Abstract
SHFV is a member of a new virus family which includes the genus arterivirus. We have cloned and sequenced 6,314 nt from the 3′ end of the SHFV genome. This sequence encompasses nine complete ORFs which is three additional ORFs as compared to the other arteriviruses. We have numbered these ORFs 2a, 2b, 3, 4, 5, 6, 7, 8 and 9. At the 5′ end of this sequence is a partial ORF (ORF 1b) of 1590 nt and at the 3′ end is a poly(A) tract preceded by a 76 nt noncoding region. The coding capacity for each of the SHFV ORFs as well as the potential mass, pI and number of N-linked glycosylation sites for each of the encoded peptides was determined.
Keywords: Arterivirus, Genome organization, 3′ genes
Abbreviations: A, adenosine; BCV, bovine coronavirus; EAV, equine arteritis virus; kb, kilobase(s); LDV, lactate dehydrogenase-elevating virus; MHV, mouse hepatitis virus; nt, nucleotide(s); ORF, open reading frame; PRRSV, porcine reproductive and respiratory syndrome virus; sgRNA(s), subgenomic mRNA(s); SHFV, simian hemorrhagic fever virus.
1. Introduction
SHFV was first isolated in 1964 from macaque monkeys in a quarantine facility at the National Institute of Health after an epizootic of hemorrhagic fever within the colony (Palmer et al., 1968). This virus was recently reclassified into a new virus family, the Arteriviridae, consisting of the genus arterivirus and, along with SHFV, includes EAV, LDV and PRRSV.
Morphologically, the arteriviruses resemble the togaviruses. Arteriviral particles are between 55 and 60 nm in diameter and contain an isometric nucleocapsid surrounded by a viral envelope (Cavanagh et al., 1994). The coronaviruses, on the other hand, are pleomorphic viruses between 60 and 200 nm in diameter; their viral envelopes surround helical nucleocapsid structures (Siddell, 1995). Although morphologically distinct, the gene order and replication strategy of the arteriviruses are similar to those of the coronaviruses (Snijder and Spaan, 1995). The single-stranded RNA genomes of EAV, LDV and PRRSV are polycistronic containing eight ORFs. The ultimate (ORF 1a) and penultimate (ORF 1b) 5′ ORFs, which make up approximately two-thirds of the viral genomes, encode the proteins necessary for viral replication. The ORF 1b genes of the corona- and arteriviruses contain four conserved domains (Godeny et al., 1993). Beginning at the N-terminal portion of the ORF 1b peptide, the domains were identified as a putative RNA-dependent RNA polymerase domain, a putative Zn-finger domain, a putative helicase domain and a carboxy-terminal ORF 1b domain; this last domain, whose function is unknown, is unique to coronaviruses and arteriviruses (Godeny et al., 1993; Snijder and Spaan, 1995). The remaining 3′ one-third of the viral genomes contain six overlapping ORFs, some of which encode the viral structural proteins (Snijder and Spaan, 1995).
The first 954 nt at the 3′ terminus of the SHFV genome were reported previously (Godeny et al., 1995). That sequence included the 76 nt 3′ noncoding region and two complete and one partial ORFs. Both of the complete ORFs overlapped their adjacent 5′ ORFs (Godeny et al., 1995). The purpose of this study was to clone and sequence the remaining ORFs at the 3′ end of the SHF viral genome and to determine their coding capacities.
SHFV has been shown to contain at least four structural proteins on the virus particle: p15, p20, p42 and p54 (Godeny et al., 1995). These peptides were named for their electrophoretic mobility by SDS-PAGE analysis. Both p42 and p54 are glycoproteins which appear as broad bands by SDS-PAGE analysis and may each represent more than one peptide. Conversely, p15 and p20 are not glycosylated and have been identified as the SHFV capsid and membrane proteins, respectively (Godeny et al., 1995). Further, the ultimate ORF at the 3′ end of the SHFV genome has been identified as the p15 gene and the penultimate ORF encodes p20 (Godeny et al., 1995).
The nucleotide sequence of the entire 3′ end of the SHFV genome is reported here. This sequence includes all of the SHFV 3′ overlapping ORFs and demonstrates that SHFV has the potential to encode three additional peptides as compared to the other arteriviruses.
2. Experimental and discussion
2.1. Cloning and sequencing the 3′ end of the SHFV genome
To obtain viral RNA template for DNA synthesis, MA-104 cells were infected with SHFV, strain LVR 42-0/M6941, at a multiplicity of infection of 0.2. The viral genome was purified from the supernatant fluid 24 h postinfection as previously described (Godeny et al., 1995). cDNA transcripts of the SHFV genome were made using a genome walking strategy. Briefly, cDNA was made by reverse transcription of the viral RNA using synthetic DNA primers complementary to identified SHFV genome sequences downstream of the sequence to be determined. The cDNA was made double-stranded as previously described (Godeny et al., 1995) and inserted into the pGEM7ZF+ plasmid vector (Promega Corp.) using standard methods (Maniatis et al., 1989). The resulting clones were amplified in Escherichia coli, strain JM107, and screened as described previously (Godeny et al., 1995). Clones were sequenced by the dideoxy-chain termination method (Sanger et al., 1977) using the Sequenase DNA sequencing kit (U.S. Biochemical Corp.). In order to procure the majority sequence of the viral genome, multiple clones for each nucleotide were sequenced. If more than one clone was not available for a particular nucleotide, than the sequence was determined directly from the SHFV RNA genome using the RT RNA sequencing kit (U.S. Biochemical Corp.). Gaps between clones were also sequenced directly from the SHFV RNA genome.
Using these methods we obtained a 6314 nt sequence representing the 3′ end of the SHFV genome (Fig. 1 ). The deduced amino acid sequences from the encoded ORFs were generated using the Translation program from the University of Wisconsin Genetics Computer Group (GCG) software. At the 5′ end of the sequence, nt 1 through 153 encode the C-terminal portion of the conserved helicase domain and the entire carboxy-terminal ORF 1b domain is encoded by nt 778 through 1053. This data suggest that the partial ORF at the 5′ end of this sequence, beginning at nt 1 and ending at nt 1590, is the 3′ end of the SHFV ORF 1b gene. Between the SHFV ORF 1b gene and the 76 nt 3′ noncoding region are nine complete ORFs (Fig. 1). This is notable because the other arteriviruses have only six ORFs in this region. Therefore, the SHFV genome encodes three additional ORFs as compared to EAV, LDV and PRRSV.
Fig. 1.

The 3′ nt sequence of the SHFV genome clones (Genbank accession No. U63121) and the deduced amino acid sequence. Position 1 is the 5′ end of the nt sequence and position 6314 is the 3′ terminus. The ORFs are numbered and the potential N-linked glycosylation sites are indicated in bold type. Stop codons are indicated by an asterisk (*). Nucleotides encoding the ORF 1b conserved domains are underlined. Putative intergenic sequences are in bold type and underlined; whereas, known intergenic sequences are in bold type and double underlined.
Using conventional nomenclature (Cavanagh et al., 1990), we numbered the SHFV 3′ ORFs 2a, 2b, 3, 4, 5, 6, 7, 8 and 9 (Fig. 1). With the exceptions of ORFs 4 and 7, each of the SHFV 3′ ORFs overlap their adjacent 5′ ORFs (Fig. 1). A unique feature of the SHFV genome is that ORF 2a overlaps ORF 1b by 41 nt. Interestingly, none of the 3′ ORFs of the arterivirus genomes overlap their respective ORF 1b genes.
The arterivirus 3′ ORFs are translated from sets of nested, 3′ coterminal, sgRNAs. Each of these sgRNAs contain an identical 5′ leader sequence which is joined to the mRNA at a conserved intergenic sequence. The intergenic sequences of the two smallest SHFV sgRNAs were previously reported (Zeng et al., 1995) and are shown in Fig. 1. Expectedly, each of the remaining 3′ ORFs contain at least one similar putative intergenic sequence within 200 nt upstream of the respective start codon (Fig. 1). Preliminary sgRNA sequence analyses suggest that the putative intergenic sequence upstream of ORF 2b may not be utilized, implying that ORFs 2a and 2b may be encoded on the same viral sgRNA (Godeny, unpublished data).
2.2. Coding capacities of the SHFV 3′ ORFs
The potential coding capacities of the SHFV 3′ ORFs are listed in Table 1 . The smallest ORF is ORF 9 which encodes a peptide of approximately 12.3 kDa and the largest ORF is ORF 2a which encodes a peptide of 31.8 kDa. ORF 7 encodes the second largest peptide with a mass of 31.3 kDa. The remaining ORFs encode peptides between 17.8 and 24.1 kDa. All of the ORFs encode neutral to basic peptides with pI values between 6.2 and 11.7, as determined by the PeptideSort computer program within the GCG software. Also, each of the nine SHFV ORFs encode peptides containing at least one N-linked glycosylation site (Fig. 1 and Table 1). The ORF 2b and 7 peptides contain the most N-linked glycosylation sites with 10 and 8, respectively. Four of the 3′ ORFs encode peptides containing only one or two potential N-linked glycosylation sites (Table 1). It has previously been reported that although the SHFV capsid and membrane proteins, which are encoded by ORFs 8 and 9, contain potential N-linked glycosylation sites, they are not glycosylated proteins (Godeny et al., 1995). Therefore, it remains to be determined which of the remaining potential SHFV peptides encoded by the 3′ ORFs are glycosylated.
