Highlights
-
•
Historical perspective and global scenario about Newcastle disease.
-
•
Characteristics about Newcastle disease virus genome.
-
•
Replication and pathogenicity of Newcastle disease virus.
-
•
Use of Newcastle disease virus as a vaccine vector.
-
•
Newcastle disease virus as an oncolytic agent.
Keywords: Newcastle disease virus, Paramyxovirus, Reverse genetics system, Pathogenicity
Abstract
Newcastle disease (ND) is one of the highly pathogenic viral diseases of avian species. ND is economically significant because of the huge mortality and morbidity associated with it. The disease is endemic in many third world countries where agriculture serves as the primary source of national income. Newcastle disease virus (NDV) belongs to the family Paramyxoviridae and is well characterized member among the avian paramyxovirus serotypes. In recent years, NDV has lured the virologists not only because of its pathogenic potential, but also for its oncolytic activity and its use as a vaccine vector for both humans and animals. The NDV based recombinant vaccine offers a pertinent choice for the construction of live attenuated vaccine due to its modular nature of transcription, minimum recombination frequency, and lack of DNA phase during replication. Our current understanding about the NDV biology is expanding rapidly because of the availability of modern molecular biology tools and high-throughput complete genome sequencing.
1. Historical perspective
Newcastle disease (ND) is a highly infectious viral disease of avian species. ND infection has been reported from a wide variety of birds with varying degree of susceptibility (Kaleta and Baldauf, 1988). ND was first reported from Indonesia in 1926 and in the New Castle upon Tyne in England in 1927 (Doyle, 1927, Kraneveld, 1926). Successive ND outbreaks were reported from different parts of the world including Korea, India, Sri Lanka, Japan, Australia, and the Philippines (Albiston and Gorrie, 1942, Crawford, 1930, Edwards, 1928, Kanno et al., 1929, Ochi and Hashimoto, 1929, Rodier, 1928). Historically, the ND is also called Ranikhet disease owing to its place of emergence in India. ND was identified with the advent of large scale commercial poultry farming toward the beginning of the 20th century. ND is a worldwide threat and is endemic in many developing parts of the world (Fig. 1 ). Based on pathogenic studies ND is categorized into three groups: lentogenic (low virulence), mesogenic (moderate virulence) and velogenic (highly virulent). The velogenic ND can either be viscerotropic or neurotropic depending on its predilection site (Alexander, 2000). Velogenic ND may result in 100% mortality in poultry leading to significant impact on trade restrictions and embargoes in the regions of its outbreak.
Fig. 1.
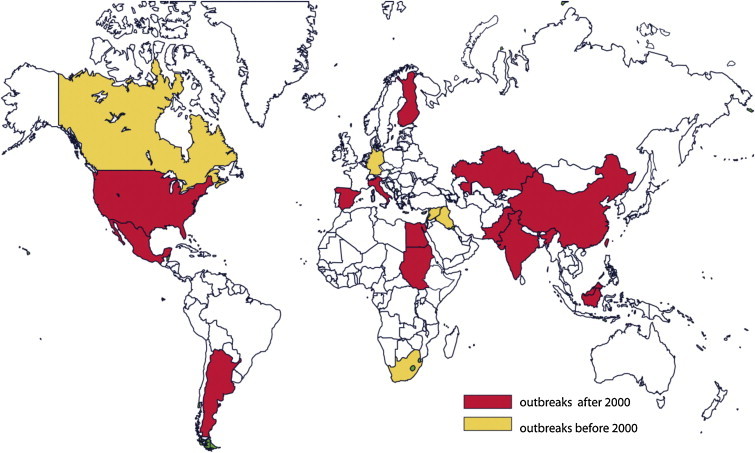
The global scenario of Newcastle disease virus (NDV) outbreaks in different parts of the world.
The causative agent of ND is a Newcastle disease virus (NDV). The devastating effect of NDV can be restricted by the use of vaccines. Many commercial vaccines both live and killed are available in the market (Mebatsion et al., 2002). Thermostable NDV vaccines are also available that can be easily preserved and transported. Vaccination protocols against NDV are outlined in the National Newcastle disease management plan 2008–2012 and are practiced globally (2013). In general, broiler chickens are vaccinated on day one through intraocular route followed by a booster on day 14 through drinking water. Layers are vaccinated first with live vaccine followed by an inactivated vaccine via intramuscular or sub-cutaneous route. The diagnosis of ND requires close monitoring of signs and symptoms as it is often misdiagnosed for salmonellosis, spirochaetosis, laryngotracheitis and hemorrhagic diseases.
2. Global scenario
Newly isolated strains of NDV are continuously being reported from all over the world (Fig. 1). Recently, NDV outbreaks have been reported from Vietnam, Indonesia, Malaysia, and Cambodia (Choi et al., 2014). In 2013, 96 NDV outbreaks were reported in poultry from Cameroon, Central African Republic, Côte d’Ivoire and Nigeria (Snoeck et al., 2013). In 2011, a velogenic NDV outbreak was reported from Israel in Little owls and African Penguins (Haddas et al., 2013). Apart from routine outbreaks, vaccination incapacity has also been reported causing emergence of new NDV strains (Chen et al., 2012). In the USA alone, virulent NDV (also called exotic NDV) isolates were reported from cormorants and gulls in the state of Minnesota, Massachusetts, Maine, New Hampshire, and Maryland (Diel et al., 2012). Successive NDV outbreaks were also reported from European continents and China (Lindh et al., 2012, Xie et al., 2012). Phylogenetic analysis of NDV isolated from different parts of the world could not provide us a clear picture of how it is crossing the topographical barrier; however one can interpret that the virulent strains of NDV isolated from Texas showed identity with the strains from hot climatic zone suggesting the prevalence of highly virulent strains in those areas (Fig. 2 ). NDV strains isolated from Egypt and Central Africa showed high identity in their genome with virulent strains such as Fontana and Texas GB suggesting its virulent pathotype (our unpublished data). Interestingly, NDV isolation was reported from mosquito pool in Jakarta (Forrester et al., 2013). Although Australia has been free of NDV outbreaks from 1932 to 1998, multiple outbreaks were reported from New South Wales from 1998 to 2002 (Westbury, 2001). The presence of NDV has also been reported from a wild bird population including Mallards (Tolf et al., 2013) and Spotted-necked dove (Liu et al., 2013). Occasionally NDV has been isolated from non-avian species such as pigs (Chen et al., 2013) and goats (Sharma et al., 2012).
Fig. 2.
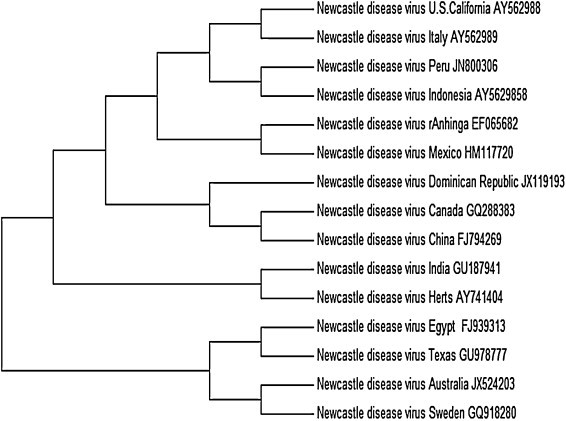
Phylogenetic analysis of Newcastle disease virus (NDV) isolated from different countries. The accession numbers against which the tree is build are as follows: GU978777, JX524203, EF065682, GU187941, FJ794269, GQ288383, FJ939313, HM117720, AY562988, JX119193, JN800306, AY562989, GU187941, AY741404, GQ918280, and AY562985.
3. Newcastle disease virus
Doyle in 1927 first identified that ND is caused by a filterable virus that is different from fowl plague later named as NDV (Doyle, 1927). NDV is classified as a member of genus Avulavirus in the subfamily Paramyxovirinae under family Paramyxoviridae (Lamb and Parks, 2007). The genus Avulavirus is divided into nine serotypes based on haemagglutination inhibition (HI) and neuraminidase inhibition (NI) assays. NDV belongs to the serotype 1 of avian paramyxoviruses (Alexander, 1998). The virus is relatively stable in nature even at sub-optimal temperature and wide range of pH, however it has been observed that the NDV becomes unstable at 56 °C (our unpublished results). NDV is sensitive to detergents, lipid solvents, formaldehyde and oxidizing agents.
NDV is a pleomorphic enveloped virus around 200–300 nm in diameter (Fig. 3 ). The genome of NDV is a non-segmented, negative sense, single stranded RNA. The NDV strains isolated from different parts of the world fall into three genome size groups: 15,186 nucleotides (nt) long in the isolates before 1960; 15,192 nt long in the isolates discovered in China, and 15,198 nt long in the avirulent strain from Germany (Czegledi et al., 2006, de Leeuw and Peeters, 1999, Huang et al., 2004a, Krishnamurthy and Samal, 1998, Romer-Oberdorfer et al., 1999). All the NDV isolates follow the “rule of six” (Calain and Roux, 1993) which means that the viral replication is most effective when the nt sequences in the genomes are in multiple of six since nucleocapsid (N) protein binds effectively with six nucleotides (Fig. 4 ). Encapsidation of the last 6 nucleotides at the 3′ end promoter by a single N protein is important for the efficient initiation of replication of paramyxovirus genome by the viral polymerase (Pelet et al., 1996). Moreover, assembly of the nascent viral genome following replication begins with the binding of N to the first 6 nucleotides at the 5′ trailer and proceeds in the 5′–3′ direction (Lamb and Parks, 2007). The typical NDV genome consists of 55 nt leader at its 3′ end and 114 nt trailer at its 5′ end, flanking six essential genes encoding nucleocapsid (N), matrix protein (M), phosphoprotein (P), fusion protein (F), haemagglutinin-neuraminidase protein (HN), and large polymerase protein (L) (Fig. 4). Each gene is marked by the presence of gene start and gene end signal sequences. Moreover the gene boundaries are separated from each other by inter genic sequences (IGS) (Lamb and Parks, 2007). Five of the six genes in the NDV code for single major protein except for P gene that encodes three proteins (P, V and W) by the phenomenon of RNA editing (Lamb and Parks, 2007, Steward et al., 1993). The N protein forms the nucleocapsid core with genomic RNA to which P and L proteins remain bound forming the herringbone like ribonucleoprotein complex (Lamb and Parks, 2007). The non-glycosylated matrix protein is present beneath the envelope and is shown to be responsible for virus assembly and budding (Pantua et al., 2006). The F and HN are the two surface glycoproteins that are virus neutralizing antigens and are responsible for virus attachment and its fusion to the host cell membrane. The F and HN spikes are around 8 nm in length and are present as a trimer and tetramer, respectively (Lamb and Parks, 2007). The V and W proteins are accessory and are present only in the virus infected cells. The V protein is interferon (IFN) antagonist and plays an important role in NDV virulence (Alamares et al., 2010, Dortmans et al., 2011, Huang et al., 2003b).
Fig. 3.
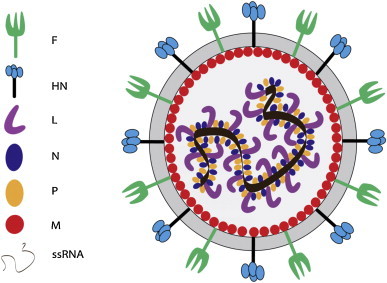
Schematic diagram of Newcastle disease virus structure.
Fig. 4.
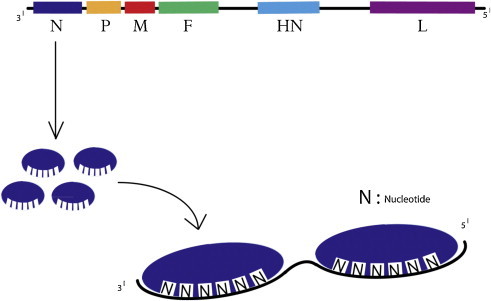
Schematic representation of Newcastle disease virus genome highlighting the rule of six essentially means that a nucleocapsid protein can bind effectively to six nucleotides. The area between the genes shown in black represents Inter Genomic Sequences that vary in size from 1 to 47 nucleotides.
3.1. Nucleocapsid (N) protein
It has a length of 489 amino acids (aa) and 55 kDa molecular weight. It coats the full-length genomic (−) sense and antigenomic (+) sense RNAs in order to protect them from nucleases (Dortmans et al., 2010). It is the most abundant protein present in the virus particle. In electron micrograph, it appears to hold a herringbone-like structure. It forms the helical nucleocapsid core structure of the virus along with the genomic RNA. The genomic RNA in association with the N, P and L proteins form the ribonucleoprotein complex (RNP), which acts as a template for RNA synthesis. The amino-terminus of N protein is responsible for its interaction with viral RNA along with its requirement in the formation of herringbone-like structure (Errington and Emmerson, 1997). It has been observed that the first few amino acids toward the amino-terminus of NDV N protein form a soluble complex with the P protein. The carboxy-terminus, on the other hand, is not required for nucleocapsid assembly but presumably plays a regulatory role in its polymerization (Kho et al., 2003). Recombinant N protein of NDV expressed in baculovirus has been reported to form herringbone-like structures which morphologically resemble the typical nucleocapsid structure (Errington and Emmerson, 1997).
3.2. Phosphoprotein (P)
The P protein of NDV consists of 395 aa. It forms multiple bands of molecular weight ranging from 50 to 55 kDa in SDS-PAGE because of different phosphorylated forms (Dortmans et al., 2010). The P protein of NDV is phosphorylated at specific serine and threonine residues and functions as a homo-oligomer. The P protein has a crucial role in viral replication and transcription. It helps in stabilizing the L protein in the P–L complex which then functions as viral RNA-dependent RNA polymerase. The P–L complex carries out genomic replication, thus synthesizing full-length plus-strand antigenomic RNA which is used as a template to synthesize minus-strand genomic RNA (Dortmans et al., 2010). A tetramer of phosphoprotein mediates the interaction between the L protein and the N-RNA template which acts as a chaperone in order to prevent the random encapsidation of non-viral RNA by the N protein. Moreover, the P protein forms a complex with unassembled N protein which regulates the shift from transcription to replication. Both amino- and carboxy-terminals of the P protein are needed for P–N interactions. Different domains of the P proteins carry out different functions of the P–N complex while interacting with N protein during virus replication. Its carboxy-terminal residue (247–291) participates in P–P and P–N interaction (Jahanshiri et al., 2005). It has also been observed that P protein has a role in virulence, which depends on the cell type and the NDV strain (Dortmans et al., 2010).
3.3. Matrix protein (M)
The M protein is about 40 kDa by weight and consists of 364 aa. It is a basic protein with a region that interacts with viral nucleic acids (17 bp long) which includes nine basic amino acids (Chambers et al., 1986). The net positive charge of M protein aids to its association with N protein during viral assembly. The M protein is hydrophobic in nature with no membrane spanning peptides and is present between nucleocapsid and lipid membrane. There are few proposed functions of M protein; primarily it controls the viral RNA synthesis, secondly it interacts with actin, and finally it helps in assembling virion on the host cell membrane (Peeples and Bratt, 1984). The M protein is highly conserved among paramyxoviruses which is evident by the presence of very few non-synonymous base substitutions following mutations in the population. The findings can be used as a basis to classify different NDV strains and isolates from different geographical locations (Seal et al., 2000). It has also been suggested that M protein might be responsible for maintaining the spherical shape of the nucleocapsid (Mebatsion et al., 1999). In addition, M protein helps in the virus budding process by interacting with the host cell plasma membrane. The M protein has its own nuclear localization sequences and therefore it does not require other NDV proteins to perform the nuclear localization function. It has also been noticed that the M protein is critically important for viral assembly when temperature sensitive mutants fail to produce the required level of M under sub-optimal temperatures (Peeples and Bratt, 1984).
3.4. Fusion protein (F)
The F protein is a surface glycoprotein present on the NDV envelope and mediates its fusion with the host cell membrane. F protein is supposed to play a major role in the virulence of NDV strains (Panda et al., 2004, Romer-Oberdorfer et al., 2003). The HN protein is known to assist F protein in its function (Morrison, 2003). F protein is synthesized as an inactive precursor F0 which is cleaved by a cellular protease into activated forms/subunits which are F1 and F2. Further F proteins are processed in the trans Golgi network inside the mammalian cell that forms a disulfide linked F1–F2 active form. The cleavage specificity is determined by the amino acid sequence present at the cleavage site and varies with the type of the strain (Glickman et al., 1988, Toyoda et al., 1987). The F protein does not need acidic pH for its cleavage and activation. The F protein fuses with the cell at neutral pH leading to multinucleate condition (syncytia) which has consequences like tissue necrosis and virus spread. Low virulent strains of NDV contain monobasic or dibasic amino acid residues at its F protein cleavage site. The presence of one or two basic amino acids renders the F proteins insensitive to the intracellular proteases and depends on extracellular proteases to get cleaved, allowing its tropism limited to respiratory and enteric tracts. For velogenic, i.e., highly virulent strains, polybasic amino acids are the preferred recognition sites for furin like proteases present in most of the cells (Glickman et al., 1988, Toyoda et al., 1987). The cleavage of F protein in a wide range of tissues is responsible for the systemic spread of NDV and also for its virulence. It has been observed that F proteins of virulent NDV strains contain lysine (K) and arginine (R) at their cleavage site (112R-R-Q-R/K-R116), and a phenylalanine at position 117 of F1. This site is recognized by intracellular proteases, furin that cleave the polybasic cleavage site forming F1 subunit which is suggested to be the contributor of neurological effects (Kattenbelt et al., 2006, Nagai et al., 1976, Toyoda et al., 1987). It has also been observed that the presence of arginine at position 113, 115, and 116 in the cleavage site would lead to variation in intracellular cleavage of virulent NDV F protein (Samal et al., 2011). The glutamine is present at 114 position which is a neutral amino acid and studies have shown that on substituting it with a basic or acidic amino acid residue would decrease the virulence of NDV (Samal et al., 2011). It was suggested that a neutral amino acid along with basic amino acids at the cleavage site is required for proper binding of furin protease and its cleavage, thus altering host-cell enzyme activity (Nagai et al., 1976). In addition, subtilisin like mammalian proteases are also the candidates for the cleavage of the F protein, e.g., PC6 and PACE4 (Sakaguchi et al., 1994).
3.5. Hemagglutinin-neuraminidase protein (HN)
The HN protein is a surface glycoprotein with a molecular weight of 74 kDa. It is a type II integral membrane protein consisting of an uncleaved signal sequence near its amino-terminal (Sergel et al., 1993b). The HN is present as a homotetramer with disulfide linked dimers within virus infected cells. The ectodomain of the HN protein consists of a long stalk supporting a terminal globular head. Earlier studies indicate that HN is a multifunctional protein involved in functions such as receptor recognition in the host cell, receptor removal, to prevent self-assembly, and interaction with F to promote fusion. Both receptor recognition and neuraminidase property of HN lie in the globular head and is highly conserved. The structure of HN protein reveals the presence of a beta propeller motif with receptor recognition and neuraminidase activity and is located in the globular head domain of its monomer. A second sialic acid-binding site on the top of the its globular head spans the membrane-distal end of the dimer interface and is oriented toward the target host membrane (Iorio and Mahon, 2008). Moreover, the globular head region is also suggested to be the antibody binding sites (Iorio et al., 1989). The HN protein is also responsible for virus specific membrane fusion. Co-expression and interaction between homologous HN and F protein are important for fusion of the virus. Experimentally two models are proposed for HN–F interaction. Earlier, it was predicted that binding of HN with the cell receptor triggers its interaction with F protein, leading to conformational changes in F for fusion (Lamb and Parks, 2007, Sergel et al., 1993a). The later model proposed that F and HN form a metastable complex prior to HN attachment. Transition of HN from binding to catalytic form releases F protein that undergoes a conformational change to initiate fusion (Stone-Hulslander and Morrison, 1997, Tong and Compans, 1999). It has been suggested that the stalk domain of HN protein confers specificity for fusion (Deng et al., 1995). The HN–F interaction is mediated by residues in the stalk region of the HN ectodomain that bind to complementary F domain (Hu et al., 1992, Melanson and Iorio, 2004). A stretch of conserved amino acids from position 74 to 110 in NDV HN protein including the heptad repeats (HR) 1 and 2 is considered to mediate its interaction. It has also been reported that substitutions in the intervening sequences between HR1 and HR2 impair fusion (Melanson and Iorio, 2004, Mirza and Iorio, 2013). It has been proposed that HN heptad repeats may interact and bind to HR1 and HR2 of F protein to keep them apart and impart flexibility to the helices in order to support conformational changes in the two proteins. This would result in disrupting HN–F interaction and thus release of fusion peptide inside the membrane being targeted (Stone-Hulslander and Morrison, 1999). Any mutation in the heptad repeats of HN protein affects fusion and destabilization of its tetrameric structures leading to alteration in biological activity of NDV.
The HN protein is responsible for the infection and the pathogenesis of NDV (Huang et al., 2004c, Khattar et al., 2009, Kim et al., 2009, Yan et al., 2009). The HN protein of NDV contains 14 cysteine residues among which 12 residues are conserved and form intramolecular disulfide linkages. A cysteine at position 123 is responsible for the covalent linkage between HN proteins and is important for its structural integrity (McGinnes and Morrison, 1994, Sheehan and Iorio, 1992). Its cytoplasmic tail possesses 26 conserved amino acids which interact with the M protein. Mutational analysis of HN protein of NDV has suggested that only first two amino acids can be removed while any other mutation affects its viability. The HN protein determines the tropism of NDV as well; however the size of HN protein does not seem to affect its enterotropism (Zhao et al., 2013). Different length of the NDV HN protein exists in nature owing to the difference in the position of the stop codon. The shortest HN of 571 amino acid is present in velogenic strains and the longest of 616 amino acid is found in lentogenic strains (Gorman et al., 1988). The length and sequence of the extended carboxy-terminus in HN protein have been marked to influence its function but their role in NDV virulence has not been clearly evaluated.
3.6. Large polymerase protein (L)
Being the largest protein of the NDV genome, L protein consists of 2204 aa with a molecular weight of 250 kDa (Lamb and Parks, 2007). The L protein synthesizes viral mRNA and assists in genomic RNA replication. It is the last gene to be transcribed during the viral replication cycle. In addition, the L protein also performs 5′ capping, methylation, and poly A polymerase activity on the newly formed mRNA (Dortmans et al., 2010, Lamb and Parks, 2007). The L and P protein together form the active viral polymerase. The helical nucleoprotein complex bound by N protein acts as a template which is recognized by the L-P complex to form active viral polymerase complex. Six extremely conserved amino acid residues QGDNQ present in domain III carry out transcription activity and are responsible for its polymerase activity (Lamb and Parks, 2007). It has been reported that L protein modulates the virulence of NDV suggesting its potent role in the virulence of the virus presumably by increasing the rate of viral RNA synthesis during replication (Rout and Samal, 2008).
3.7. V protein
The V protein of NDV has a molecular weight of 36 kDa and is 239 aa long (Lamb and Parks, 2007). The insertion of one G residue in the P protein ORF at consensus sequence “394AAAAGGG401” during RNA editing codes for V protein (Fig. 5 ). The V protein modulates the viral RNA replication by interacting with N protein (Horikami et al., 1996). The V protein is an additional virulence factor of NDV (Park et al., 2003). The V protein of NDV in particular to its carboxy-terminal domain inhibits the IFN alpha/beta (IFN-α/β) response which is important for its virulence. Successful NDV replication requires dodging of proapoptotic cascade in order to accomplish efficient virus production and spread of viral progeny. The V protein of NDV has been observed to inhibit host IFN response in two ways. It inhibits IFN signaling by targeting STAT1 for its degradation by its carboxy-terminal domain. Alternatively, carboxy-terminal domain of the V protein of NDV interacts with MDA5 and inhibits IFNβ response (Park et al., 2003). Occasionally, insertion of two G residues at the RNA editing site in the P gene encodes W mRNA. Although W mRNA has been detected in NDV infected cells, W protein in an NDV infected cell has never been reported. Therefore, the role of W protein in the NDV replication cycle has not yet been established. The V and W proteins are similar to P protein toward its amino-terminal while they differ in its carboxy-terminus.
Fig. 5.
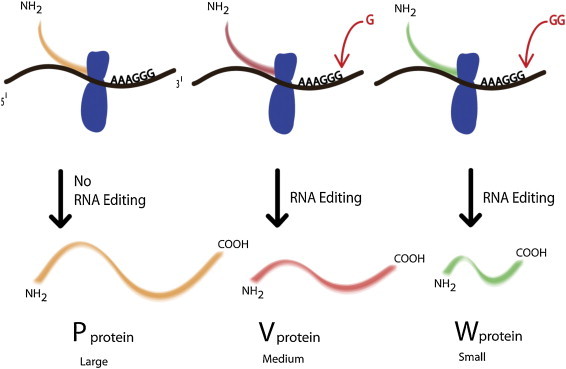
Phenomenon of RNA editing in Newcastle disease virus. Incorporation of single G or two G residues at the RNA editing site could give rise to V or W, respectively.
4. Replication and pathogenicity
NDV attacks respiratory epithelium cells by binding to sialic acid-containing compounds such as gangliosides and N-glycoproteins receptors by its surface glycoprotein. NDV infection occurs primarily via pH-independent fashion where virus envelope fuses with the host cell membrane. In addition infection can also occur by receptor mediated endocytosis and sometimes through caveolae-dependent endocytosis (Cantin et al., 2007). Following entry into the host cell cytoplasm, the negative sense RNA genome transcribes into positive sense mRNA which then translates into viral proteins. Transcription begins at the extreme 3′ leader sequence and synthesizes the mRNA of individual genes from gene-start to gene-end sequences. Reinitiation of transcription to a downstream gene at the gene-start site is not uniform, which leads to a gradient of mRNA population that decreases according to the distance from the 3′ end of the viral genome (Fig. 6 ). The proteins N, P and L are essential for nucleocapsid assembly. The positive sense RNA is then used as a template for the synthesis of negative sense genomic RNA. Assembly and budding of mature NDV depends on the M protein and lipid raft over the cell membrane (Fig. 6). NDV infectivity is dependent on the cleavage of F protein precursor F0 to F1–F2 subunits. The amino acid sequence of the F protein cleavage site is thus postulated as the primary determinant of infection (Panda et al., 2004, Peeters et al., 1999, Samal et al., 2011). The HN protein with receptor recognition and neuraminidase activities of the virus determines and also contributes to virulence of NDV (de Leeuw et al., 2005, Huang et al., 2004c). It has also been suggested that F and HN proteins in NDV are determinants of NDV virulence in chicken and infected macrophages (Cornax et al., 2013, Romer-Oberdorfer et al., 2003). NDV is also pathogenic to humans and can cause conjunctivitis if exposed to the eyes (Swayne and King, 2003).
Fig. 6.
Schematic representation of Newcastle disease virus replication. Entry of the virus into the host cell system is mediated by the interaction of glycoproteins (F, HN) on viral surface and binding to sialic acid-containing compounds such as gangliosides and N-glycoproteins receptors on cell surface, resulting in fusion of the virus to host cells. The viral nucleocapsid is then pushed into the host cytoplasm where the negative sense viral RNA is transcribed to produce the structural mRNAs, with the help of virus associated RNA dependent RNA polymerase in a gradient fashion. The respective proteins are translated and folded using host cell machinery. Once a threshold of the first transcribed N mRNA is reached, the negative sense genomic RNA is converted to positive sense anti-genome template for the synthesis of new negative sense RNA genome. This newly formed genomic RNA is then wrapped in N, P and L proteins to form the nucleocapsid that is assembled with matrix and surface glycoproteins and released from the host cell.
5. Recent advancement in understanding NDV virulence and pathogenicity
Many studies following the development of reverse genetics system of different NDV strains enlightened our understanding of NDV virulence and pathogenicity. The reverse genetics system offers the flexibility to mutate the individual genes or replace the same with identical homolog rendering the NDV genome quite manipulative. Although the major players of NDV virulence and pathogenicity are F and HN, but its gradients are largely multigenic. The F being the major antigenic determinant in NDV changed our views of considering HN as a major protective antigen (Kumar et al., 2011). We showed earlier that F is a better antigen than HN and incorporation of F in place of HN will provide better and sterile immunity against NDV infection (Kumar et al., 2011). It has been made evident that the conserved glutamine residue in the F protein cleavage site is important for NDV replication and pathogenicity and mutation of Q114R can attenuate the virus (Samal et al., 2011). Furthermore, the NDV could also be attenuated by substituting valine to isoleucine at position 118 around the fusion cleavage site (Samal et al., 2011). In another study, it has been shown that changing glycosylation site of F protein can increase the virulence and pathogenicity of NDV (Samal et al., 2012). It is also evident that mutation in the cytoplasmic domain of F protein can lead to the production of a hyperfusogenic virus that could ensure increased viral replication and pathogenesis in chickens (Samal et al., 2013).
The formation of a complex between homologous HN and F proteins is required for NDV-induced membrane fusion and is influenced by receptor binding and removal properties of HN. It has been shown that substitution of residues F220, S222, and L224 in the membrane proximal part of dimer interface of the HN globular domain weakens its association with its receptors thereby affecting intermonomer sub-unit interaction of F and HN (Corey et al., 2003). In addition, substitution of amino acid residue (L94) in the region intervening two heptad repeats in HN stalk structure affects fusion by hindering interaction with F protein (Melanson and Iorio, 2004). Moreover, virulence and tissue tropism in chimeric viruses recovered by shuffling HN genes of different NDV strains were found to be function of its amino acid sequence (Huang et al., 2004c). Recently, it has been reported that carboxy-terminal extension of the HN gene of a virulent Indonesian NDV strain from 571 amino acids to 577 and 616 amino acids reduces viral pathogenicity in 1-day-old and 3-week-old chickens (Kim et al., 2014).
6. Newcastle disease virus as a vaccine vector
NDV is an attractive vaccine vector candidate for both human and animal use (Samal, 2011). It presents a promising candidate for rational design of live attenuated vaccine and vaccine vectors because of its modular nature of transcription, minimum recombination frequency and an absence of DNA phase during replication. The genome of NDV is quite easy to manipulate using reverse genetics system (Bukreyev and Collins, 2008, Huang et al., 2003a, Krishnamurthy et al., 2000, Peeters et al., 2001). Live attenuated vaccines and bivalent vaccines are economically very popular for the poultry industry. The lentogenic strain of NDV appears to be a good vaccine. Both live attenuated and recombinant viruses are explored as vaccine and vaccine vectors with various degrees of success. The recombinant NDV expressing foreign protein is explored as a viral vector by many scientists around the globe. Following properties of NDV can be attributed for its credibility as a viral vector:
-
I.
NDV grows with high titers in embryonated chicken eggs, cell culture and respiratory tract of avian and non-avian species.
-
II.
NDV infects naturally via the respiratory tract and is thus useful to deliver protective antigens derived from respiratory pathogens. In addition, it induces both local and systemic immune responses.
-
III.
NDV elicits both humoral and cellular immune response.
-
IV.
NDV has a modular genome with only six essential and two accessory genes that is easy to manipulate.
-
V.
NDV does not integrate with host genome as it replicates in host cytoplasm and shows least genetic recombination.
-
VI.
Recombinant NDV expressing the foreign antigen shows quite a high and stable expression of foreign protein after many passages both in vitro and in vivo (Huang et al., 2003a).
-
VII.
NDV can be attenuated using reverse genetics for the development of stable vaccine and vaccine vector. Development of deletion mutants (V, W) and mutants with altered F cleavage site may result in more immunogenic and attenuated recombinant virus at the same time (Huang et al., 2003a, Samal et al., 2011).
For construction of vaccine vector, the foreign gene (∼3.8 kb) must be flanked by NDV specific gene-start and gene-end sequences at the gene junction or intergenic sequence without perturbing the rule of six (Bukreyev et al., 2006, DiNapoli et al., 2007, Huang et al., 2001). The level of foreign gene expression is found to be more in case of its insertion at the 3′ end of the genome (Carnero et al., 2009, Huang et al., 2001).
Recombinant NDV acts as a potential vaccine vector for humans because of its attenuation due to natural host range restriction (Samal, 2011). NDV offers an important vaccine candidate in terms of safety, efficacy and cost effectiveness. A number of NDV based vaccines are generated for treating various human viral infections. Recombinant NDV confers immunogenic response against different antigenic challenges like human influenza virus heamagglutinin protein (HA) (DiNapoli et al., 2010, Ge et al., 2007), HIV and SIV Gag protein (Lawrence et al., 2013, Nakaya et al., 2001, Nakaya et al., 2004), HIV glycoproteins (Khattar et al., 2011, Khattar et al., 2013), F glycoprotein of human respiratory syncytial virus (Martinez-Sobrido et al., 2006), HN protein of human parainfluenza virus 3 (Bukreyev et al., 2005), and spike glycoprotein of SARS-CoV (DiNapoli et al., 2007).
NDV acts as an excellent vaccine vector for veterinary pathogens and is successfully marketed. Recombinant NDV expressing VP2 protein is used as a dual vaccine against NDV and infectious Bursal disease infection in chickens (Huang et al., 2004b). Recombinant NDV expressing the HA protein of H5N1 (Ge et al., 2007, Nayak et al., 2009, Park et al., 2006, Romer-Oberdorfer et al., 2008) and H7N7 influenza virus (Park et al., 2006, Schroer et al., 2009) protects against both NDV and influenza virus infection in chickens. Recombinant NDV expressing gN and gG glycoprotein of Rift Valley fever virus is shown to protect mice and lambs, against viral infection by eliciting effective antibody response (Kortekaas et al., 2010). Recombinant NDV expressing gD glycoprotein partially protects cattle against bovine herpes virus infection (Khattar et al., 2010). In another study, recombinant NDV expressing the G and F proteins of Nipah virus induces neutralizing antibody responses in mice and pigs (Kong et al., 2012).
Many studies have been conducted around the world to improve the vaccine efficiency of NDV. It has been shown that recombinant NDV expressing the H5N1 HA protein in mule ducks showed sterile protection in the absence of maternally-derived antibodies (Ferreira et al., 2012). Recombinant NDV expressing soluble trimeric HA protein against highly pathogenic H5N1 influenza virus provides better efficacy than the membrane anchored HA suggesting that the type of antigenic foreign protein could be modulated for better protection (Cornelissen et al., 2010).
7. Newcastle disease virus as an oncolytic agent
The use of oncolytic viruses has emerged as potent cancer therapeutics based on its anti-cancer potency, tumor-specificity, efficacy and safety. Virotherapy for cancer treatment began before 1950s and since then a number of viruses including adenovirus, polio virus, herpes simplex virus, hepatitis A virus, influenza A virus, and measles virus have been explored for their oncolytic potential. The NDV has emerged as an important area of research in the late 20th century not only because of its economic importance but also for its oncolytic potential (Elankumaran, 2013, Reichard et al., 1992). Cassel and Garrett first observed the oncolytic effect of NDV in 1965, since then NDV has been used extensively as an oncolytic agent in both preclinical and clinical studies (Cassel and Garrett, 1965, Schirrmacher et al., 2001). NDV has been identified and tested in various animal and human models for cancer treatment (Lam et al., 2011, Shobana et al., 2013). Oncolytic properties of NDV emanate from its inherent ability to grow in IFN deficient cells such as tumor cells (Stojdl et al., 2000). NDV is now labeled as complementary and alternative medicine by National Cancer Institute and many clinical trials are in different phases of completion (Freeman et al., 2006, Hotte et al., 2007, Lorence et al., 1994, Lorence et al., 2003). Both lytic and non-lytic strains of NDV are proven to be potent anti-cancer agents. The mesogenic and velogenic NDV strains behave as lytic while the lentogenic behaves as the non-lytic strain. The NDV strains which are most widely used for treatment of human cancer are PV701, 73-T, MTH-68 and Ulster. The NDV strains such as PV701, 73-T and MTH-68 are lytic in nature whereas Ulster is non-lytic.
NDV is non-pathogenic to humans and thus relatively safe with no side effects. It has good cell binding ability via its HN glycoprotein and enters the cell through pH independent direct fusion of its envelope to host membrane and also via receptor mediated endocytosis (Cantin et al., 2007, Sanchez-Felipe et al., 2014). NDV replicates around 104 times faster in tumor cells due to host restriction of V protein and virus induced cytokines IFN-γ and TNF-α. NDV efficiently induces synthesis of cytokines like IFN and tumor necrosis factor as well as stimulates production of heat shock proteins, adrenocorticotropic hormones and tissue inhibitor of metalloproteases (Lam et al., 2011, Ravindra et al., 2009). NDV also has pleiotropic immuno-stimulatory effect as it can augment the production of cytokines such IFN-α, IFN-β, TNF-α, and interleukin 1 which further activates NK cells, macrophages and sensitized T cells for oncolysis. NDV infection results in potent up-regulation of antigen presentation by MHC I and expression of cell adhesion molecules such as ICAM-I and LFA-3 causing co-stimulation of T cells (Ten et al., 1993). The expression of viral HN molecules leads to increased production of IFN-α and TNF-related apoptosis-inducing ligand which further up-regulates monocyte tumoricidal activity and apoptosis (Batliwalla et al., 1998, Lam et al., 2011). In addition, NDV replication and de-novo viral protein synthesis induces caspase dependent apoptosis in cancerous cells by both intrinsic and extrinsic pathways (Elankumaran et al., 2006, Ravindra et al., 2008a). Moreover, NDV can also up-regulate the expression of pro-apoptotic P53 and Bax and down-regulate anti-apoptotic Bcl-2 gene in target cells (Molouki and Yusoff, 2012, Ravindra et al., 2009).
Oncolytic property of NDV can be enhanced by using recombinant NDV expressing immunostimulatory molecules like complement regulatory protein, IFNs, IL-2, GM-CSF, IgG antibody and TNF-α (Biswas et al., 2012, Janke et al., 2007, Puhler et al., 2008, Vigil et al., 2007, Zamarin et al., 2009). Interestingly, recombinant NDV with hyper-cleavable F protein has been shown to have a higher therapeutic index attributed to its tumor restricted growth and enhanced fusogenicity (Altomonte et al., 2010, Ch’ng et al., 2013). Recombinant NDV sensitive to type I interferon has been reported to augment innate immune responses resulting in a selective oncolysis (Elankumaran et al., 2010). Moreover, development of trackable recombinant NDV as cancer virotherapy tool has been reported with or without therapeutic transgenes (Elankumaran et al., 2006). The contribution of individual NDV genes on tumor cell has also been studied with HN shown to be the major player (Ghrici et al., 2013, Ravindra et al., 2008b). The combination of HN gene along with VP3 gene of chicken anemia virus showed promising results in reducing mouse osteosarcoma (Zhang et al., 2013).
8. Conclusion
ND is a major threat to the poultry industry around the globe. The disease is endemic in many developing countries while the disease free countries are prone to accidental outbreaks. NDV strains having varying degree of virulence circulate among avian species. The topographical distribution of NDV is not well understood and regular sporadic cases are reported throughout the years from endemic areas. Research priorities are toward the improved diagnostics and better vaccine development. The reverse genetics system offers the opportunity to engineer NDV as a vaccine vector for human and animal uses. Moreover, NDV as an oncolytic agent has gained major attention and would provide an alternate therapy for the treatment of cancer in the years to come.
Acknowledgements
The corresponding author is grateful to Dr Siba K. Samal at the University of Maryland for providing an opportunity to work with avian paramyxoviruses. We are thankful to Dr. Monika Koul at the National Research Center on Pig (NRCP) and Mr. Bansi Lal Koul for proof reading the manuscript. The NDV research in our laboratory is currently supported by the Department of Biotechnology (NER-BPMC/2013/134), the Department of Science and Technology (IFA-LSBM-34), and the Board of Research on Nuclear Sciences (2012/20/37B/067/BRNS).
References
- Anon. Department of Primary Industries Parks Water and Environment; 2013. Vaccination Against Newcastle Disease. [Google Scholar]
- Alamares J.G., Elankumaran S., Samal S.K., Iorio R.M. The interferon antagonistic activities of the V proteins from two strains of Newcastle disease virus correlate with their known virulence properties. Virus Res. 2010;147(1):153–157. doi: 10.1016/j.virusres.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albiston H., Gorrie C. Newcastle disease on Victoria. Aust. Vet. J. 1942;18:75–79. [Google Scholar]
- Alexander D.J. Newcastle disease and other avian paramyxoviruses. In: Swayne D.E., Glisson J., Jackwood M.W., Pearson J.E., Reed W.M., editors. A Laboratory Manual for the Isolation and Identification of Avian Pathogens. 4th ed. American Association of Avian Pathologists; Philadelphia: 1998. [Google Scholar]
- Alexander D.J. Newcastle disease and other avian paramyxoviruses. Rev. Sci. Tech. 2000;19(2):443–462. doi: 10.20506/rst.19.2.1231. [DOI] [PubMed] [Google Scholar]
- Altomonte J., Marozin S., Schmid R.M., Ebert O. Engineered Newcastle disease virus as an improved oncolytic agent against hepatocellular carcinoma. Mol. Ther. 2010;18(2):275–284. doi: 10.1038/mt.2009.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batliwalla F.M., Bateman B.A., Serrano D., Murray D., Macphail S., Maino V.C., Ansel J.C., Gregersen P.K., Armstrong C.A. A 15-year follow-up of AJCC stage III malignant melanoma patients treated postsurgically with Newcastle disease virus (NDV) oncolysate and determination of alterations in the CD8T cell repertoire. Mol. Med. 1998;4(12):783–794. [PMC free article] [PubMed] [Google Scholar]
- Biswas M., Johnson J.B., Kumar S.R., Parks G.D., Elankumarana S. Incorporation of host complement regulatory proteins into Newcastle disease virus enhances complement evasion. J. Virol. 2012;86(23):12708–12716. doi: 10.1128/JVI.00886-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukreyev A., Collins P.L. Newcastle disease virus as a vaccine vector for humans. Curr. Opin. Mol. Ther. 2008;10(1):46–55. [PubMed] [Google Scholar]
- Bukreyev A., Huang Z., Yang L., Elankumaran S., St Claire M., Murphy B.R., Samal S.K., Collins P.L. Recombinant Newcastle disease virus expressing a foreign viral antigen is attenuated and highly immunogenic in primates. J. Virol. 2005;79(21):13275–13284. doi: 10.1128/JVI.79.21.13275-13284.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukreyev A., Skiadopoulos M.H., Murphy B.R., Collins P.L. Nonsegmented negative-strand viruses as vaccine vectors. J. Virol. 2006;80(21):10293–10306. doi: 10.1128/JVI.00919-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calain P., Roux L. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J. Virol. 1993;67(8):4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantin C., Holguera J., Ferreira L., Villar E., Munoz-Barroso I. Newcastle disease virus may enter cells by caveolae-mediated endocytosis. J. Gen. Virol. 2007;88(Pt 2):559–569. doi: 10.1099/vir.0.82150-0. [DOI] [PubMed] [Google Scholar]
- Carnero E., Li W., Borderia A.V., Moltedo B., Moran T., Garcia-Sastre A. Optimization of human immunodeficiency virus gag expression by newcastle disease virus vectors for the induction of potent immune responses. J. Virol. 2009;83(2):584–597. doi: 10.1128/JVI.01443-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassel W.A., Garrett R.E. Newcastle disease virus as an antineoplastic agent. Cancer. 1965;18:863–868. doi: 10.1002/1097-0142(196507)18:7<863::aid-cncr2820180714>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Ch’ng W.C., Stanbridge E.J., Yusoff K., Shafee N. The oncolytic activity of Newcastle disease virus in clear cell renal carcinoma cells in normoxic and hypoxic conditions: the interplay between von Hippel-Lindau and interferon-beta signaling. J. Interferon Cytokine Res. 2013;33(7):346–354. doi: 10.1089/jir.2012.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers P., Millar N.S., Platt S.G., Emmerson P.T. Nucleotide sequence of the gene encoding the matrix protein of Newcastle disease virus. Nucleic Acids Res. 1986;14(22):9051–9061. doi: 10.1093/nar/14.22.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Liu J., Liu D., Yan Z., Ji J., Qin J., Li H., Ma J., Bi Y., Xie Q. Complete genome sequence of a Newcastle disease virus strain isolated from broiler breeder flocks in China. J. Virol. 2012;86(22):12461–12462. doi: 10.1128/JVI.02314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Hao H., Wang X., Du E., Liu H., Yang T., Liu Y., Fu X., Zhang P., Yang Z. Genomic characterisation of a lentogenic Newcastle disease virus strain HX01 isolated from sick pigs in China. Virus Genes. 2013;46(2):264–270. doi: 10.1007/s11262-012-0844-4. [DOI] [PubMed] [Google Scholar]
- Choi K.S., Kye S.J., Kim J.Y., To T.L., Nguyen D.T., Lee Y.J., Choi J.G., Kang H.M., Kim K.I., Song B.M., Lee H.S. Molecular epidemiology of Newcastle disease viruses in Vietnam. Trop. Anim. Health Prod. 2014;46(1):271–277. doi: 10.1007/s11250-013-0475-3. [DOI] [PubMed] [Google Scholar]
- Corey E.A., Mirza A.M., Levandowsky E., Iorio R.M. Fusion deficiency induced by mutations at the dimer interface in the Newcastle disease virus hemagglutinin-neuraminidase is due to a temperature-dependent defect in receptor binding. J. Virol. 2003;77(12):6913–6922. doi: 10.1128/JVI.77.12.6913-6922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornax I., Diel D.G., Rue C.A., Estevez C., Yu Q., Miller P.J., Afonso C.L. Newcastle disease virus fusion and haemagglutinin-neuraminidase proteins contribute to its macrophage host range. J. Gen. Virol. 2013;94(Pt 6):1189–1194. doi: 10.1099/vir.0.048579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen L.A., de Vries R.P., de Boer-Luijtze E.A., Rigter A., Rottier P.J., de Haan C.A. A single immunization with soluble recombinant trimeric hemagglutinin protects chickens against highly pathogenic avian influenza virus H5N1. PLoS ONE. 2010;5(5):e10645. doi: 10.1371/journal.pone.0010645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford M. Govt. Vet. Surgeon; Colombo, Ceylon: 1930. Rannikhet. Ann. Rept. [Google Scholar]
- Czegledi A., Ujvari D., Somogyi E., Wehmann E., Werner O., Lomniczi B. Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Res. 2006;120(1–2):36–48. doi: 10.1016/j.virusres.2005.11.009. [DOI] [PubMed] [Google Scholar]
- de Leeuw O., Peeters B. Complete nucleotide sequence of Newcastle disease virus: evidence for the existence of a new genus within the subfamily Paramyxovirinae. J. Gen. Virol. 1999;80(Pt 1):131–136. doi: 10.1099/0022-1317-80-1-131. [DOI] [PubMed] [Google Scholar]
- de Leeuw O.S., Koch G., Hartog L., Ravenshorst N., Peeters B.P. Virulence of Newcastle disease virus is determined by the cleavage site of the fusion protein and by both the stem region and globular head of the haemagglutinin-neuraminidase protein. J. Gen. Virol. 2005;86(Pt 6):1759–1769. doi: 10.1099/vir.0.80822-0. [DOI] [PubMed] [Google Scholar]
- Deng R., Wang Z., Mirza A.M., Iorio R.M. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology. 1995;209(2):457–469. doi: 10.1006/viro.1995.1278. [DOI] [PubMed] [Google Scholar]
- Diel D.G., Miller P.J., Wolf P.C., Mickley R.M., Musante A.R., Emanueli D.C., Shively K.J., Pedersen K., Afonso C.L. Characterization of Newcastle disease viruses isolated from cormorant and gull species in the United States in 2010. Avian Dis. 2012;56(1):128–133. doi: 10.1637/9886-081111-Reg.1. [DOI] [PubMed] [Google Scholar]
- DiNapoli J.M., Kotelkin A., Yang L., Elankumaran S., Murphy B.R., Samal S.K., Collins P.L., Bukreyev A. Newcastle disease virus, a host range-restricted virus, as a vaccine vector for intranasal immunization against emerging pathogens. Proc. Natl. Acad. Sci. U. S. A. 2007;104(23):9788–9793. doi: 10.1073/pnas.0703584104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNapoli J.M., Nayak B., Yang L., Finneyfrock B.W., Cook A., Andersen H., Torres-Velez F., Murphy B.R., Samal S.K., Collins P.L., Bukreyev A. Newcastle disease virus-vectored vaccines expressing the hemagglutinin or neuraminidase protein of H5N1 highly pathogenic avian influenza virus protect against virus challenge in monkeys. J. Virol. 2010;84(3):1489–1503. doi: 10.1128/JVI.01946-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortmans J.C., Koch G., Rottier P.J., Peeters B.P. Virulence of Newcastle disease virus: what is known so far? Vet. Res. 2011;42:122–130. doi: 10.1186/1297-9716-42-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortmans J.C., Rottier P.J., Koch G., Peeters B.P. The viral replication complex is associated with the virulence of Newcastle disease virus. J. Virol. 2010;84(19):10113–10120. doi: 10.1128/JVI.00097-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle T. A hitherto unrecorded disease of fowls due to filter passing virus. J. Comp. Pathol. Therap. 1927:144–169. [Google Scholar]
- Edwards J. Vet. Res. Mukteshwar; 1928. A New Fowl Disease. March 31st, pp. 14–15. [Google Scholar]
- Elankumaran S. Genetically engineered Newcastle disease virus for prostate cancer: a magic bullet or a misfit. Expert Rev. Anticancer Ther. 2013;13(7):769–772. doi: 10.1586/14737140.2013.811062. [DOI] [PubMed] [Google Scholar]
- Elankumaran S., Chavan V., Qiao D., Shobana R., Moorkanat G., Biswas M., Samal S.K. Type I interferon-sensitive recombinant newcastle disease virus for oncolytic virotherapy. J. Virol. 2010;84(8):3835–3844. doi: 10.1128/JVI.01553-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elankumaran S., Rockemann D., Samal S.K. Newcastle disease virus exerts oncolysis by both intrinsic and extrinsic caspase-dependent pathways of cell death. J. Virol. 2006;80(15):7522–7534. doi: 10.1128/JVI.00241-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington W., Emmerson P.T. Assembly of recombinant Newcastle disease virus nucleocapsid protein into nucleocapsid-like structures is inhibited by the phosphoprotein. J. Gen. Virol. 1997;78(Pt 9):2335–2339. doi: 10.1099/0022-1317-78-9-2335. [DOI] [PubMed] [Google Scholar]
- Ferreira H.L., Pirlot J.F., Reynard F., van den Berg T., Bublot M., Lambrecht B. Immune responses and protection against H5N1 highly pathogenic avian influenza virus induced by the Newcastle disease virus H5 vaccine in ducks. Avian Dis. 2012;56(4 Suppl.):940–948. doi: 10.1637/10148-040812-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Forrester N.L., Widen S.G., Wood T.G., Travassos da Rosa A.P., Ksiazek T.G., Vasilakis N., Tesh R.B. Identification of a new Newcastle disease virus isolate from Indonesia represents an ancestral lineage of class II genotype XIII. Virus Genes. 2013;47(1):168–172. doi: 10.1007/s11262-013-0900-8. [DOI] [PubMed] [Google Scholar]
- Freeman A.I., Zakay-Rones Z., Gomori J.M., Linetsky E., Rasooly L., Greenbaum E., Rozenman-Yair S., Panet A., Libson E., Irving C.S., Galun E., Siegal T. Phase I/II trial of intravenous NDV-HUJ oncolytic virus in recurrent glioblastoma multiforme. Mol. Ther. 2006;13(1):221–228. doi: 10.1016/j.ymthe.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Ge J., Deng G., Wen Z., Tian G., Wang Y., Shi J., Wang X., Li Y., Hu S., Jiang Y., Yang C., Yu K., Bu Z., Chen H. Newcastle disease virus-based live attenuated vaccine completely protects chickens and mice from lethal challenge of homologous and heterologous H5N1 avian influenza viruses. J. Virol. 2007;81(1):150–158. doi: 10.1128/JVI.01514-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghrici M., El Zowalaty M., Omar A.R., Ideris A. Induction of apoptosis in MCF-7 cells by the hemagglutinin-neuraminidase glycoprotein of Newcastle disease virus Malaysian strain AF2240. Oncol. Rep. 2013;30(3):1035–1044. doi: 10.3892/or.2013.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman R.L., Syddall R.J., Iorio R.M., Sheehan J.P., Bratt M.A. Quantitative basic residue requirements in the cleavage-activation site of the fusion glycoprotein as a determinant of virulence for Newcastle disease virus. J. Virol. 1988;62(1):354–356. doi: 10.1128/jvi.62.1.354-356.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J.J., Nestorowicz A., Mitchell S.J., Corino G.L., Selleck P.W. Characterization of the sites of proteolytic activation of Newcastle disease virus membrane glycoprotein precursors. J. Biol. Chem. 1988;263(25):12522–12531. [PubMed] [Google Scholar]
- Haddas R., Meir R., Perk S., Horowitz I., Lapin E., Rosenbluth E., Lublin A. Newcastle disease virus in Little Owls (Athene noctua) and African Penguins (Spheniscus demersus) in an Israeli Zoo. Transbound Emerg. Dis. 2013 doi: 10.1111/tbed.12064. [DOI] [PubMed] [Google Scholar]
- Horikami S.M., Smallwood S., Moyer S.A. The Sendai virus V protein interacts with the NP protein to regulate viral genome RNA replication. Virology. 1996;222(2):383–390. doi: 10.1006/viro.1996.0435. [DOI] [PubMed] [Google Scholar]
- Hotte S.J., Lorence R.M., Hirte H.W., Polawski S.R., Bamat M.K., O’Neil J.D., Roberts M.S., Groene W.S., Major P.P. An optimized clinical regimen for the oncolytic virus PV701. Clin. Cancer Res. 2007;13(3):977–985. doi: 10.1158/1078-0432.CCR-06-1817. [DOI] [PubMed] [Google Scholar]
- Hu X.L., Ray R., Compans R.W. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J. Virol. 1992;66(3):1528–1534. doi: 10.1128/jvi.66.3.1528-1534.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Wan H.Q., Liu H.Q., Wu Y.T., Liu X.F. Genomic sequence of an isolate of Newcastle disease virus isolated from an outbreak in geese: a novel six nucleotide insertion in the non-coding region of the nucleoprotein gene. Brief report. Arch. Virol. 2004;149(7):1445–1457. doi: 10.1007/s00705-004-0297-8. [DOI] [PubMed] [Google Scholar]
- Huang Z., Elankumaran S., Panda A., Samal S.K. Recombinant Newcastle disease virus as a vaccine vector. Poult. Sci. 2003;82(6):899–906. doi: 10.1093/ps/82.6.899. [DOI] [PubMed] [Google Scholar]
- Huang Z., Elankumaran S., Yunus A.S., Samal S.K. A recombinant Newcastle disease virus (NDV) expressing VP2 protein of infectious bursal disease virus (IBDV) protects against NDV and IBDV. J. Virol. 2004;78(18):10054–10063. doi: 10.1128/JVI.78.18.10054-10063.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Krishnamurthy S., Panda A., Samal S.K. High-level expression of a foreign gene from the most 3’-proximal locus of a recombinant Newcastle disease virus. J. Gen. Virol. 2001;82(Pt 7):1729–1736. doi: 10.1099/0022-1317-82-7-1729. [DOI] [PubMed] [Google Scholar]
- Huang Z., Krishnamurthy S., Panda A., Samal S.K. Newcastle disease virus V protein is associated with viral pathogenesis and functions as an alpha interferon antagonist. J. Virol. 2003;77(16):8676–8685. doi: 10.1128/JVI.77.16.8676-8685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Panda A., Elankumaran S., Govindarajan D., Rockemann D.D., Samal S.K. The hemagglutinin-neuraminidase protein of Newcastle disease virus determines tropism and virulence. J. Virol. 2004;78(8):4176–4184. doi: 10.1128/JVI.78.8.4176-4184.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio R.M., Glickman R.L., Riel A.M., Sheehan J.P., Bratt M.A. Functional and neutralization profile of seven overlapping antigenic sites on the HN glycoprotein of Newcastle disease virus: monoclonal antibodies to some sites prevent viral attachment. Virus Res. 1989;13(3):245–261. doi: 10.1016/0168-1702(89)90019-1. [DOI] [PubMed] [Google Scholar]
- Iorio R.M., Mahon P.J. Paramyxoviruses: different receptors – different mechanisms of fusion. Trends Microbiol. 2008;16(4):135–137. doi: 10.1016/j.tim.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshiri F., Eshaghi M., Yusoff K. Identification of phosphoprotein:phosphoprotein and phosphoprotein:nucleocapsid protein interaction domains of the Newcastle disease virus. Arch. Virol. 2005;150(3):611–618. doi: 10.1007/s00705-004-0439-z. [DOI] [PubMed] [Google Scholar]
- Janke M., Peeters B., de Leeuw O., Moorman R., Arnold A., Fournier P., Schirrmacher V. Recombinant Newcastle disease virus (NDV) with inserted gene coding for GM-CSF as a new vector for cancer immunogene therapy. Gene Ther. 2007;14(23):1639–1649. doi: 10.1038/sj.gt.3303026. [DOI] [PubMed] [Google Scholar]
- Kaleta E., Baldauf C. Newcastle disease in free-living and pet birds. In: Alexander D.J., editor. Newcastle Disease. Kluwer Acedemic Press; Boston, MA: 1988. [Google Scholar]
- Kanno T., Ochi Y., Hashimoto K. Dtsch. Tierarzhl. Wshr.; 1929. Neue Geflugelscuche in Korea. [Google Scholar]
- Kattenbelt J.A., Stevens M.P., Gould A.R. Sequence variation in the Newcastle disease virus genome. Virus Res. 2006;116(1–2):168–184. doi: 10.1016/j.virusres.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Khattar S.K., Collins P.L., Samal S.K. Immunization of cattle with recombinant Newcastle disease virus expressing bovine herpesvirus-1 (BHV-1) glycoprotein D induces mucosal and serum antibody responses and provides partial protection against BHV-1. Vaccine. 2010;28(18):3159–3170. doi: 10.1016/j.vaccine.2010.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattar S.K., Samal S., Devico A.L., Collins P.L., Samal S.K. Newcastle disease virus expressing human immunodeficiency virus type 1 envelope glycoprotein induces strong mucosal and serum antibody responses in Guinea pigs. J. Virol. 2011;85(20):10529–10541. doi: 10.1128/JVI.05050-11. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Khattar S.K., Samal S., Labranche C.C., Montefiori D.C., Collins P.L., Samal S.K. Comparative immunogenicity of HIV-1 gp160, gp140 and gp120 expressed by live attenuated newcastle disease virus vector. PLoS ONE. 2013;8(10):e78521. doi: 10.1371/journal.pone.0078521. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Khattar S.K., Yan Y., Panda A., Collins P.L., Samal S.K. A Y526Q mutation in the Newcastle disease virus HN protein reduces its functional activities and attenuates virus replication and pathogenicity. J. Virol. 2009;83(15):7779–7782. doi: 10.1128/JVI.00536-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho C.L., Tan W.S., Tey B.T., Yusoff K. Newcastle disease virus nucleocapsid protein: self-assembly and length-determination domains. J. Gen. Virol. 2003;84(Pt 8):2163–2168. doi: 10.1099/vir.0.19107-0. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Xiao S., Paldurai A., Collins P.L., Samal S.K. Role of C596 in the C-terminal extension of the haemagglutinin-neuraminidase protein in replication and pathogenicity of a highly virulent Indonesian strain of Newcastle disease virus. J. Gen. Virol. 2014;95(Pt 2):331–336. doi: 10.1099/vir.0.055285-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kim S.H., Yan Y., Samal S.K. Role of the cytoplasmic tail amino acid sequences of Newcastle disease virus hemagglutinin-neuraminidase protein in virion incorporation, cell fusion, and pathogenicity. J. Virol. 2009;83(19):10250–10255. doi: 10.1128/JVI.01038-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D., Wen Z., Su H., Ge J., Chen W., Wang X., Wu C., Yang C., Chen H., Bu Z. Newcastle disease virus-vectored Nipah encephalitis vaccines induce B and T cell responses in mice and long-lasting neutralizing antibodies in pigs. Virology. 2012;432(2):327–335. doi: 10.1016/j.virol.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Kortekaas J., de Boer S.M., Kant J., Vloet R.P., Antonis A.F., Moormann R.J. Rift valley fever virus immunity provided by a paramyxovirus vaccine vector. Vaccine. 2010;28(27):4394–4401. doi: 10.1016/j.vaccine.2010.04.048. [DOI] [PubMed] [Google Scholar]
- Kraneveld F. Over een in Ned-Indie heerschende Ziete under het pluimves. Ned. Indisch Bl. Diergeneesk. 1926;38:448–450. [Google Scholar]
- Krishnamurthy S., Huang Z., Samal S.K. Recovery of a virulent strain of Newcastle disease virus from cloned cDNA: expression of a foreign gene results in growth retardation and attenuation. Virology. 2000;278(1):168–182. doi: 10.1006/viro.2000.0618. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S., Samal S.K. Nucleotide sequences of the trailer, nucleocapsid protein gene and intergenic regions of Newcastle disease virus strain Beaudette C and completion of the entire genome sequence. J. Gen. Virol. 1998;79(Pt 10):2419–2424. doi: 10.1099/0022-1317-79-10-2419. [DOI] [PubMed] [Google Scholar]
- Kumar S., Nayak B., Collins P.L., Samal S.K. Evaluation of the Newcastle disease virus F and HN proteins in protective immunity by using a recombinant avian paramyxovirus type 3 vector in chickens. J. Virol. 2011;85(13):6521–6534. doi: 10.1128/JVI.00367-11. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lam H.Y., Yeap S.K., Rasoli M., Omar A.R., Yusoff K., Suraini A.A., Alitheen N.B. Safety and clinical usage of newcastle disease virus in cancer therapy. J. Biomed. Biotechnol. 2011;2011:718710. doi: 10.1155/2011/718710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R., Parks G. Paramyxoviridae: the viruses and their replication. In: Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., Straus S.E., editors. Fields Virology. 5th ed. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1449–1496. [Google Scholar]
- Lawrence T.M., Wanjalla C.N., Gomme E.A., Wirblich C., Gatt A., Carnero E., Garcia-Sastre A., Lyles D.S., McGettigan J.P., Schnell M.J. Comparison of heterologous prime-boost strategies against human immunodeficiency virus type 1 gag using negative stranded RNA viruses. PLoS ONE. 2013;8(6):e67123. doi: 10.1371/journal.pone.0067123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindh E., Ek-Kommonen C., Vaananen V.M., Alasaari J., Vaheri A., Vapalahti O., Huovilainen A. Molecular epidemiology of outbreak-associated and wild-waterfowl-derived newcastle disease virus strains in Finland, including a novel class I genotype. J. Clin. Microbiol. 2012;50(11):3664–3673. doi: 10.1128/JCM.01427-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zhang P., Wu P., Chen S., Mu G., Duan X., Hao H., Du E., Wang X., Yang Z. Phylogenetic characterization and virulence of two Newcastle disease viruses isolated from wild birds in China. Infect. Genet. Evol. 2013;20C:215–224. doi: 10.1016/j.meegid.2013.08.021. [DOI] [PubMed] [Google Scholar]
- Lorence R.M., Katubig B.B., Reichard K.W., Reyes H.M., Phuangsab A., Sassetti M.D., Walter R.J., Peeples M.E. Complete regression of human fibrosarcoma xenografts after local Newcastle disease virus therapy. Cancer Res. 1994;54(23):6017–6021. [PubMed] [Google Scholar]
- Lorence R.M., Pecora A.L., Major P.P., Hotte S.J., Laurie S.A., Roberts M.S., Groene W.S., Bamat M.K. Overview of phase I studies of intravenous administration of PV701, an oncolytic virus. Curr. Opin. Mol. Ther. 2003;5(6):618–624. [PubMed] [Google Scholar]
- Martinez-Sobrido L., Gitiban N., Fernandez-Sesma A., Cros J., Mertz S.E., Jewell N.A., Hammond S., Flano E., Durbin R.K., Garcia-Sastre A., Durbin J.E. Protection against respiratory syncytial virus by a recombinant Newcastle disease virus vector. J. Virol. 2006;80(3):1130–1139. doi: 10.1128/JVI.80.3.1130-1139.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnes L.W., Morrison T.G. The role of the individual cysteine residues in the formation of the mature, antigenic HN protein of Newcastle disease virus. Virology. 1994;200(2):470–483. doi: 10.1006/viro.1994.1210. [DOI] [PubMed] [Google Scholar]
- Mebatsion T., Koolen M.J., de Vaan L.T., de Haas N., Braber M., Romer-Oberdorfer A., van den Elzen P., van der Marel P. Newcastle disease virus (NDV) marker vaccine: an immunodominant epitope on the nucleoprotein gene of NDV can be deleted or replaced by a foreign epitope. J. Virol. 2002;76(20):10138–10146. doi: 10.1128/JVI.76.20.10138-10146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebatsion T., Weiland F., Conzelmann K.K. Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J. Virol. 1999;73(1):242–250. doi: 10.1128/jvi.73.1.242-250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melanson V.R., Iorio R.M. Amino acid substitutions in the F-specific domain in the stalk of the Newcastle disease virus HN protein modulate fusion and interfere with its interaction with the F protein. J. Virol. 2004;78(23):13053–13061. doi: 10.1128/JVI.78.23.13053-13061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza A.M., Iorio R.M. A mutation in the stalk of the newcastle disease virus hemagglutinin-neuraminidase (HN) protein prevents triggering of the F protein despite allowing efficient HN–F complex formation. J. Virol. 2013;87(15):8813–8815. doi: 10.1128/JVI.01066-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molouki A., Yusoff K. NDV-induced apoptosis in absence of Bax; evidence of involvement of apoptotic proteins upstream of mitochondria. Virol. J. 2012;9:179. doi: 10.1186/1743-422X-9-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison T.G. Structure and function of a paramyxovirus fusion protein. Biochim. Biophys. Acta. 2003;1614(1):73–84. doi: 10.1016/s0005-2736(03)00164-0. [DOI] [PubMed] [Google Scholar]
- Nagai Y., Klenk H.D., Rott R. Proteolytic cleavage of the viral glycoproteins and its significance for the virulence of Newcastle disease virus. Virology. 1976;72(2):494–508. doi: 10.1016/0042-6822(76)90178-1. [DOI] [PubMed] [Google Scholar]
- Nakaya T., Cros J., Park M.S., Nakaya Y., Zheng H., Sagrera A., Villar E., Garcia-Sastre A., Palese P. Recombinant Newcastle disease virus as a vaccine vector. J. Virol. 2001;75(23):11868–11873. doi: 10.1128/JVI.75.23.11868-11873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya Y., Nakaya T., Park M.S., Cros J., Imanishi J., Palese P., Garcia-Sastre A. Induction of cellular immune responses to simian immunodeficiency virus gag by two recombinant negative-strand RNA virus vectors. J. Virol. 2004;78(17):9366–9375. doi: 10.1128/JVI.78.17.9366-9375.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak B., Rout S.N., Kumar S., Khalil M.S., Fouda M.M., Ahmed L.E., Earhart K.C., Perez D.R., Collins P.L., Samal S.K. Immunization of chickens with Newcastle disease virus expressing H5 hemagglutinin protects against highly pathogenic H5N1 avian influenza viruses. PLoS ONE. 2009;4(8):e6509. doi: 10.1371/journal.pone.0006509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi Y., Hashimoto K. Govt. Inst. Vet. Research (Chosen); 1929. Uber eine neue Geflugelseuche in Korea. 6th Rept. August 20th, 16 pp. [Google Scholar]
- Panda A., Huang Z., Elankumaran S., Rockemann D.D., Samal S.K. Role of fusion protein cleavage site in the virulence of Newcastle disease virus. Microb. Pathog. 2004;36(1):1–10. doi: 10.1016/j.micpath.2003.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantua H.D., McGinnes L.W., Peeples M.E., Morrison T.G. Requirements for the assembly and release of Newcastle disease virus-like particles. J. Virol. 2006;80(22):11062–11073. doi: 10.1128/JVI.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.S., Garcia-Sastre A., Cros J.F., Basler C.F., Palese P. Newcastle disease virus V protein is a determinant of host range restriction. J. Virol. 2003;77(17):9522–9532. doi: 10.1128/JVI.77.17.9522-9532.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.S., Steel J., Garcia-Sastre A., Swayne D., Palese P. Engineered viral vaccine constructs with dual specificity: avian influenza and Newcastle disease. Proc. Natl. Acad. Sci. U. S. A. 2006;103(21):8203–8208. doi: 10.1073/pnas.0602566103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeples M.E., Bratt M.A. Mutation in the matrix protein of Newcastle disease virus can result in decreased fusion glycoprotein incorporation into particles and decreased infectivity. J. Virol. 1984;51(1):81–90. doi: 10.1128/jvi.51.1.81-90.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters B.P., de Leeuw O.S., Koch G., Gielkens A.L. Rescue of Newcastle disease virus from cloned cDNA: evidence that cleavability of the fusion protein is a major determinant for virulence. J. Virol. 1999;73(6):5001–5009. doi: 10.1128/jvi.73.6.5001-5009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters B.P., de Leeuw O.S., Verstegen I., Koch G., Gielkens A.L. Generation of a recombinant chimeric Newcastle disease virus vaccine that allows serological differentiation between vaccinated and infected animals. Vaccine. 2001;19(13-14):1616–1627. doi: 10.1016/s0264-410x(00)00419-9. [DOI] [PubMed] [Google Scholar]
- Pelet T., Delenda C., Gubbay O., Garcin D., Kolakofsky D. Partial characterization of a Sendai virus replication promoter and the rule of six. Virology. 1996;224(2):405–414. doi: 10.1006/viro.1996.0547. [DOI] [PubMed] [Google Scholar]
- Puhler F., Willuda J., Puhlmann J., Mumberg D., Romer-Oberdorfer A., Beier R. Generation of a recombinant oncolytic Newcastle disease virus and expression of a full IgG antibody from two transgenes. Gene Ther. 2008;15(5):371–383. doi: 10.1038/sj.gt.3303095. [DOI] [PubMed] [Google Scholar]
- Ravindra P.V., Tiwari A.K., Ratta B., Chaturvedi U., Palia S.K., Subudhi P.K., Kumar R., Sharma B., Rai A., Chauhan R.S. Induction of apoptosis in Vero cells by Newcastle disease virus requires viral replication, de-novo protein synthesis and caspase activation. Virus Res. 2008;133(2):285–290. doi: 10.1016/j.virusres.2008.01.010. [DOI] [PubMed] [Google Scholar]
- Ravindra P.V., Tiwari A.K., Sharma B., Chauhan R.S. Newcastle disease virus as an oncolytic agent. Indian J. Med. Res. 2009;130(5):507–513. [PubMed] [Google Scholar]
- Ravindra P.V., Tiwari A.K., Sharma B., Rajawat Y.S., Ratta B., Palia S., Sundaresan N.R., Chaturvedi U., Kumar G.B., Chindera K., Saxena M., Subudhi P.K., Rai A., Chauhan R.S. HN protein of Newcastle disease virus causes apoptosis in chicken embryo fibroblast cells. Arch. Virol. 2008;153(4):749–754. doi: 10.1007/s00705-008-0057-2. [DOI] [PubMed] [Google Scholar]
