Abstract
Objectives
To measure the reproducibility of radiomic features in pancreatic parenchyma and ductal adenocarcinomas (PDAC) in patients who underwent consecutive contrast enhanced computed tomography (CECT) scans.
Methods
In this IRB-approved and HIPAA-compliant retrospective study, 37 pairs of scans from 37 unique patients who underwent CECTs within a two-week interval were included in the analysis of the reproducibility of features derived from pancreatic parenchyma, and a subset of 18 pairs of scans were further analyzed for the reproducibility of features derived from PDAC. In each patient, pancreatic parenchyma and pancreatic tumor (when present) were manually segmented by two radiologists independently. A total of 266 radiomic features were extracted from the pancreatic parenchyma and tumor region, and also the volume and diameter of the tumor. The concordance correlation coefficient (CCC) was calculated to assess feature reproducibility for each patient in 3 scenarios: 1) different radiologists, same CECT, 2) same radiologist, different CECTs, and 3) different radiologists, different CECTs.
Results
Among pancreatic parenchyma-derived features, using a threshold of CCC > 0.90, 58/266 (21.8%) and 48/266 (18.1%) features met the threshold for scenario 1, 14/266 (5.3%) and 15/266 (5.6%) for scenario 2, and 14/266 (5.3%) and 10/266 (3.8%) for scenario 3. Among pancreatic tumor-derived features, 11/268 (4.1%) and 17/268 (6.3%) features for scenario 1, 1/268 (0.4%) and 5/268 (1.9%) features for scenario 2, and no features for scenario 3 met the threshold, respectively.
Conclusions
Variations between CECT scans affected radiomic feature reproducibility to a greater extent than variation in segmentation. A smaller number of pancreatic tumor-derived radiomic features were reproducible compared with pancreatic parenchyma-derived radiomic features under the same conditions.
Keywords: Reproducibility of Results; Carcinoma, Pancreatic Ductal; Tomography, X-Ray Computed; Radiomics; Texture Analysis
Introduction
Pancreatic cancer is the fourth leading cause of cancer death in the United States with an overall five-year survival of 8% [1]. The outcomes of patients with pancreatic cancer have not improved significantly during the past three decades [2]. Although surgical resection is the only potentially curative therapy for pancreatic cancer, patients who undergo curative-intent resection have a five‑year survival rate of only 15–20% [2]. There has thus been great interest in developing novel biomarkers to better stratify patients with pancreatic cancers for personalized treatment decisions and consequently improve survival outcomes.
Radiomics involves the high-throughput extraction of quantitative imaging features from medical images and has the potential to yield imaging biomarkers that enable precision diagnosis and treatment [3–5]. The potential of radiomics in pancreatic cancers have been shown in various studies for the evaluation of resectability, treatment response, and prognosis [6–11]. To ensure the successful application of these radiomic biomarkers, there is a further need to investigate radiomic feature reproducibility [12–15], a key step in the development of clinically relevant radiomics algorithms [16, 17].
Previous studies in lung cancer as well as head and neck cancers have shown that various factors, e.g., acquisition and reconstruction parameters as well as inter-reader variability in segmentation, can degrade radiomic feature reproducibility [18, 19]. However, the reproducibility of radiomic features extracted from contrast-enhanced computed tomography (CECT) of the pancreas remains unknown. Thus, the aim of our study was to investigate the reproducibility of radiomic features in pancreatic parenchyma and pancreatic ductal adenocarcinomas (PDAC) in patients who underwent consecutive CECT scans within a two-week period.
Materials and Methods
Patients
This HIPAA-compliant retrospective study was approved by the Institutional Review Board, and the need for patient informed consent was waived. Figure 1 shows the patient flowchart for this study. A cohort of eligible patients was identified through a database query for patients with a diagnosis code of pancreatic malignancy between June 2009 and October 2015. This patient population was narrowed to include all consecutive patients who underwent two CECT scans within an interval of two weeks, with available Digital Imaging and Communications in Medicine (DICOM) images on Picture Archiving and Computer System (Centricity PACS, GE Healthcare), with CECT obtained during the portal venous phase, and with slice reconstruction thickness of 5 mm or less. To achieve a clean test-retest dataset to study radiomic feature reproducibility in the pancreatic parenchyma, we included the following exclusion criteria: (1) any systemic or loco-regional treatment (i.e., chemotherapy, surgical procedure, and radiation therapy) directed to the tumor during the time interval between the two consecutive CECTs or within one month preceding the first CT and (2) non-evaluable CECTs (e.g., with imaging artifacts over the pancreatic parenchyma or the tumor). According to these inclusion and exclusion criteria, the study cohort consisted of 37 pairs of scans from 37 unique patients in the analysis of the reproducibility of radiomic features derived from the pancreatic parenchyma. To identify a subset of patients to study radiomic feature reproducibility in the pancreatic tumor, we applied the following additional exclusion criteria: (1) not a PDAC as confirmed by electronic medical record review, and (2) size of the tumor smaller than 2 cm or not perceptible by a radiologist for segmentation. A subset of 18 pair of scans were further analyzed for the reproducibility of radiomic features derived from PDAC. For patients with more than one pair of CECT scans during the inclusion period, we included only the first pair for analysis.
Figure 1.
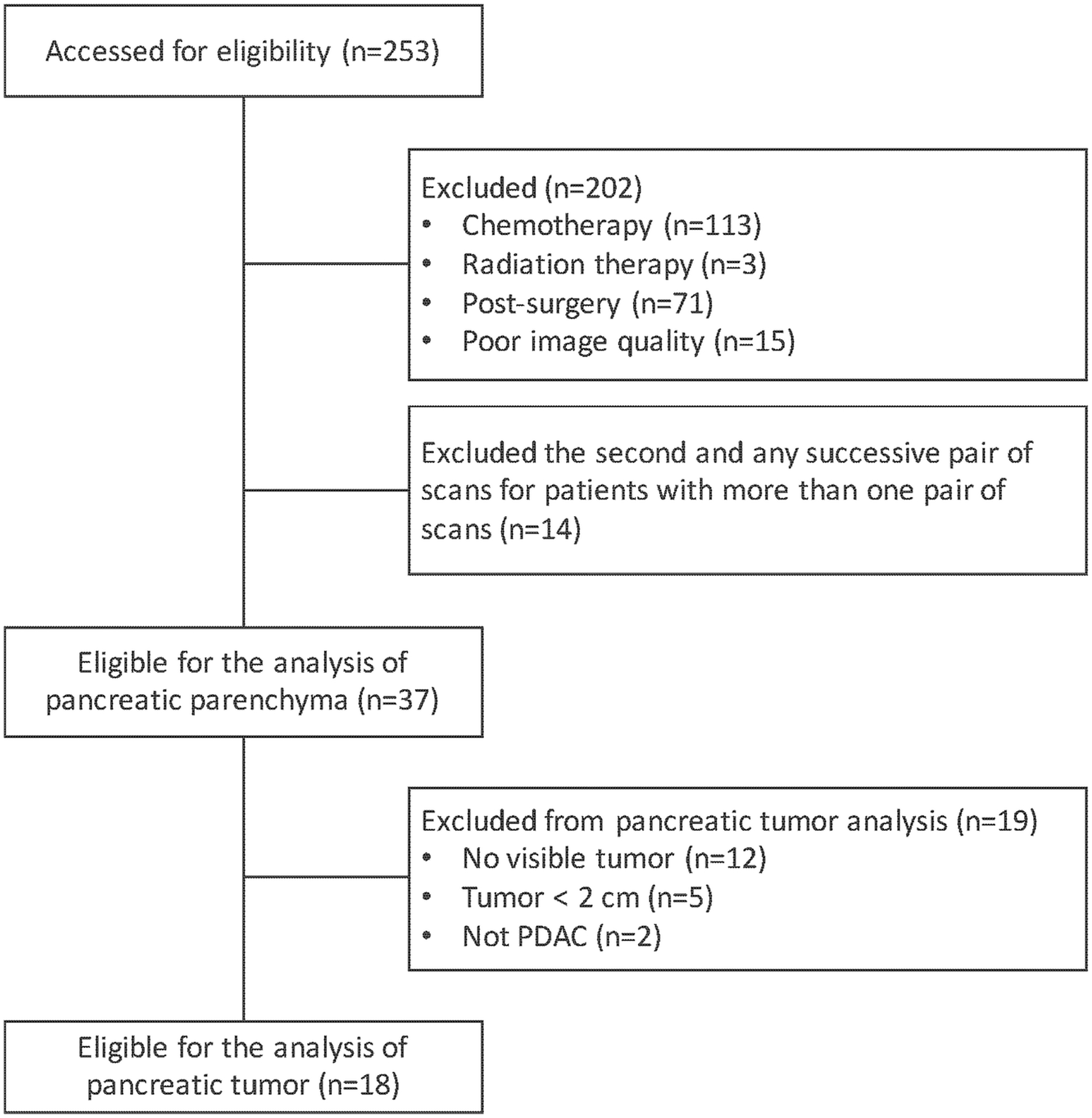
Study flowchart. Note that n is the number of pairs of scans.
Abbreviations: PDAC; pancreatic ductal adenocarcinoma
CT scan parameters
Acquisition and reconstruction parameters were extracted from the DICOM headers and electronic medical records for each CECT examination including scan institution, scanner model, scan protocol, slice thickness, pixel spacing, reconstruction diameter, tube voltage, tube current, exposure time, exposure, noise index, adaptive statistical iterative reconstruction (ASiR) value, convolution kernel, contrast injection dose and administration rate. For this study, we also calculated the time interval in days between the two scans.
Segmentation
CT images were downloaded to a workstation. Pancreatic parenchyma and pancreatic tumor (when present) were manually segmented by T.P. and M.A.K., and then edited and confirmed by two radiologists (R.Y. and N.H. with 10 and 5 years of experience in abdominal CT interpretation, respectively), independently, using a commercially available segmentation software (Scout Liver, Pathfinder Technologies Inc.). The individual scans were randomized so that while annotating any given scan, the reader was blinded to the other time point for the same patient to mitigate recall bias.
Feature extraction
In total, 266 radiomic features were extracted from the pancreatic parenchyma and pancreatic tumor, for each patient and each scan. The feature sets comprised 12 well-known shape-based features [20] that describe shape of an object, and 254 widely used texture-based features that represent heterogeneity of a lesion [13], including 19 features from gray-level co-occurrence matrix (GLCM) [21–23], 11 from run length matrix (RLM) [24], 5 from intensity histogram (IH), 127 from local binary patterns (LBP) [25–27], 54 from fractal dimension (FD) [28, 29], and 38 from two angle co-occurrence matrices (ACM) [30–32]. Two additional features, tumor volume and maximal diameter, were also calculated for the pancreatic tumors. Each feature was extracted from all slices in which the pancreas or tumor appeared and averaged to form a single value. Electronic Supplementary Material 1 lists all imaging features included in this study. All quantitative image analysis was implemented in MATLAB version R2015a (MathWorks).
Statistical analysis
To evaluate inter-reader agreement of the segmentations performed independently by the two radiologists, we calculated the Dice coefficient which measured the spatial overlap between two segmentations, X and Y, and was defined as Dice (X,Y) = 2 (X ∩ Y) / (X + Y) where ∩ is the intersection [33].
Reproducibility was defined as the amount of agreement between measures under all possible conditions on identical subjects for whom measurements are taken [34]. Image acquisition and reconstruction parameters as well as scanner models (Discovery vs LightSpeed, GE Medical Systems) were summarized using the frequency and percentage for categorical variables and compared between the first and second CECT scans using McNemar’s test. The median and range for continuous covariates were compared between the first and second CECT scans using Wilcoxon Sign Rank tests. Concordance correlation coefficients (CCCs) were used to quantify feature reproducibility for the following six sets of analyses in three different conditions.
-
CCC between radiologists segmenting the same CECT
S1R1/R2: reader 1 on scan 1 vs reader 2 on scan 1
S2R1/R2: reader 1 on scan 2 vs reader 2 on scan 2
-
CCC between paired CECTs segmented by the same radiologist
R1S1/S2: reader 1 on scan 1 vs reader 1 on scan 2
R2S1/S2: reader 2 on scan 1 vs reader 2 on scan 2
-
CCC between paired CECTs segmented by different radiologists
R1S2/R2S1: reader 1 on scan 2 vs reader 2 on scan 1
R1S1/R2S2: reader 1 on scan 1 vs reader 2 on scan 2
The number of reproducible features were calculated for thresholds of CCC > 0.80, 0.85, and 0.90, respectively [13].
To assess the influence of scan-related parameters on the agreement of each radiologist performing segmentations on paired CECT scans from the same patient (i.e., R1S1/S2 and R2S1/S2), intraclass correlation coefficient (ICC) was calculated using a mixed effects linear model method by treating CECT scans as a random effect [35]. This allows the flexibility to adjust for covariates in the model. Five univariate mixed effect models were constructed for each radiologist performing the segmentation on paired CECT scans and included the following scan-related parameters as a fixed effect: 1) scanner models (Discovery vs LightSpeed); 2) pixel spacing resolution; 3) exposure; 4) administration rate; 5) gap time. Three of these parameters are routinely modified by CT technologists: CT scanner model choice is based on availability of the scanner at the time of scheduling, pixel resolution varies depending on the size of the scan field of view, and contrast injection rate is modified based on the type of venous access available [13]. Due to the small sample size, it was not feasible to construct a multivariable model and to account for other potential confounders. Differences in the number of reproducible features between unadjusted ICC and adjusted ICC were assessed according to radiomic features groups. All analyses were done in SAS (version 9.3, Inc.). All P-values were two-sided. P-values of <0.05 were considered to indicate statistical significance.
Results
Study cohort and scan parameters
The study cohort consisted of 37 unique patients, 20 males and 17 females (median age = 66 years, range 35–86). Table 1 summarizes the scan parameters between pairs of CECTs in the entire cohort (Electronic Supplementary Material 2 details the scan parameters for all the scans). CECTs were performed either at our institution (n = 71) or at an outside institution (n = 3). The tube voltage was constant at 120 kVp across all the scans. The convolution kernel was “Standard” for all the scans. There were no significant differences between the first and seconds CECTs in the entire cohort. Similarly, distributions of scan parameters were similar in the subset of 18 patients with pancreatic tumor (data not shown). The time interval between the two scans ranged from 2 to 14 days (median 9 days) for the analysis of pancreatic parenchyma and from 3 to 14 days (median 10 days) for the analysis of the pancreatic tumor.
Table 1.
Imaging reconstruction and acquisition variables for the analysis of pancreatic parenchyma
| Scan 1 | Scan 2 | p-value* | |
|---|---|---|---|
| (n=37) | (n=37) | ||
| Model, n (%) | 0.32 | ||
| Discovery (CT750 HD/STE) | 8 (21.6) | 11 (29.7) | |
| LightSpeed(VCT/16) | 29 (78.4) | 26 (70.3) | |
| Pixel resolution (mm) | 0.78 (0.65–0.97) | 0.80 (0.60–0.9) | 0.77 |
| Slice thickness (mm) | 5 (3.75–5) | 5 (2.5–5) | 0.43 |
| Reconstruction diameter (mm) | 400 (351–500) | 410 (334–470) | 0.71 |
| Exposure time (ms) | 724 (500–1040) | 800 (500–1389) | 0.42 |
| Exposure (mAs) | 29 (6–85) | 30 (9–85) | 0.35 |
| Tube current (mA) | 260 (119–645) | 263 (172–441) | 0.26 |
| Contrast dose (cc) | 150 (100–150) | 150 (100–150) | 0.99 |
| Injection Rate (cc/s) | 2 (1–4) | 2 (1–3.5) | 0.32 |
| Scan Institution, n (%) | 0.56 | ||
| Internal | 35 (94.6) | 36 (97.3) | |
| External | 2 (5.4) | 1 (2.3) | |
| Protocol, n (%) | 0.65 | ||
| Monophasic | 34 (91.8) | 33 (89.2) | |
| Triphasic | 3 (8.2) | 4 (10.8) | |
| Noise Index | 12.5 (8–16.6) | 12.5 (8–16.0) | 0.37 |
| ASiR | 0 (0–40) | 0 (0–40) | 0.32 |
Note: The distributions of continuous and categorical variables were compared between scans using Wilcoxon singed rank test and McNemar’s test, respectively. The median and range are presented unless otherwise specified.
Inter-reader variability in segmentation
The median Dice coefficient was 0.851 (interquartile range [IQR], 0.086) for scan 1 and 0.845 (IQR, 0.068) for scan 2 for segmentation of the pancreatic parenchyma. The median Dice coefficient was 0.707 (IQR, 0.121) for scan 1 and 0.686 (IQR, 0.160) for scan 2 for segmentation of the pancreatic tumor (Supplemental Figure 2).
Radiomic feature reproducibility
For pancreatic parenchyma-derived radiomic features, using a threshold CCC > 0.90, 58/266 (21.5%) and 48/266 (18.1%) features met the threshold for S1R1/R2 and S2R1/R2, respectively (same CECT, different radiologists), 14/266 (5.3%) and 15/266 (5.6%) features for R1S1/S2 and R2S1/S2, respectively (same radiologist, different CECTs), and 14/266 (5.3%) and 10/266 (3.8%) features for R1S2/R2S1 and R1S1/R2S2, respectively (different radiologists, different CECTs). Figure 2 shows the number of pancreatic parenchyma-derived features meeting different thresholds.
Figure 2.
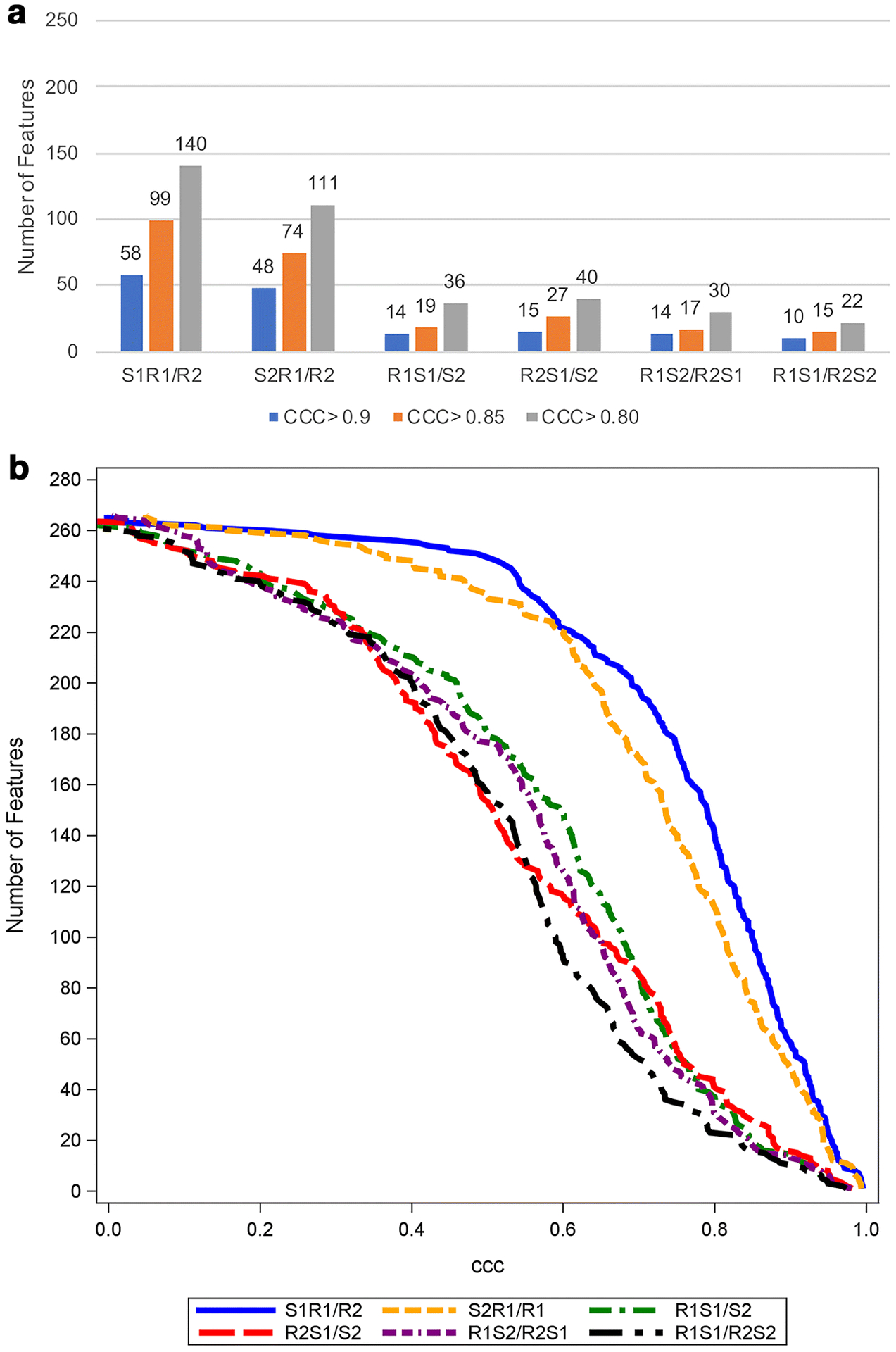
The number of pancreatic parenchyma-derived features with CCC > 0.9, 0.85, and 0.8 (a) and cumulative distribution function plot of the CCC (b) in six sets of analyses for pancreatic parenchyma (n=37). The total number of features examined is 266.
Abbreviations: CCC; concordance correlation coefficient, S1R1/R2; reader 1 on scan 1 vs reader 2 on scan 1, S2R1/R2; reader 1 on scan 2 vs reader 2 on scan 2, R1S1/S2; reader 1 on scan 1 vs reader 1 on scan 2, R2S1/S2; reader 2 on scan 1 vs reader 2 on scan 2, R1S2/R2S1; reader 1 on scan 2 vs reader 2 on scan 1, R1S1/R2S2; reader 1 on scan 1 vs reader 2 on scan 2
For pancreatic tumor-derived radiomic features, using a threshold CCC > 0.90, 11/268 (4.1%) met the threshold for S1R1/R2, 17/268 (6.3%) for S2R1/R2, 1/268 (0.4%) for R1S1/S2, 5/268 (1.9%) for R2S1/S2, and no features (0/268) for both R1S2/R2S1 and R1S1/R2S2. Figure 3 shows the number of pancreatic tumor-derived features meeting different thresholds.
Figure 3.
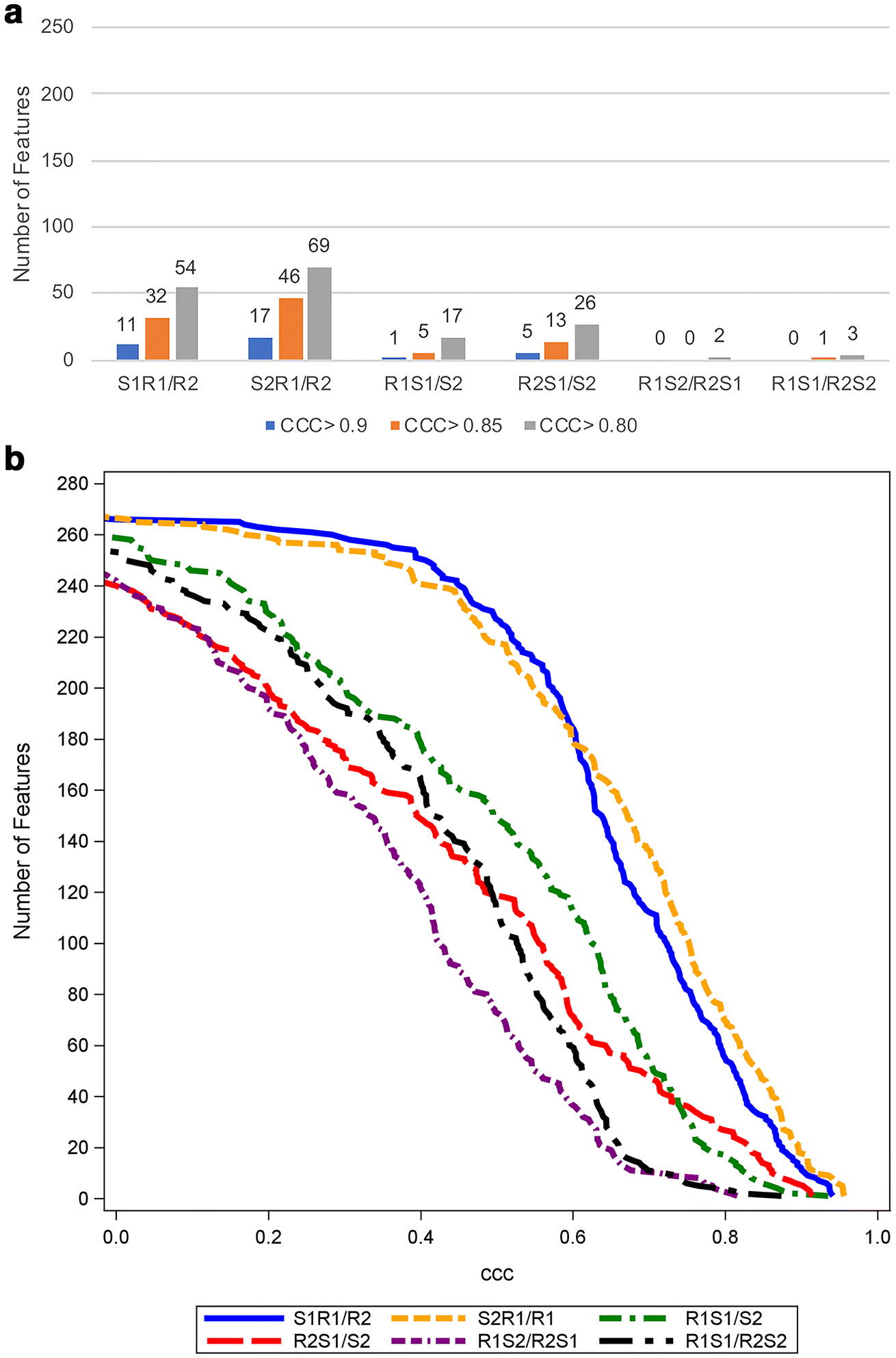
The number of pancreatic tumor-derived features with CCC > 0.9, 0.85, and 0.8 (a) and cumulative distribution function plot of the CCC (b) in six sets of analyses for pancreatic tumor (n=18). The total number of features examined is 268.
Abbreviations: CCC; concordance correlation coefficient, S1R1/R2; reader 1 on scan 1 vs reader 2 on scan 1, S2R1/R2; reader 1 on scan 2 vs reader 2 on scan 2, R1S1/S2; reader 1 on scan 1 vs reader 1 on scan 2, R2S1/S2; reader 2 on scan 1 vs reader 2 on scan 2, R1S2/R2S1; reader 1 on scan 2 vs reader 2 on scan 1, R1S1/R2S2; reader 1 on scan 1 vs reader 2 on scan 2
The FD feature group derived from the pancreatic parenchyma tended to be more robust across all six sets of analyses compared with other feature groups (Figure 4, Supplemental Table 1). On the other hand, only one FD feature (FD1_6) derived from the pancreatic tumor had a CCC that was consistently larger than 0.8, and no feature had a CCC value larger than 0.85 (Figure 5).
Figure 4.
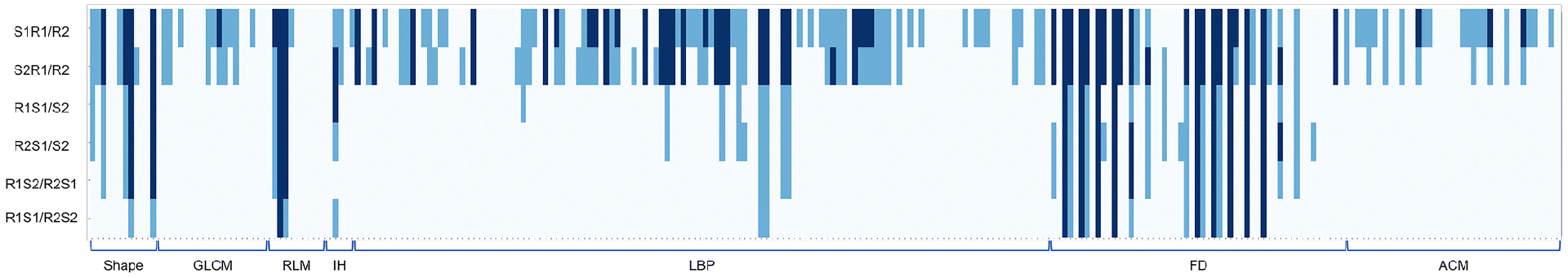
Heatmap demonstrating pancreatic parenchyma-derived reproducible features with thresholds of CCC > 0.8 and CCC > 0.9, where each row and column represents each comparison dataset and radiomic feature, respectively. Pale blue indicates CCC ≤ 0.8, light blue indicates 0.8 < CCC ≤ 0.9, and dark blue indicates 0.9 < CCC.
Abbreviations: CCC; concordance correlation coefficient, GLCM; gray-level co-occurrence matrix, RLM; run length matrix, IH; intensity histogram, LBP; local binary pattern, FD; fractal dimension, ACM; angle co-occurrence matrix, S1R1/R2; reader 1 on scan 1 vs reader 2 on scan 1, S2R1/R2; reader 1 on scan 2 vs reader 2 on scan 2, R1S1/S2; reader 1 on scan 1 vs reader 1 on scan 2, R2S1/S2; reader 2 on scan 1 vs reader 2 on scan 2, R1S2/R2S1; reader 1 on scan 2 vs reader 2 on scan 1, R1S1/R2S2; reader 1 on scan 1 vs reader 2 on scan 2
Figure 5.
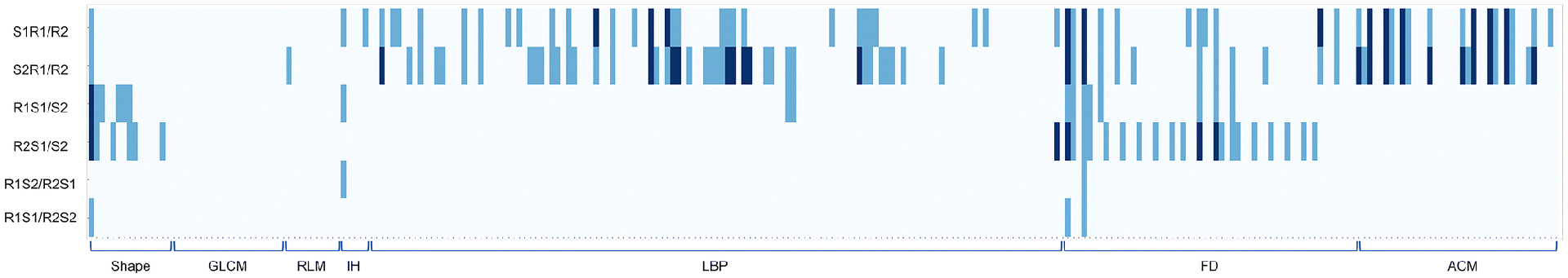
Heatmap demonstrating pancreatic tumor-derived reproducible features with thresholds of CCC > 0.8 and CCC > 0.9, where each row and column represents each comparison dataset and radiomic feature, respectively. Pale blue indicates CCC ≤ 0.8, light blue indicates 0.8 < CCC ≤ 0.9, and dark blue indicates 0.9 < CCC.
Abbreviations: CCC; concordance correlation coefficient, GLCM; gray-level co-occurrence matrix, RLM; run length matrix, IH; intensity histogram, LBP; local binary pattern, FD; fractal dimension, ACM; angle co-occurrence matrix, S1R1/R2; reader 1 on scan 1 vs reader 2 on scan 1, S2R1/R2; reader 1 on scan 2 vs reader 2 on scan 2, R1S1/S2; reader 1 on scan 1 vs reader 1 on scan 2, R2S1/S2; reader 2 on scan 1 vs reader 2 on scan 2, R1S2/R2S1; reader 1 on scan 2 vs reader 2 on scan 1, R1S1/R2S2; reader 1 on scan 1 vs reader 2 on scan 2
Influence of scan-related parameters on radiomic feature reproducibility
Figure 6 shows the reproducibility of radiomic features between paired CECT scans and between paired CECTs after controlling for individual scan-related parameters. Using the mixed effects linear model to account for potential covariates, reproducibility was found to be less impacted by exposure or gap time for both radiologists, and the number of radiomic features was not substantially different at any given ICC (Figure 6aC/6aE; Figure 6bC/6bE). On the contrary, for both radiologists, reproducibility improved slightly after taking into account variabilities between scanner models (Figure 6aA/Figure6bA) and contrast administration rate (Figure6aD/Figure6bD). For R1 only, ICC improved when taking into account the variability in either the pixel spacing or the contrast administration rate from scan 1 to scan 2; the number of radiomic features with ICC > = 0.8 increased from 36 to 49 after adjusting for pixel spacing and improved from 36 to 51 after adjusting for contrast administration rate. This was not observed for R2 (Figure 6aB; Figure 6bB). In terms of radiomic feature groups, LBP, FD, and ACM tended to be relatively influenced by machine model, pixel spacing, and injection rate compared with other feature groups (Supplemental Figure 1).
Figure 6.
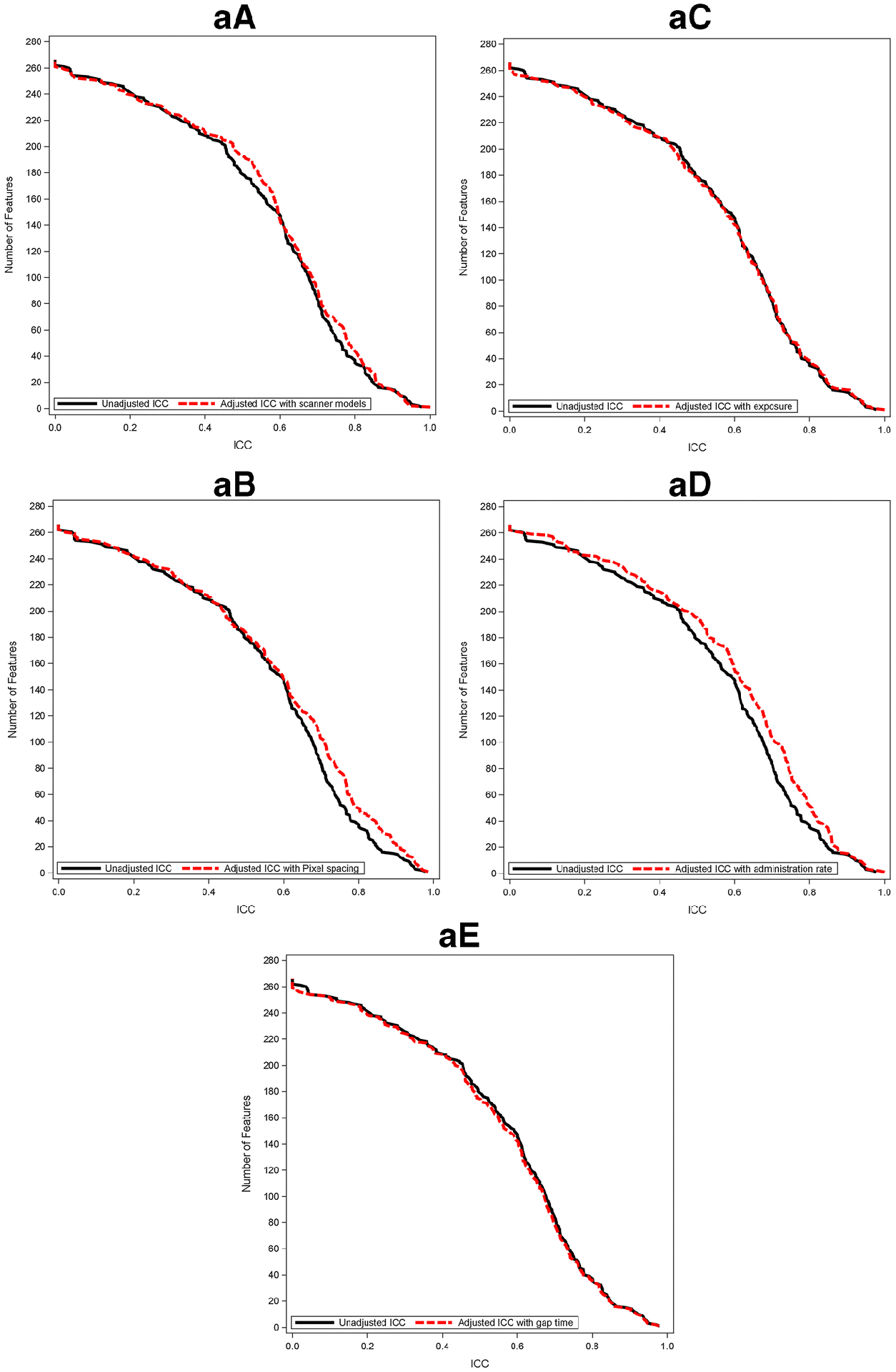
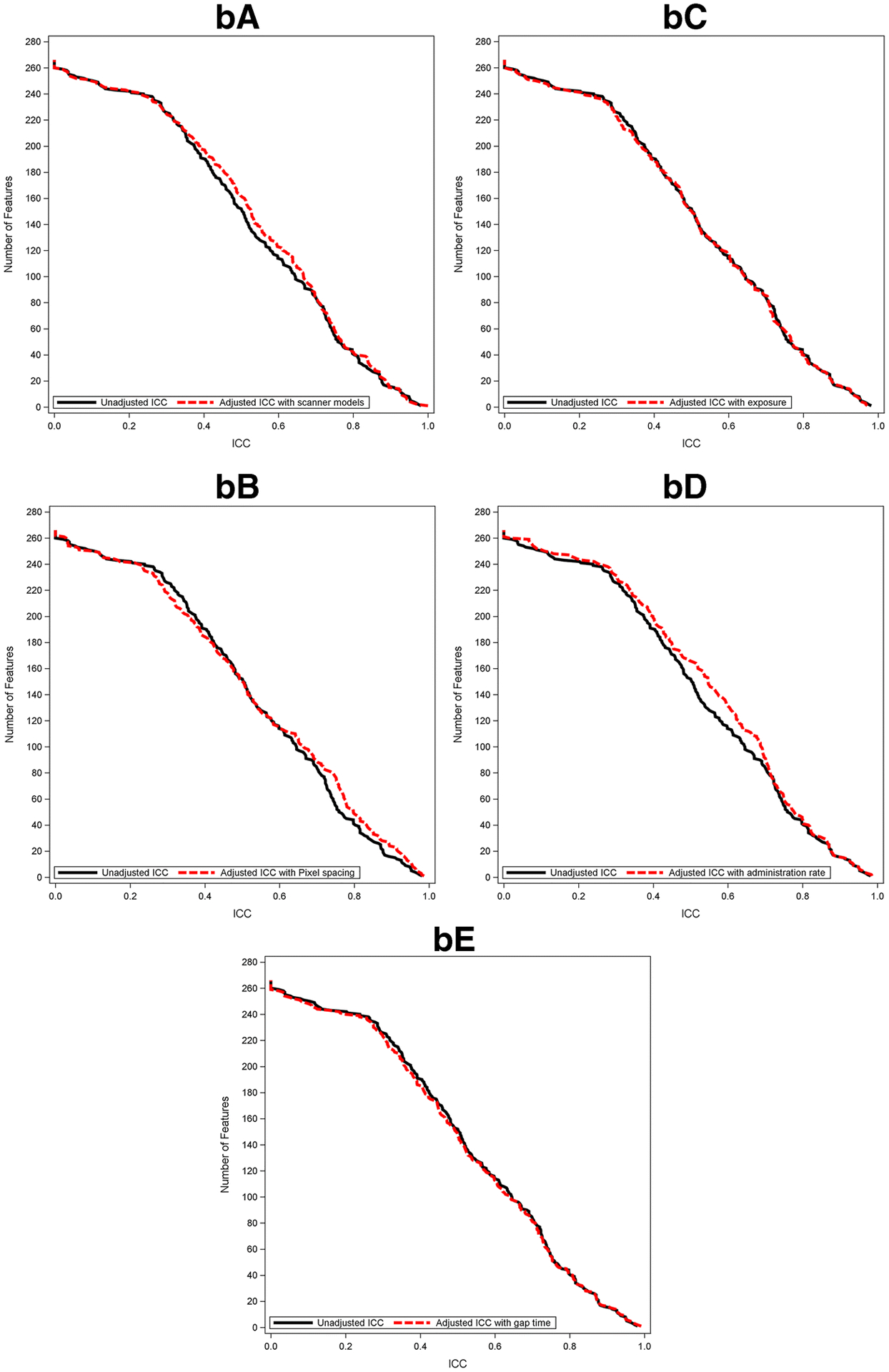
Cumulative distribution function plots of adjusted ICC value of pancreatic parenchyma-derived (a) and tumor-derived (b) features with scanner model (A), pixel spacing (B), exposure (C), administration rate (D), and gap time (E) along with that of unadjusted ICC.
Abbreviations: ICC; intraclass correlation coefficient
Discussion
Our results show that routine variations between CECT scans obtained within a two-week period affected the number of reproducible pancreatic radiomic features to a greater extent than variations in segmentation from different radiologists. When both the CECT scan and the radiologist performing segmentation were different, the number of reproducible features were further decreased. When comparing pancreatic tumor-derived radiomic features and pancreatic parenchyma-derived features under the same conditions, we found that a smaller number of pancreatic tumor-derived features were reproducible; this could be due to the larger inter-reader variability in segmentations of pancreatic tumor compared with the parenchyma or the smaller size of the pancreatic tumors.
While there were no significant differences between acquisition variables such as scanner model, pixel spacing, and contrast administration rate, the number of reproducible features was increased slightly after accounting for variabilities in the scanner model, pixel spacing, and contrast administration rate. The improvement was more noticeable with the first radiologist when either pixel spacing or administration rate was accounted for in the regression model. Improvement was not observed in the readings by the second radiologist. This suggested that some radiologists were more sensitive to the scanner-related parameters, demonstrating the importance to adjusting for these variables. Our results are consistent with previous research by Perrin et al. [13] and highlight the potential hurdles in the use of quantitative radiomic features derived from contrast-enhanced CT images of the pancreas.
Compared with prior studies in other malignancies, our study found a lower number of reproducible radiomic features for the pancreatic malignancy. Balagurunathan et al. [36, 37] assessed the CCC of 219 radiomic features from test–retest non contrast-enhanced CT in lung cancer patients acquired 15 minutes apart; they found 66 features with a CCC > 0.9. The lower number of reproducible pancreas features in our study compared with prior lung radiomics investigations, including the Balagurunathan study with the RIDER data set [38], may be due to our inclusion of CECT scans separated by two weeks rather than using a coffee-break type design as used in these studies. A number of scan-related parameters and normal patient physiologic variability may affect feature reproducibility in CECT obtained days apart. For example, contrast dose and injection rate may change based on renal function and venous access on different days, and additional physiologic patient variations from week to week, such as cardiac output and hydration state, could further affect radiomics reproducibility in CECT. Timmeren et al. [14] compared radiomic feature reproducibility between a clinical scenario-derived rectal cancer test-retest data set and a coffee-break lung cancer RIDER test-retest data set [38]. In total, only 9 features out of 542 were reproducible in the clinical scenario-derived rectal cancer data set, whereas 234 features were reproducible in the RIDER data set. Perrin et al. [13] assessed the reproducibility of 254 texture features in 38 patients with liver tumors in CECT acquired within a two-week interval; 35 features were found to have CCC > 0.9, which is also a higher proportion compared to our results in the pancreas. In addition, inter-reader variability in manual segmentation of pancreatic tumors could be larger compared with other malignancies because of the poorly defined borders of pancreatic tumors. However, as noted above, the variability in pancreas segmentation appeared to affect reproducibility to a lesser extent than short-term scan variability.
Our study had several limitations. First, the number of subjects was small. This small sample size also limited the number of variables that can be included in the regression analyses, so we were not able to clearly identify which imaging variables most affected reproducibility. Second, the number of radiologists involved in our study was limited to two. Third, while we used different threshold CCC > 0.9, 0.85, and 0.8 to illustrate the number of reproducible features at each cut-off, it is unclear which threshold would be clinically meaningful in future pancreatic radiomic feature biomarker development. Further investigation on whether the reproducibility of individual feature has significant impact on the overall performance of a predictive model that combines several features remains to be determined. It is possible that combinations of features in a predictive model will obviate the need to assess individual feature reproducibility. Finally, this study was conducted based on an assumption that biological conditions for pancreatic parenchyma and tumor were stable within a two-week period. A shorter interval may improve the number of reproducibility of pancreatic radiomic features but would have further limited the sample size.
Although prospective studies would be the optimal way to properly address the influence of each individual acquisition and reconstruction parameter on radiomic feature reproducibility, our retrospective results are still informative since our cohort could reflect real world variations in scanning patients for biomarker trials in terms of variations in acquisition parameters, scanner hardware, and institution specific CT protocols that are expected to have an impact on radiomic feature reproducibility. Further investigations into the effects of segmentations by larger number of radiologists may be less fruitful with the eventual development of automated segmentation algorithm for pancreatic parenchyma and tumors. The development of alternatives to improve radiomic feature reproducibility are also anticipated, such as image preprocessing techniques. Emerging initiatives, such as Image Biomarker Standardization Initiative [39], are essential to foster clinical translation of radiomic research with the aim of precision medicine and may provide further guidance in improving reproducibility of radiomic features.
In conclusion, our study shows the potential hurdles in the development of reproducible pancreatic-derived radiomic features from CECT. Further studies that inform the creation of reproducible and clinically relevant pancreatic radiomic features are warranted.
Supplementary Material
Key Points.
For pancreatic-derived radiomic features from contrast-enhanced CT (CECT), fewer than 25% are reproducible (with a threshold of CCC < 0.9) in a clinical heterogeneous dataset.
Variations between CECT scans affected the number of reproducible radiomic features to a greater extent than variations in radiologist segmentation.
A smaller number of pancreatic tumor-derived radiomic features were reproducible compared with pancreatic parenchyma-derived radiomic features under the same conditions.
Acknowledgements
We thank Luz Adriana Escobar Hoyos and Juliana Brooke Schilsky for their support in figure creation and Joanne Chin for editorial assistance.
Funding:
This study was funded in part through the 2016 Society of Abdominal Radiology Wylie J. Dodds Research Award, the National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30 CA008748, and the Japan Society Promotion of Science Overseas Research Fellowships JSPS/OT/290125.
Abbreviations and acronyms
- ACM
angle co-occurrence matrix
- CCC
concordance correlation coefficient
- CECT
contrast-enhanced computed tomography
- DICOM
digital imaging and communications in medicine
- FD
fractal dimension
- GLCM
gray-level co-occurrence matrix
- ICC
intraclass correlation coefficient
- IH
intensity histogram
- IQR
interquartile range
- LBP
local binary pattern
- PDAC
pancreatic ductal adenocarcinoma
- RLM
run length matrix
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Guarantor:
The scientific guarantor of this publication is Richard K. G. Do, MD, PhD.