Table 1.
Characteristics of the deduced peptides encoded by the SHFV 3′ ORFs
| ORF | Amino acids | Peptide mass(kDa) | pI | Glycosylation sites | Identity |
|---|---|---|---|---|---|
| 2a | 281 | 31.8 | 10.2 | 1 | ? |
| 2b | 204 | 22.7 | 6.7 | 10 | ? |
| 3 | 205 | 23.1 | 6.2 | 4 | ? |
| 4 | 214 | 24.1 | 9.5 | 2 | ? |
| 5 | 179 | 19.1 | 6.2 | 6 | ? |
| 6 | 182 | 19.6 | 7.5 | 4 | ? |
| 7 | 278 | 31.3 | 8.2 | 8 | ? |
| 8 | 162 | 17.8 | 11.3 | 2 | p20 |
| 9 | 111 | 12.3 | 11.7 | 2 | p15 |
2.3. Comparison of the SHFV, arterivirus and coronavirus genomes
Members of the family Arteriviridae share genome organizations and replication strategies with members of the family Coronaviridae. The EAV, LDV and PRRSV genomes are approximately 12 700 (Den Boon et al., 1991), 14 200 (Godeny et al., 1993) and 15 100 nt in length (Meulenberg et al., 1993), respectively. On the other hand, the genomes of the coronaviruses are approximately 30 000 nt in length (Siddell, 1995), twice as long as the genomes of the arteriviruses. ORFs 1a and 1b are located at the 5′ end of the coronavirus and arterivirus genomes and encompass approximately two-thirds of the genomes. The gene products of these two ORFs are non-structural proteins which are necessary for viral replication. The remaining one-third of the genomes contain the 3′ ORFs, some of which encode structural proteins found on the viral particle. The 3′ SHFV ORFs, ORFs 2a through 9, and the 3′ noncoding region are encoded on a total of 4754 nt. Assuming that these ORFs make up one-third of the viral genome, it can be deduced that the SHFV genome is approximately 14 300 nt in length. Indeed, biochemical analysis of purified SHFV RNA suggested that the genome is 15 000 nt in length (Sagripanti, 1984)
The genome organization of the 3′ ORFs of MHV, the prototype coronavirus, EAV, the prototype arterivirus, and SHFV are shown in Fig. 2 . The 3′ end of the SHFV genome contains nine ORFs. Therefore, based on genome organization, SHFV appears more similar to MHV, which also has nine 3′ ORFs, than to EAV, which has only six 3′ ORFs (Fig. 2).
Fig. 2.
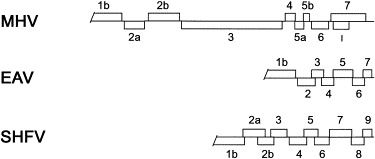
Schematic representations of the genome organizations of the 3′ ORFs of MHV, the prototype coronavirus, EAV, the prototype arterivirus, and SHFV. The sizes of the genes and genomes are drawn approximately to scale.
Some of the proteins encoded by the 3′ ORFs on the MHV genome have been identified [reviewed in Lai (1990)and Luytjes (1995)]. ORF 3 encodes the large-surface protein or spike protein, S. ORFs 5b and 6 encode the small-membrane, sM, and membrane, M, proteins, respectively. The nucleocapsid protein (N) gene is ORF 7 and ORF 2b is the hemagglutinin-esterase (HE) gene. Among the coronaviruses, the HE protein is unique to MHV and BCV (Lai, 1990). ORF 2a encodes a 30 kDa peptide which is believed to be a nonstructural protein. The functions of the gene products produced by ORFs 4, 5a and I have not been identified but may also be nonstructural in nature (Luytjes, 1995). Interestingly, it has been shown that the ORF 2a and 4 gene products of MHV are nonessential for virus replication [reviewed in Luytjes (1995)].
The peptides encoded by the arterivirus ORFs have not yet been fully identified; however, they show little sequence homology to those of the coronaviruses. ORF 7 of EAV, LDV and PRRSV has been shown to encode the viral capsid, C, protein and ORF 6 encodes the membrane, M, protein (Godeny et al., 1990; De Vries et al., 1992; Muelenberg et al., 1995). ORFs 2 and 5 of EAV and LDV were reported to encode the small (GS) and large (GL) envelope glycoproteins (De Vries et al., 1992; Faaberg and Plagemann, 1995). ORF 5 of PRRSV was also shown to encode a viral envelope glycoprotein. However, the PRRSV ORF 2 gene product could not be detected in purified virus particles (Muelenberg et al., 1995). It has been reported that the gene products of ORFs 3 and 4 of PRRSV are also envelope glycoproteins but, because high concentrations of virus were needed for detection, they are probably present on the virion in low copy number (Van Nieuwstadt et al., 1996). These gene products in EAV and LDV have not been identified.
Few of the gene products of SHFV have been identified. We previously showed that the ultimate, ORF 9, and penultimate, ORF 8, 3′ ORFs encode the SHFV capsid, p15, and membrane, p20, proteins, respectively (Godeny et al., 1995). The ORF 7 gene likely encodes one of the envelope glycoproteins based on hydrophobicity analysis (Wang et al., 1995) and the number of potential N-linked glycosylation sites (Table 1) on the encoded peptide. Computer analysis of the SHFV ORF 7 peptide using the GAP program within the GCG software package shows a 53% and 57% amino acid sequence similarity between this peptide and the ORF 5 products of LDV and LV, respectively, further suggesting that the SHFV ORF 7 encodes the SHFV large envelope glycoprotein, p54. The gene products of the SHFV ORFs 2a through 6 remain to be identified. Computer analyses of the potential products from these ORFs reveal a low amino acid sequence similarity (39% to 49%) with the ORFs 2 through 4 products of EAV, LDV and LV (data not shown). Presently, there is no evidence that any of the arterivirus 3′ ORFs encode nonstructural proteins. However, since there are three additional ORFs on the SHFV genome as compared to the genomes of the other arteriviruses, it is possible that some, or all, of these additional SHFV ORFs may encode nonstructural peptides. Due to packaging constraints on isometric viruses, it is unlikely, however, that any of the SHFV 3′ ORFs encode nonessential peptides as do the coronaviruses.
Peptides encoded by the 3′ ORFs of the arteriviruses and coronaviruses are translated from nested sgRNAs which are 3′ coterminal. Since only one ORF is translated from each of the arterivirus sgRNAs and SHFV contains nine ORFs at the 3′ end of the genome, it is expected that SHFV produces nine sgRNAs during replication. However, Northern blot analyses show that SHFV produces a set of six sgRNAs during replication (Godeny et al., 1995; Zeng et al., 1995). Therefore, further studies are underway to determine whether (1) more than one ORF is translated from some of the SHFV sgRNA, (2) some of the SHFV ORFs are silent, or (3) some of the SHFV sgRNAs are produced at levels below the detection limits of Northern analyses.
3. Conclusion
We have cloned and sequence 6314 nt from the 3′ end of the SHF viral genome. This sequence, beginning in ORF 1b and ending with the 3′ poly(A) tract, includes all of the SHFV 3′ ORFs and the 3′ noncoding region. Based on morphology, gene order, genome characteristics and replication strategy, SHFV should be included in the genus arterivirus. However, the SHFV ORF 2a overlaps ORF 1b which is a unique feature of this virus as compared to the other arteriviruses. Also, the SHFV genome contains three additional ORFs as compared to EAV, LDV and PRRSV and therefore appears to be more complex than these other arteriviruses. The identity of the gene products encoded by all of the 3′ SHFV ORFs is currently in progress.
Acknowledgements
We thank Dr. Matthew Philpott for technical assistance. This research was supported by the Public Health Service grant RR06841 from NCRR.
J.A. Engler
References
- Cavanagh, D., D.A. Brian, M. Brinton, L. Enjuanes, K.V. Holmes, M.C. Horzinek, M.M.C. Lai, P.G.W. Plagemann, S. Siddell, W.J.M. Spaan, F. Taguchi and P.J. Talbot, 1994. Revision of the taxonomy of the Coronavirus, Torovirus and Arterivirus genera. Arch. Virol. 135, 227–237. [DOI] [PMC free article] [PubMed]
- Cavanagh, D., D.A. Brian, L. Enjuanes, K.V. Holmes, M.M.C. Lai, H. Laude, S.G. Siddell, W. Spaan, F. Taguchi and P.J. Talbot, 1990. Recommendations of the coronavirus study group for the nomenclature of the structural proteins, mRNAs, and genes of coronaviruses. Virology 176, 306-307. [DOI] [PMC free article] [PubMed]
- De Vries, A.A.F., E.D. Chirnside, M.C. Horzinek and P.J.M. Rottier, 1992. Structural proteins of equine arteritis virus. J. Virol. 66, 6294-6303. [DOI] [PMC free article] [PubMed]
- Den Boon, J., E.J. Snijder, E.D. Chirnside, A.A.F. de Vries, M.C. Horzinek and W.J.M. Spaan, 1991. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike family. J. Virol. 65, 2910-2920. [DOI] [PMC free article] [PubMed]
- Faaberg, K.S. and P.G.W. Plagemann, 1995. The envelope proteins of lactate dehydrogenase-elevating virus and their membrane topography. Virology 212, 512-525. [DOI] [PubMed]
- Godeny, E.K., L. Chen, S.N. Kumar, S.L. Methven, E.V. Koonin and M.A. Brinton, 1993. Complete genomic sequence and phylogenetic analysis of the lactate dehydrogenase-elevating virus (LDV). Virology 194, 585-596. [DOI] [PMC free article] [PubMed]
- Godeny, E.K., D.W. Speicher and M.A. Brinton, 1990. Map location of lactate dehydrogenase- elevating virus (LDV) capsid protein (Vp1) gene. Virology 177, 768-771. [DOI] [PMC free article] [PubMed]
- Godeny, E.K., L. Zeng, S.L. Smith and M.A. Brinton, 1995. Molecular characterization of the 3′ terminus of the simian hemorrhagic fever virus genome. J. Virol. 69, 2679-2683. [DOI] [PMC free article] [PubMed]
- Lai, M.M.C., 1990. Coronavirus: organization, replication and expression of genome. Annu. Rev. Microbiol. 44, 303-333. [DOI] [PubMed]
- Luytjes, W., 1995. Coronavirus gene expression: Genome organization and protein synthesis. In: S.G. Siddell (Ed.), The Coronaviridae. Plenum Press, New York, pp. 33-54.
- Maniatis, T., E.F. Fritsch and J. Sambrook, 1989. Molecular Cloning: A Laboratory Manual. Cold Springs Harbor Laboratory, Cold Springs Harbor, NY.
- Meulenberg, J.J.M., M.M. Hulst, E.J. de Meijer, P.L.J.M. Moonen, A. den Besten, E.P. de Kluyver, G. Wensvoort and R.J.M. Moormann, 1993. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS) is related to LDV and EAV. Virology 192, 62-72. [DOI] [PMC free article] [PubMed]
- Muelenberg, J.J.M., A. Petersen-den Besten, E.P. de Kluyver, R.J.M. Moormann, W.M.M. Schaaper and G. Wensvoort, 1995. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad Virus. Virology 206, 155-163. [DOI] [PMC free article] [PubMed]
- Palmer, A.E., A.M. Allen, N.M. Tauraso and A. Shelokov, 1968. Simian hemorrhagic fever. I. Clinical and epizootiologic aspects of an outbreak among quarantined monkeys. Am. J. Trop. Med. Hyg. 17, 404-412. [PubMed]
- Sagripanti, J.L., 1984. The genome of simian hemorrhagic fever virus. Arch. Virol. 82, 61-72. [DOI] [PubMed]
- Sanger, F., S. Nicklen and A.R. Coulson, 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74, 5463-5467. [DOI] [PMC free article] [PubMed]
- Siddell, S.G., 1995. The Coronaviridae: An Introduction. In: S.G. Siddell (Ed.), The Coronaviridae. New York, Plenum Press, New York, pp. 1-10.
- Snijder, E.J. and W.J.M. Spaan, 1995. The coronaviruslike superfamily. In: S.G. Siddell (Ed.), The Coronaviridae. Plenum Press, New York, pp. 239-255.
- Van Nieuwstadt, A.P., J.J.M. Meulenberg, A. van Essen-Zandbergen, A. Petersen-den Besten, R.J. Bende, R.J.M. Moormann and G. Wensvoort, 1996. Proteins encoded by open reading frames 3 and 4 of the genome of Lelystad virus (Arteriviridae) are structural proteins of the virion. J. Virol. 70, 4767-4772. [DOI] [PMC free article] [PubMed]
- Wang, X.C., S.L. Smith and E.K. Godeny, 1995. Characterization of the 3′ terminal end of the simian hemorrhagic fever virus (SHFV) genome and the viral structural proteins. American Society for Virology, Austin, TX.
- Zeng, L., E.K. Godeny, S.L. Methven and M.A. Brinton, 1995. Analysis of simian hemorrhagic fever virus (SHFV) subgenomic RNAs, junction sequences, and 5′ leader. Virology 207, 543-548. [DOI] [PubMed]