- Reichard K.W., Lorence R.M., Cascino C.J., Peeples M.E., Walter R.J., Fernando M.B., Reyes H.M., Greager J.A. Newcastle disease virus selectively kills human tumor cells. J. Surg. Res. 1992;52(5):448–453. doi: 10.1016/0022-4804(92)90310-v. [DOI] [PubMed] [Google Scholar]
- Rodier E. Philippines fowl disease. Proc. Soc. Exp. Biol. Med. 1928;25:781–783. [Google Scholar]
- Romer-Oberdorfer A., Mundt E., Mebatsion T., Buchholz U.J., Mettenleiter T.C. Generation of recombinant lentogenic Newcastle disease virus from cDNA. J. Gen. Virol. 1999;80(Pt 11):2987–2995. doi: 10.1099/0022-1317-80-11-2987. [DOI] [PubMed] [Google Scholar]
- Romer-Oberdorfer A., Veits J., Helferich D., Mettenleiter T.C. Level of protection of chickens against highly pathogenic H5 avian influenza virus with Newcastle disease virus based live attenuated vector vaccine depends on homology of H5 sequence between vaccine and challenge virus. Vaccine. 2008;26(19):2307–2313. doi: 10.1016/j.vaccine.2008.02.061. [DOI] [PubMed] [Google Scholar]
- Romer-Oberdorfer A., Werner O., Veits J., Mebatsion T., Mettenleiter T.C. Contribution of the length of the HN protein and the sequence of the F protein cleavage site to Newcastle disease virus pathogenicity. J. Gen. Virol. 2003;84(Pt 11):3121–3129. doi: 10.1099/vir.0.19416-0. [DOI] [PubMed] [Google Scholar]
- Rout S.N., Samal S.K. The large polymerase protein is associated with the virulence of Newcastle disease virus. J. Virol. 2008;82(16):7828–7836. doi: 10.1128/JVI.00578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi T., Fujii Y., Kiyotani K., Yoshida T. Correlation of proteolytic cleavage of F protein precursors in paramyxoviruses with expression of the fur, PACE4 and PC6 genes in mammalian cells. J. Gen. Virol. 1994;75(Pt 10):2821–2827. doi: 10.1099/0022-1317-75-10-2821. [DOI] [PubMed] [Google Scholar]
- Samal S., Khattar S.K., Kumar S., Collins P.L., Samal S.K. Coordinate deletion of N-glycans from the heptad repeats of the fusion F protein of Newcastle disease virus yields a hyperfusogenic virus with increased replication, virulence, and immunogenicity. J. Virol. 2012;86(5):2501–2511. doi: 10.1128/JVI.06380-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samal S., Khattar S.K., Paldurai A., Palaniyandi S., Zhu X., Collins P.L., Samal S.K. Mutations in the cytoplasmic domain of the Newcastle disease virus fusion protein confer hyperfusogenic phenotypes modulating viral replication and pathogenicity. J. Virol. 2013;87(18):10083–10093. doi: 10.1128/JVI.01446-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samal S., Kumar S., Khattar S.K., Samal S.K. A single amino acid change, Q114R, in the cleavage-site sequence of Newcastle disease virus fusion protein attenuates viral replication and pathogenicity. J. Gen. Virol. 2011;92(Pt 10):2333–2338. doi: 10.1099/vir.0.033399-0. [DOI] [PubMed] [Google Scholar]
- Samal S.K. In: Newcastle Disease and Related Avian Paramyxoviruses. Vol. I. The Biology of Paramyxoviruses. Samal S.K., editor. Caister Academic Press; Norfolk, United Kingdom: 2011. [Google Scholar]
- Sanchez-Felipe L., Villar E., Munoz-Barroso I. Entry of Newcastle disease virus into the host cell: role of acidic pH and endocytosis. Biochim. Biophys. Acta. 2014;1838(1):300–309. doi: 10.1016/j.bbamem.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirrmacher V., Griesbach A., Ahlert T. Antitumor effects of Newcastle disease virus in vivo: local versus systemic effects. Int. J. Oncol. 2001;18(5):945–952. doi: 10.3892/ijo.18.5.945. [DOI] [PubMed] [Google Scholar]
- Schroer D., Veits J., Grund C., Dauber M., Keil G., Granzow H., Mettenleiter T.C., Romer-Oberdorfer A. Vaccination with Newcastle disease virus vectored vaccine protects chickens against highly pathogenic H7 avian influenza virus. Avian Dis. 2009;53(2):190–197. doi: 10.1637/8416-072308-Reg.1. [DOI] [PubMed] [Google Scholar]
- Seal B.S., King D.J., Meinersmann R.J. Molecular evolution of the Newcastle disease virus matrix protein gene and phylogenetic relationships among the paramyxoviridae. Virus Res. 2000;66(1):1–11. doi: 10.1016/s0168-1702(99)00119-7. [DOI] [PubMed] [Google Scholar]
- Sergel T., McGinnes L.W., Morrison T.G. The fusion promotion activity of the NDV HN protein does not correlate with neuraminidase activity. Virology. 1993;196(2):831–834. doi: 10.1006/viro.1993.1541. [DOI] [PubMed] [Google Scholar]
- Sergel T., McGinnes L.W., Peeples M.E., Morrison T.G. The attachment function of the Newcastle disease virus hemagglutinin-neuraminidase protein can be separated from fusion promotion by mutation. Virology. 1993;193(2):717–726. doi: 10.1006/viro.1993.1180. [DOI] [PubMed] [Google Scholar]
- Sharma B., Pokhriyal M., Rai G.K., Saxena M., Ratta B., Chaurasia M., Yadav B.S., Sen A., Mondal B. Isolation of Newcastle disease virus from a non-avian host (sheep) and its implications. Arch. Virol. 2012;157(8):1565–1567. doi: 10.1007/s00705-012-1317-8. [DOI] [PubMed] [Google Scholar]
- Sheehan J.P., Iorio R.M. A single amino acid substitution in the hemagglutinin-neuraminidase of Newcastle disease virus results in a protein deficient in both functions. Virology. 1992;189(2):778–781. doi: 10.1016/0042-6822(92)90605-o. [DOI] [PubMed] [Google Scholar]
- Shobana R., Samal S.K., Elankumaran S. Prostate-specific antigen-retargeted recombinant Newcastle disease virus for prostate cancer virotherapy. J. Virol. 2013;87(7):3792–3800. doi: 10.1128/JVI.02394-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoeck C.J., Owoade A.A., Couacy-Hymann E., Alkali B.R., Okwen M.P., Adeyanju A.T., Komoyo G.F., Nakoune E., Le Faou A., Muller C.P. High genetic diversity of Newcastle disease virus in poultry in West and Central Africa: cocirculation of genotype XIV and newly defined genotypes XVII and XVIII. J. Clin. Microbiol. 2013;51(7):2250–2260. doi: 10.1128/JCM.00684-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward M., Vipond I.B., Millar N.S., Emmerson P.T. RNA editing in Newcastle disease virus. J. Gen. Virol. 1993;74(Pt 12):2539–2547. doi: 10.1099/0022-1317-74-12-2539. [DOI] [PubMed] [Google Scholar]
- Stojdl D.F., Lichty B., Knowles S., Marius R., Atkins H., Sonenberg N., Bell J.C. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat. Med. 2000;6(7):821–825. doi: 10.1038/77558. [DOI] [PubMed] [Google Scholar]
- Stone-Hulslander J., Morrison T.G. Detection of an interaction between the HN and F proteins in Newcastle disease virus-infected cells. J. Virol. 1997;71(9):6287–6295. doi: 10.1128/jvi.71.9.6287-6295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone-Hulslander J., Morrison T.G. Mutational analysis of heptad repeats in the membrane-proximal region of Newcastle disease virus HN protein. J. Virol. 1999;73(5):3630–3637. doi: 10.1128/jvi.73.5.3630-3637.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne D.E., King D.J. Avian influenza and Newcastle disease. J. Am. Vet. Med. Assoc. 2003;222(11):1534–1540. doi: 10.2460/javma.2003.222.1534. [DOI] [PubMed] [Google Scholar]
- Ten R.M., Blank V., Le Bail O., Kourilsky P., Israel A. Two factors, IRF1 and KBF1/NF-kappa B, cooperate during induction of MHC class I gene expression by interferon alpha beta or Newcastle disease virus. C. R. Acad. Sci. III. 1993;316(5):496–501. [PubMed] [Google Scholar]
- Tolf C., Wille M., Haidar A.K., Avril A., Zohari S., Waldenstrom J. Prevalence of avian paramyxovirus type 1 in Mallards during autumn migration in the western Baltic Sea region. Virol. J. 2013;10(1):285. doi: 10.1186/1743-422X-10-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., Compans R.W. Alternative mechanisms of interaction between homotypic and heterotypic parainfluenza virus HN and F proteins. J. Gen. Virol. 1999;80(Pt 1):107–115. doi: 10.1099/0022-1317-80-1-107. [DOI] [PubMed] [Google Scholar]
- Toyoda T., Sakaguchi T., Imai K., Inocencio N.M., Gotoh B., Hamaguchi M., Nagai Y. Structural comparison of the cleavage-activation site of the fusion glycoprotein between virulent and avirulent strains of Newcastle disease virus. Virology. 1987;158(1):242–247. doi: 10.1016/0042-6822(87)90261-3. [DOI] [PubMed] [Google Scholar]
- Vigil A., Park M.S., Martinez O., Chua M.A., Xiao S., Cros J.F., Martinez-Sobrido L., Woo S.L., Garcia-Sastre A. Use of reverse genetics to enhance the oncolytic properties of Newcastle disease virus. Cancer Res. 2007;67(17):8285–8292. doi: 10.1158/0008-5472.CAN-07-1025. [DOI] [PubMed] [Google Scholar]
- Westbury H. Newcastle disease virus: an evolving pathogen? Avian Pathol. 2001;30(1):5–11. doi: 10.1080/03079450020023131. [DOI] [PubMed] [Google Scholar]
- Xie Z., Xie L., Chen A., Liu J., Pang Y., Deng X., Fan Q. Complete genome sequence analysis of a Newcastle disease virus isolated from a wild egret. J. Virol. 2012;86(24):13854–13855. doi: 10.1128/JVI.02669-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Rout S.N., Kim S.H., Samal S.K. Role of untranslated regions of the hemagglutinin-neuraminidase gene in replication and pathogenicity of newcastle disease virus. J. Virol. 2009;83(11):5943–5946. doi: 10.1128/JVI.00188-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamarin D., Martinez-Sobrido L., Kelly K., Mansour M., Sheng G., Vigil A., Garcia-Sastre A., Palese P., Fong Y. Enhancement of oncolytic properties of recombinant Newcastle disease virus through antagonism of cellular innate immune responses. Mol. Ther. 2009;17(4):697–706. doi: 10.1038/mt.2008.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.Y., Wang L.Q., Fu C.F., Li X., Cui Z.L., Zhang J.Y., Xue S.H., Sun N., Xu F. Combination of targeting gene-viro therapy with recombinant Fowl-pox viruses with HN and VP3 genes on mouse osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2013;17(6):767–776. [PubMed] [Google Scholar]
- Zhao W., Hu H., Zsak L., Yu Q., Yang Z. HN gene C-terminal extension of Newcastle disease virus is not the determinant of the enteric tropism. Virus Genes. 2013;47(1):27–33. doi: 10.1007/s11262-013-0903-5. [DOI] [PubMed] [Google Scholar]


