rec. 1984;114:279. doi: 10.1136/vr.114.11.279-a. [DOI] [PubMed] [Google Scholar]
- 22.Guu T.S.Y., Liu Z., Ye Q., Mata D.A., Li K., Yin C. Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc. Natl Acad. Sci. USA. 2009;106:12992–12997. doi: 10.1073/pnas.0904848106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeager M., Berriman J.A., Baker T.S., Bellamy A.R. Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J. 1994;13:1011–1018. doi: 10.1002/j.1460-2075.1994.tb06349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conway J.F., Steven A.C. Methods for reconstructing density maps of “single” particles from cryoelectron micrographs to subnanometer resolution. J. Struct. Biol. 1999;128:106–118. doi: 10.1006/jsbi.1999.4168. [DOI] [PubMed] [Google Scholar]
- 25.Baker T.S., Cheng R.H. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 26.Yan X., Dryden K.A., Tang J., Baker T.S. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J. Struct. Biol. 2007;157:211–225. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carrillo-Tripp M., Shepherd C.M., Borelli I.A., Venkataraman S., Lander G., Natarajan P. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic Acid Res. 2009;37:D436–D442. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludtke S.J., Baldwin P.R., Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 29.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials