Abstract
The diagnosis of bacterial infections relies on isolation of the bacterium, which is rarely achieved when needed for patient management. Furthermore, culture is poorly suited to the diagnosis of polymicrobial infections. Finally, a syndromic approach should target both bacteria and viruses causing the same syndrome. The detection of specific DNA sequences in clinical specimen, using DNA microarrays, is an alternative. Microarrays were first used as a diagnostic tool in 1993, to identify a hantavirus associated with an outbreak of acute respiratory diseases. The main advantage of microarrays is multiplexing, enabling exploration of the microbiota and pathogen detection in bacteremia, respiratory infections, and digestive infections: circumstance in which DNA arrays may lack sensitivity and provide false negatives. Enrichment of sampling can increase sensitivity. Furthermore, chips allow typing Streptococcus pneumoniae and detecting resistance in Staphylococcus aureus (MRSA) and Mycobacterium tuberculosis (rifampicin, isoniazid, fluoroquinolones). However, the cost and high technical requirements remain a problem for routine use of this bacterial infection diagnostic technology.
Keywords: Diagnosis, DNA microarray, Infectious diseases
Résumé
Le diagnostic des infections bactériennes repose sur l’isolement du pathogène, qui est rarement réalisé dans le temps du soin. La culture est mal adaptée au diagnostic des infections polymicrobiennes. Enfin, une approche syndromique doit cibler en parallèle les bactéries et les virus responsables d’un même syndrome. Une alternative est la détection de séquences ADN spécifiques dans l’échantillon clinique par les puces à ADN, dont la première application en 1993 était l’identification d’un hantavirus associé à une épidémie de maladies respiratoires. L’avantage essentiel des puces à ADN est le multiplexage, permettant l’exploration des microbiotes et la détection des pathogènes au cours des bactériémies, des infections respiratoires et des infections digestives, situation dans laquelle les puces à ADN peuvent manquer de sensibilité et donner des faux-négatifs. Une étape d’enrichissement du prélèvement peut palier cette limite. Également, les puces permettent le typage de Streptococcus pneumoniae et la détection de la résistance chez Staphylococcus aureus (méthicilline) et Mycobacterium tuberculosis (rifampicine, izoniaside, fluoroquinolones). Le coût et la technicité demeurent deux freins au déploiement en routine de cette technologie pour le diagnostic des infections bactériennes.
Mots clés: Diagnostic, Infections, Puces à ADN
1. Introduction
The diagnosis of bacterial infections in the laboratory relies on isolation of the bacterium, its identification, and antibiograms. This direct diagnostic approach is limited by: the delay before obtaining culture results, sometimes after caregiving; the presence of non-cultivable bacteria in the sample because of the presence of inhibitors such as antibiotics or because of inappropriate culture conditions; and the capacity to isolate and differentiate the various colonies of a polymicrobial sample. For example, around 75% of bacterial species in the digestive microbiotium, cannot be identified routinely with culture techniques [1]. An alternative to this direct diagnostic is the detection de universal gene sequences 16S rRNA or rpoB [2], [3] by PCR followed by hybridization of a fluorescent oligonucleotide probe, with real time PCR [4]. As an alternative, the product of PCR amplification may be hybridized on a solid base fixing a great number of oligonucleotide probes, using the DNA microarray technique which is reviewed in this article. Searching for “DNA microarray” in the NCBI search engine (http://www.ncbi.nlm.nih.gov/pubmed/) gives 60,630 references (May 2012), showing the importance of the molecular biology tool described for the first time in 1995 [5] (Fig. 1 ). The “DNA microarray and infectious diseases” combination, gives 721 references (May 2012); the first reported use of this tool, for the diagnosis of a hantavirus infection, was published in 1993 [6].
Fig. 1.
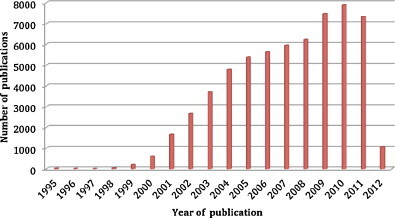
Histogram of “DNA microarray” topics published in the PubMed database from 1995 to 2012 (May 2012).
Histogramme des publications concernant le sujet « DNA microarray » parues dans la banque de données Pubmed de 1995 à 2012 (mai 2012).
2. Principle of DNA microarrays
The DNA microarray technique was adapted from the Southern blot by replacing the nylon base by a glass or silicon slide allowing covalent fixation of several thousand oligonucleotide probes. The probes correspond to: universal genes 16S rRNA [2] or rpoB [3]; or to genus, species, or serotype specific genes; to genes coding for resistance to antibiotics; or to toxinic genes. The DNA microarray technique includes several steps, such as manual or automatic extraction of nucleic acids from the clinical sample, their amplification by PCR, the labeling of amplification products by a fluorescent cyanine (Cy3- or Cy5-dCTP): a monochrome labeling is less expensive but a two-colored one has a better reproducibility [7], [8]; then hybridization of labeled nucleic acids for 24 to 50 hours on the DNA microarray which is later scanned to measure the specific probe/DNA interactions of the sample. These interactions are measured by fluorescence as numerical data (Fig. 2 ). The total time for the procedure is inferior to 4 hours, not including time for hybridization.
Fig. 2.
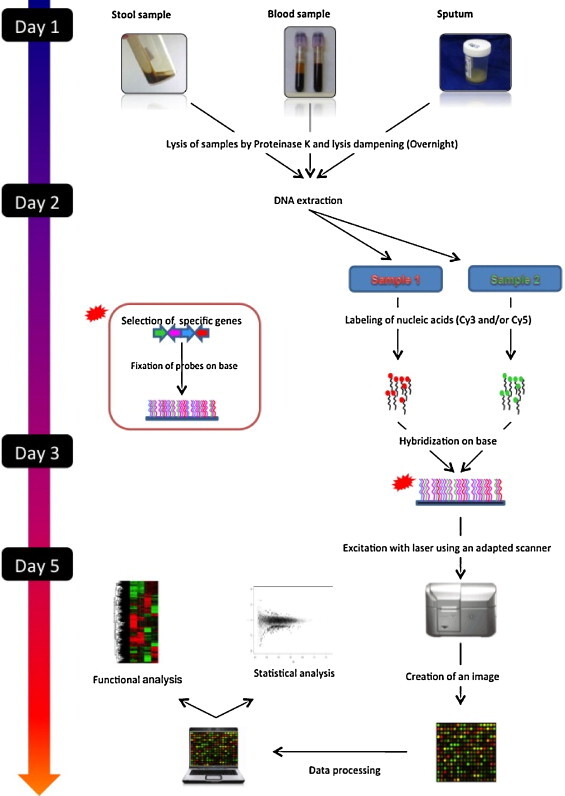
Protocol for the DNA microarray analysis of clinical samples.
Protocole d’analyse des échantillons cliniques par puce à ADN.
There are few DNA microarrays available on the market for the diagnosis of bacterial infections. Two firms, Agilent (http://www.agilent.com) and Affymetrix (http://www.affymetrix.com/estore/), currently market custom made or prefabricated chips available in France. The Phylochip™ (Affymetrix; Santa Clara, California) chip carries 1.1 million probes for 25 nucleotides allowing the detection of the 16S rRNA gene for 60,000 bacterial species [9]. The CapitalBio Corporation firm (Beijing, China; http://www.capitalbio.com) markets a DNA chip for the identification and determination of resistance profile of 17 mycobacterial species, the most frequently isolated in laboratories. Finally, Prove-it™ Sepsis StripArray (Mobidiag, Helsinki, Finland) is an automated system coupling a PCR stage and a DNA microarray analysis for the detection of bacteria responsible for bacteremia.
3. Study of the microbiota
The human body carries interacting microorganisms called microbiota, containing ten times more cells than the human body and 100 times more genes than the human genome [10]. The DNA Phylochip™ (Affymetrix) allows investigating the human microbiota [9] (Fig. 3 ). Among these, the intestinal microbiota counting 1011 to 1014 bacteria [11] has a major role in the individual's homeostasis and in some diseases [12]. Three teams studied the intestinal microbiota with DNA microarrays carrying the probes of 25 to 40 bases targeting the 16S rRNA gene and detecting from 500 to 1140 bacterial species [11], [13], [14]. These authors reported that the intestinal microbiota includes a part found in all individuals belonging to phyla Actinobacteria, Firmicutes, and Bacteroidetes, and a part specific to each individual [13]. DNA microarrays also allowed observing a decrease of some Firmicutes and an increase of Enterococcus, Clostridium difficile, Escherichia coli, Shigella flexneri, and Listeria spp. species in patients presenting with Crohn's disease compared to healthy individuals [14].
Fig. 3.
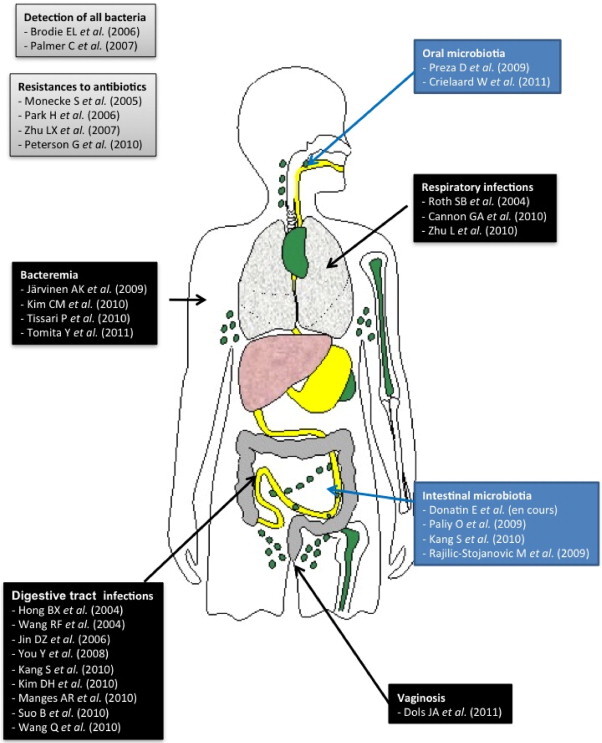
DNA microarrays used to investigate various human microbiota, or for diagnostic purposes.
Les puces à ADN utilisées pour l’exploration des différents microbiotes humains, ou à visée diagnostique.
The oral microbiota was also investigated with DNA microarrays detecting the gene 16S rRNA with probes carrying 18 to 20 nucleotides [15], [16]. The Human Oral Microbe Identification Microarray (HOMIM) allowed investigating explorer the oral microbiota in five patients presenting with oral cancer, five patients presenting with pre-cancerous oral lesions, and ten healthy controls [15]. The authors of the second study analyzed the oral microbiota in 74 children 3 to 18 years of age, in four different groups according to the development stage of their teeth: (1) milk teeth, (2) early mixed dentition, (3) late mixed dentition, and (4) final dentition. The results of the two studies prove that Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria are the most prevalent phyla in the oral microbiota depending on the individual's dentition: there are more proteobacteria than bacteroidetes in the final dentition group, contrary to the 3 other groups. The prevalence of Prevotella increases with age [16]. Finally, Porphyromonas catoniae and Neisseiria flavescens are significantly correlated to the presence of carries [16].
4. Diagnosis of bacterial infections
4.1. Respiratory infections
Three DNA microarrays were designed for the diagnosis of respiratory infections. A chip targeting the variable regions of the gyrB and parE of the bacterial genes detects Corynebacterium diphteriae, Fusobacterium necrophorum, Haemophilus influenza, Legionella pneumophila, Moraxella catarrhalis, Mycoplasma pneumoniae, Staphylococcus aureus, Streptococcus pneumoniae and Streptococcus pyogenes. Comparing with culture of 65 middle ear samples and 29 throat samples proved a better sensibility of the chip [17].
An other chip detecting Adenovirus, Bocavirus, Coronavirus type 229E, OC43, NL63, HKU1, Human Metapneumovirus types A and B, Influenza A-C, Para-influenza 1-4, Respiratory Syncytial Virus types A and B, Rhinovirus, Chlamydia pneumoniae and M. pneumoniae was used on 50 throat samples from adults during the winter 2007–2008 in Ireland [18]. The results proved that the chip gave a reliable diagnosis within one day. A third chip detecting 17 species of mycobacteria including Mycobacterium tuberculosis allowed directly analyzing 195 sputum samples from patients suspected to present with pulmonary tuberculosis, along with Ziehl staining and culture on agar [19]. Hundred and sixteen samples were found positive by the chip and 79 negative. The 116 positive samples were also positive in culture, but the chip identified a culture-negative sample later. The chip detected Mycobacterium intracellulare in three patients confirmed by sequencing of the strains [19].
These various studies confirm the sensibility and specificity of the DNA microarray compatible with immediate use on respiratory tract samples.
4.2. Digestive infections
Two DNA microarrays target the variable regions of the bacterial ribosome sub-units 16S and 23S to identify 15 bacterial species frequently associated with food infections, such as E. coli O157:H7, Salmonella enterica, Shigella dysenteriae, and Vibrio cholerae [20], [21]. A second kind of DNA microarray targets the virulence genes of enteropathogenic bacteria, for a combined detection of E. coli, V. cholerae, Vibrio parahaemolyticus, S. enterica, Campylobacter jejuni, S. dysenteriae, S. flexneri, Shigella sonnei, Yersinia enterocolitica, and L. monocytogenes [22]; or of enterotoxinogenic E. coli [ETEC], enterohemorrhagic E. coli [EHEC], S. enterica serovar Enteritidis, S. enterica serovar Typhimurium, S. flexneri, S. sonnei, V. cholerae, V. parahaemolyticus, Vibrio vulnificus, and Y. enterocolitica [23]; or of the 19 most frequent serogroups of enterotoxic E. coli [24]. A common limitation of all these chips is sensibility of detection. Indeed, some enteropathogens such as Salmonella spp. or Campylobacter spp. are presents in low inoculum less or equal to 103 organisms/mL of diarrhea stools [25], [26]. These inoculums are at the DNA microarray's threshold of detection [20], [21], because of interference with some commensal species. A PCR amplification stage of pathogens before hybridization partly decreases this limitation [21].
One DNA microarray detects enteropathogenic bacteria E. coli O157:H7, S. enterica, L. monocytogenes, and C. jejuni in fresh food samples [27]. Twelve identical hybridization zones allow analyzing 12 different samples on a single slide so as to reduce test cost.
4.3. Bacteremia
Three DNA microarrays were described for the diagnosis of bacteremia. A chip targeting the bacterial gyrB and parE detects Acinetobacter baumanii, Enterococcus faecalis, Enterococcus faecium, H. influenza, Klebsiella pneumoniae, L. monocytogenes, N. meningitidis, S. aureus, S. epidermidis, Streptococcus agalactiae, S. pneumoniae and S. pyogenes, and the mecA gene associated with methicillin resistance in S. aureus [28].
It was tested on 146 positive hemoculture and 40 negative hemocultures, with a sensibility of 96% and a specificity of 98% [28].
Another DNA microarray targeting the internal transcribed spacer (ITS) region located between the ribosomal sub-units 16S and 23S, carries a universal positive probe control, two probes for the detection of Gram-positive bacteria, 1 probe for the detection of Gram-negative bacteria, nine genus specific probes (Enterococcus, Listeria, Staphylococcus, Streptococcus, Bacteroides, Enterobacter/Klebsiella, Haemophilus, Pseudomonas and Serratia) and 30 species specific probes [29]. Its sensibility was 85.8% on 825 blood samples within 1 hour [29]. The Prove-it™ Sepsis StripArray includes PCR targeting the genes gyrB and parE to identify 50 Gram-positive and Gram-negative bacteria responsible for bacteremia, and the mecA gene associated with methicillin resistance in less than 3.5 hours [28], [30]. It was tested on 3318 blood samples 63.5% of which were positive in culture, a proved a sensibility of 94.7% and a specificity of 98.8% [30]. The results of these studies prove that DNA microarrays are perfectly adapted for the diagnosis of bacteremia and the detection of methicillin resistance in S. aureus with a sensibility and a specificity of 100%, with faster results than standard methods relying on culture; indeed these two studies include a hybridization stage which lasts less than 1 hour.
4.4. Serotyping
A DNA microarray targeting the genes of glycosyltransferase (GT) [31] allows typing S. pneumoniae strains correlated with serotyping of this bacterium which includes 90 different serotypes [32]. Indeed, 25.6% of these serotypes are responsible for more than 90% of S. pneumoniae infections [33]. This chip, the first to use GT genes as molecular target, allows determining the serogroup of the S. pneumoniae strain in a single step.
4.5. Detection of resistance to antibiotics
4.5.1. Non-specific chip
Peterson et al. developed a DNA microarray allowing identification of 43 pathogenic bacteria (human and animal) targeting genes of resistance to antibiotics and genes of virulence [34]. The chip carries specific 227 probes for 90 bacterial genes of resistance, 99 probes targeting the genes of resistance to 20 metals, 113 specific probes for genes of virulence, 31 probes targeting transferable elements and seven probes corresponding to positive controls. The specificity of the chip was assessed on cultures of S. enterica Typhimurium, Fusobacterium necrosum, E. faecalis, and E. coli O157:H7. Little non-specific hybridizations was observed during the test. The tests made with this chip allowed confirming the results of previous studies with chips detecting only the msrC gene in strains of methicillin resistant E. faecium [35] and only the pbp5 gene in strains of penicillin resistant E. faecium [36]. All the results obtained with the chip were confirmed by PCR.
4.5.2. Staphylococcus aureus
A DNA microarray was developed, detecting five specific S. aureus genes by targeting the gene 23S rRNA, 23 genes of resistance to antibiotics (macrolides, lincosamides, streptogramins, tetracyclin, cotrimoxazole, and aminoglycosides) and ten genes coding for toxins. The chip was tested by analyzing 100 methicillin resistant S. aureus strains [37]. A second chip identifying S. aureus versus S. non-aureus (gene 16S rRNA) and detecting the genes mecA and blaZ for methicillin and penicillin resistance, aac(6′)-Ie-aph(2″) for resistance to aminoglycosides, ermA and ermC for resistance to streptogramins, and msrA for resistance to macrolides, has a sensibility of 94.8 to 99% and a specificity of 69.3 to 99.2% according to molecular targets [38]. Nevertheless, some atypical results have been observed: some isolates have given signals of hybridization for the gene blaZ even though no β-lactamase activity was detected for these strains. One isolate, negative for the detection of the gene blaZ, had a β-lactamase activity. The authors explained this by a proved sequence variation at the gene blaZ level [37]. In this study, a decreased hybridization time from 90 to 30 minutes decreased the intensity of fluorescence by 10 to 30% [38].
4.5.3. Tuberculosis
M. tuberculosis resistance to rifampicin is related to the mutations in a region of 81 base pairs of the gene rpoB which codes for the RNA polymerase sub-unit β [39], [40], [41], [42], [43]. Isoniazid resistance is partly associated to a mutation of the gene katG which codes for a catalase-peroxidase and partly to a mutation of the regulator gene inhA [41], [42]. A DNA microarray detecting these various mutations showed a sensibility of 93% and a specificity of 98.4% for the detection of rifampicin resistance; and a sensibility of 71.4% and a specificity of 97.6% for the detection of isoniazid resistance [43]. The authors recommend using chips to screen for multi-drug resistant (MDR) bacteria if tuberculosis is suspected.
5. Conclusions
DNA microarrays for the diagnosis of infectious diseases were first described in 1993 [6] and have been the topic of a great number of publications (Fig. 1). Indeed, DNA microarrays allow a microbiological diagnosis within 48 hours (including hybridization time) compatible with the delay for medical management of patients. DNA microarrays also allow multiplexed detection adapted to a syndromic approach of infectious diseases diagnosis and to the diagnosis of polymicrobial infections extended to viruses. All authors agree that DNA microarrays have a sensibility and specificity at least equal to reference tests.
Nevertheless, designing the chip is a crucial step to obtain reliable results. The various chosen probes should have a very similar length and fusion temperature to optimize hybridization conditions, the current limiting stage. Several teams have solved the problem of cross-reaction by fixing, on the slide, specific probes for the same pathogen, in a redundant manner, and the current standard is a triplicate test. The sample preparation and the labeling of nucleic acids should be even more simplified for a routine use in bacteriological laboratories, to decrease time and cost of the test, and to integrate DNA microarrays among available techniques for the diagnostic point-of-care of infectious diseases [44]. Indeed, the currently available techniques, real time PCR and immunochromatographic methods are rapid diagnostic methods for a single pathogen per test. DNA microarrays have the advantage to allow multiplexed diagnosing perfectly adapted to a syndromic approach of infectious diseases diagnosis.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
References
- 1.Candela M., Consolandi C., Severgnini M., Biagi E., Castiglioni B., Vitali B., et al. High taxonomic level fingerprint of the human intestinal microbiota by Ligase Detection Reaction–Universal Array approach. BMC Microbiol. 2010;10:116. doi: 10.1186/1471-2180-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drancourt M., Berger P., Raoult D. Systematic 16S rRNA gene sequencing of atypical clinical isolates identified 27 new bacterial species associated with humans. J Clin Microbiol. 2004;42:2197–2202. doi: 10.1128/JCM.42.5.2197-2202.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adékambi T., Drancourt M., Raoult D. The rpoB gene as a tool for clinical microbiologists. Trends Microbiol. 2009;17:37–45. doi: 10.1016/j.tim.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Arya M., Shergill I.S., Williamson M., Gommersall L., Arya N., Patel H.R. Basic principles of real-time quantitative PCR. Expert Rev Mol Diagn. 2005;5:209–219. doi: 10.1586/14737159.5.2.209. [DOI] [PubMed] [Google Scholar]
- 5.Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 6.Nichol S.T., Spiropoulou C.F., Morzunov S., Rollin P.E., Ksiasek T.G., Feldmann H., et al. Genetic identification of a Hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262:914–917. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- 7.Jaluria P., Konstantopoulos K., Betenbaugh M., Shiloach J. A perspective on microarrays: current applications, pitfalls, and potential uses. Microb Cell Fact. 2007;6:4. doi: 10.1186/1475-2859-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leroy Q., Raoult D. Review of microarray studies for host-intracellular pathogen interactions. J Microbiol Methods. 2010;81:81–95. doi: 10.1016/j.mimet.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Kellogg C.A., Piceno Y.M., Tom L.M., DeSantis T.Z., Zawada D.G., Andersen G.L. PhyloChip™ microarray comparison of sampling methods used for coral microbial ecology. J Microbiol Methods. 2012;88:103–109. doi: 10.1016/j.mimet.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 10.Gill S.R., Pop M., Deboy R.T., Eckburg P.B., Turnbaugh P.J., Samuel B.S., et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paliy O., Kenche H., Abernathy F., Michail S. High-throughput quantitative analysis of the human intestinal microbiota with a phylogenetic microarray. Appl Environ Microbiol. 2009;75:3572–3579. doi: 10.1128/AEM.02764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mai V., Morris J.G., Jr. Colonic bacterial flora: changing understandings in the molecular age. J Nutr. 2004;134:459–464. doi: 10.1093/jn/134.2.459. [DOI] [PubMed] [Google Scholar]
- 13.Rajilic-Stojanovic M., Heilig H.G., Molenaar D., Kajander K., Surakka A., Smidt H., et al. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol. 2009;11:1736–1751. doi: 10.1111/j.1462-2920.2009.01900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang S., Denman S.E., Morrison M., Yu Z., Dore J., Leclerc M., et al. Dysbiosis of fecal microbiota in Crohn's disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis. 2010;16:2034–2042. doi: 10.1002/ibd.21319. [DOI] [PubMed] [Google Scholar]
- 15.Ahn J., Yang L., Paster B.J., Ganly I., Morris L., Pei Z., et al. Oral microbiome profiles: 16S rRNA pyrosequencing and microarray assay comparison. PLoS One. 2011;6:e22788. doi: 10.1371/journal.pone.0022788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crielaard W., Zaura E., Schuller A.A., Huse S.M., Montjin R.C., Keijser B.F.J. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genomics. 2011;4:22. doi: 10.1186/1755-8794-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth S.B., Jalava J., Ruuskanen O., Ruohola A., Nikkari S. Use of an oligonucleotide array for laboratory diagnosis of bacteria responsible for acute upper respiratory infections. J Clin Microbiol. 2004;42:4268–4274. doi: 10.1128/JCM.42.9.4268-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cannon G.A., Carr M.J., Yandle Z., Schaffer K., Kidney R., Hosny G., et al. A low density oligonucleotide microarray for the detection of viral and atypical bacterial respiratory pathogens. J Virol Methods. 2010;163:17–24. doi: 10.1016/j.jviromet.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu L., Jiang G., Wang S., Wang C., Li Q., Yu H., et al. Biochip system for rapid and accurate identification of mycobacterial species from isolates and sputum. J Clin Microbiol. 2010;48:3654–3660. doi: 10.1128/JCM.00158-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong B.X., Jiang L.F., Hu Y.S., Fang D.Y., Guo H.Y. Application of oligonucleotide array technology for the rapid detection of pathogenic bacteria of foodborne infections. J Microbiol Methods. 2004;58:403–411. doi: 10.1016/j.mimet.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Jin D.Z., Wen S.Y., Chen S.H., Lin F., Wang S.Q. Detection and identification of intestinal pathogens in clinical specimens using DNA microarrays. Mol Cell Probes. 2006;20:337–347. doi: 10.1016/j.mcp.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 22.You Y., Fu C., Zeng X., Fang D., Yan X., Sun B., et al. A novel DNA microarray for rapid diagnosis of enteropathogenic bacteria in stool specimens of patients with diarrhea. J Microbiol Methods. 2008;75:566–571. doi: 10.1016/j.mimet.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Kim D.H., Lee B.K., Kim Y.D., Rhee S.K., Kim Y.C. Detection of representative enteropathogenic bacteria, Vibrio spp., pathogenic Escherichia coli, Salmonella spp., Shigella spp., and Yersinia enterocolitica, using a virulence factor gene-based oligonucleotide microarray. J Microbiol. 2010;48:682–688. doi: 10.1007/s12275-010-0119-5. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q., Wang S., Beutin L., Cao B., Feng L., Wang L. Development of a DNA microarray for detection and serotyping of Enterotoxigenic Escherichia coli. J Clin Microbiol. 2010;48:2066–2074. doi: 10.1128/JCM.02014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blaser M.J., Newman L.S. A review of human salmonellosis: I. Infective dose. Rev Infect Dis. 1982;4:1096–1106. doi: 10.1093/clinids/4.6.1096. [DOI] [PubMed] [Google Scholar]
- 26.Wallis M.R. The pathogenosis of Campylobacter jejuni. Br J Biomed Sci. 1994;51:57–64. [PubMed] [Google Scholar]
- 27.Suo B., He Y., Paoli G., Gehring A., Tu S.I., Shi X. Development of an oligonucleotide-based microarray to detect multiple foodborne pathogens. Mol Cell Probes. 2010;24:77–86. doi: 10.1016/j.mcp.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Järvinen A.K., Laakso S., Piiparinen P., Aittakorpi A., Lindfors M., Huopaniemi L., et al. Rapid identification of bacterial pathogens using a PCR- and microarray-based assay. BMC Microbiol. 2009;9:161. doi: 10.1186/1471-2180-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim C.M., Song E.S., Jang H.J., Kim H.J., Lee S., Shin J.H., et al. Development and evaluation of oligonucleotide chip based on the 16S-23S rRNA gene spacer region for detection of pathogenic microorganisms associated with sepsis. J Clin Microbiol. 2010;48:1578–1583. doi: 10.1128/JCM.01130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tissari P., Zumla A., Tarkka E., Mero S., Savolainen L., Vaara M., et al. Accurate and rapid identification of bacterial species from positive blood cultures with a DNA-based microarray platform: an observational study. Lancet. 2010;375:224–230. doi: 10.1016/S0140-6736(09)61569-5. [DOI] [PubMed] [Google Scholar]
- 31.Tomita Y., Okamoto A., Yamada K., Yagi T., Hasegawa Y., Ohta M. A new microarray system to detect Streptococcus pneumoniae serotypes. J Biomed Biotechnol. 2011;2011:352736. doi: 10.1155/2011/352736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henrichsen J. Six newly recognized types of Streptococcus pneumonia. J Clin Microbiol. 1995;33:2759–2762. doi: 10.1128/jcm.33.10.2759-2762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein J.O. The epidemiology of pneumococcal disease in infants and children. Rev Infect Dis. 1981;3:246–253. doi: 10.1093/clinids/3.2.246. [DOI] [PubMed] [Google Scholar]
- 34.Peterson G., Bai J., Nagaraja T.G., Narayanan S. Diagnostic microarray for human and bacterial diseases and their virulence and antimicrobial resistance genes. J Microbiol Methods. 2010;80:223–230. doi: 10.1016/j.mimet.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 35.Portillo A., Ruiz-Larrez F., Zarazaga M., Alonso A., Martinez J.L., Torres C. Macrolide resistance genes in Enterococcus spp. Antimicrob Agents Chemother. 2000;44:967–971. doi: 10.1128/aac.44.4.967-971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fontana R., Cerini R., Longoni P., Grossato A., Canepari P. Identification of a streptococcal penicillin-binding protein that reacts very slowly with penicillin. J Bacteriol. 1983;155:1343–1350. doi: 10.1128/jb.155.3.1343-1350.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monecke S., Ehricht R. Rapid genotyping of methicillin-resistant Staphylococcus aureus (MRSA) isolates using miniaturized oligonucleotide arrays. Clin Microbiol Infect. 2005;11:825–833. doi: 10.1111/j.1469-0691.2005.01243.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhu L.X., Zhang Z.W., Wang C., Yang H.W., Jiang D., Zhang Q., et al. Use of a DNA microarray for simultaneous detection of antibiotic resistance genes among Staphylococcal clinical isolates. J Clin Microbiol. 2007;45:3514–3521. doi: 10.1128/JCM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Musser J.M. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev. 1995;8:496–514. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramaswamy S., Musser J.M. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis. Tuber Lung Dis. 1998;79:3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 41.Rattan A., Kalia A., Ahmad N. Multidrug-resistant Mycobacterium tuberculosis: molecular perspectives. Emerg Infect Dis. 1998;4:195–209. doi: 10.3201/eid0402.980207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mdluli K., Sherman D.R., Hickey M.J., Kreiswirth B.N., Morris S., Stover C.K., et al. Biochemical and genetic data suggest that inhA is not the primary target for activated isoniazid in Mycobacterium tuberculosis. J Infect Dis. 1996;174:1085–1090. doi: 10.1093/infdis/174.5.1085. [DOI] [PubMed] [Google Scholar]
- 43.Park H., Song E.J., Song E.S., Lee E.Y., Kim C.M., Jeong S.H., et al. Comparison of a conventional antimicrobial susceptibility assay to an oligonucleotide chip system for detection of drug resistance in Mycobacterium tuberculosis isolates. J Clin Microbiol. 2006;44:1619–1624. doi: 10.1128/JCM.44.5.1619-1624.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ninove L., Nougairede A., Gazin C., Zandotti C., Drancourt M., de Lamballerie X., et al. POC tests: from antigen detection to molecular methods. Future trends. J Clin Virol. 2010;4:304–305. doi: 10.1016/j.jcv.2010.08.012. [DOI] [PubMed] [Google Scholar]