Conflict of Interest:
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and Biometry:
Two of the authors (Joanne Chou and Mithat Gonen, PhD) have significant statistical expertise.
Informed Consent:
Written informed consent was waived by the Institutional Review Board.
Ethical Approval:
Institutional Review Board approval was obtained.
- retrospective
- cross sectional study
- performed at one institution
References
- 1.Siegel RL, Miller KD, Jemal A (2018) Cancer statistics, 2018. CA Cancer J Clin 68:7–30. doi: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2.Garrido-Laguna I, Hidalgo M (2015) Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nat Rev Clin Oncol 12:319–334. doi: 10.1038/nrclinonc.2015.53 [DOI] [PubMed] [Google Scholar]
- 3.Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: Images Are More than Pictures, They Are Data. Radiology 278:563–577. doi: 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambin P, Rios-Velazquez E, Leijenaar R, et al. (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48:441–446. doi: 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aerts HJWL Velazquez ER, Leijenaar RTH, et al. (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006. doi: 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandrasegaran K, Lin Y, Asare-Sawiri M, et al. (2018) CT texture analysis of pancreatic cancer. Eur Radiol. doi: 10.1007/s00330-018-5662-1 [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Oshima K, Schott D, et al. (2017) Assessment of treatment response during chemoradiation therapy for pancreatic cancer based on quantitative radiomic analysis of daily CTs: An exploratory study. PLoS One 12:e0178961. doi: 10.1371/journal.pone.0178961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakraborty J, Langdon-Embry L, Cunanan KM, et al. (2017) Preliminary study of tumor heterogeneity in imaging predicts two year survival in pancreatic cancer patients. PLoS One 12:e0188022. doi: 10.1371/journal.pone.0188022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim BR, Kim JH, Ahn SJ, et al. (2018) CT prediction of resectability and prognosis in patients with pancreatic ductal adenocarcinoma after neoadjuvant treatment using image findings and texture analysis. Eur Radiol. doi: 10.1007/s00330-018-5574-0 [DOI] [PubMed] [Google Scholar]
- 10.Yun G, Kim YH, Lee YJ, et al. (2018) Tumor heterogeneity of pancreas head cancer assessed by CT texture analysis: association with survival outcomes after curative resection. Sci Rep 8:7226. doi: 10.1038/s41598-018-25627-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Attiyeh MA, Chakraborty J, Doussot A, et al. (2018) Survival prediction in pancreatic ductal adenocarcinoma by quantitative computed tomography image analysis. Ann Surg Oncol 25:1034–1042. doi: 10.1245/s10434-017-6323-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berenguer R, Pastor-Juan MDR, Canales-Vázquez J, et al. (2018) Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology 172361. doi: 10.1148/radiol.2018172361 [DOI] [PubMed] [Google Scholar]
- 13.Perrin T, Midya A, Yamashita R, et al. (2018) Short-term reproducibility of radiomic features in liver parenchyma and liver malignancies on contrast-enhanced CT imaging. Abdom Radiol (NY) 1–8. doi: 10.1007/s00261-018-1600-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Timmeren JE, Leijenaar RTH, van Elmpt W, et al. (2016) Test-Retest Data for Radiomics Feature Stability Analysis: Generalizable or Study-Specific? Tomography 2:361–365. doi: 10.18383/j.tom.2016.00208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Summers RM (2017) Texture analysis in radiology: Does the emperor have no clothes? Abdom Radiol (NY) 42:342–345. doi: 10.1007/s00261-016-0950-1 [DOI] [PubMed] [Google Scholar]
- 16.Kumar V, Gu Y, Basu S, et al. (2012) Radiomics: the process and the challenges. Magn Reson Imaging 30:1234–1248. doi: 10.1016/j.mri.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambin P, Leijenaar RTH, Deist TM, et al. (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749–762. doi: 10.1038/nrclinonc.2017.141 [DOI] [PubMed] [Google Scholar]
- 18.Zhao B, Tan Y, Tsai W-Y, et al. (2016) Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep 6:23428. doi: 10.1038/srep23428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavic M, Bogowicz M, Würms X, et al. (2018) Influence of inter-observer delineation variability on radiomics stability in different tumor sites. Acta Oncol 1–5. doi: 10.1080/0284186X.2018.1445283 [DOI] [PubMed] [Google Scholar]
- 20.Mingqiang Y, Kidiyo K, Joseph R (2008) A survey of shape feature extraction techniques. Pattern recognition techniques, technology and applications. doi: 10.5772/6237 [DOI] [Google Scholar]
- 21.Haralick RM, Shanmugam K, Dinstein I (1973) Textural Features for Image Classification. IEEE Trans Syst Man Cybern 3:610–621. doi: 10.1109/TSMC.1973.4309314 [DOI] [Google Scholar]
- 22.Yang X, Tridandapani S, Beitler JJ, et al. (2012) Ultrasound GLCM texture analysis of radiation-induced parotid-gland injury in head-and-neck cancer radiotherapy: an in vivo study of late toxicity. Med Phys 39:5732–5739. doi: 10.1118/1.4747526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banik S, Rangayyan RM, Desautels JEL (2013) Measures of angular spread and entropy for the detection of architectural distortion in prior mammograms. Int J Comput Assist Radiol Surg 8:121–134. doi: 10.1007/s11548-012-0681-x [DOI] [PubMed] [Google Scholar]
- 24.Tang Xiaoou (1998) Texture information in run-length matrices. IEEE Trans on Image Process 7:1602–1609. doi: 10.1109/83.725367 [DOI] [PubMed] [Google Scholar]
- 25.Ojala T, Pietikäinen M, Harwood D (1996) A comparative study of texture measures with classification based on featured distributions. Pattern Recognit 29:51–59. doi: 10.1016/0031-3203(95)00067-4 [DOI] [Google Scholar]
- 26.Pietikäinen M, Hadid A, Zhao G, Ahonen T (2011) Local binary patterns for still images Computer vision using local binary patterns. Springer London, London, pp 13–47 [Google Scholar]
- 27.Mehta R, Egiazarian K (2013) Rotated Local Binary Pattern (RLBP): Rotation invariant texture descriptor. Institute of Electrical and Electronics Engineers IEEE, p pp 497–502 [Google Scholar]
- 28.Al-Kadi OS, Watson D (2008) Texture analysis of aggressive and nonaggressive lung tumor CE CT images. IEEE Trans Biomed Eng 55:1822–1830. doi: 10.1109/TBME.2008.919735 [DOI] [PubMed] [Google Scholar]
- 29.Costa AF, Humpire-Mamani G, Traina AJM (2012) An efficient algorithm for fractal analysis of textures. 2012 25th SIBGRAPI Conference on Graphics, Patterns and Images. IEEE, pp 39–46 [Google Scholar]
- 30.Chakraborty J, Rangayyan RM, Banik S, et al. (2012) Statistical measures of orientation of texture for the detection of architectural distortion in prior mammograms of interval-cancer. J Electron Imaging 21:033010–033011. doi: 10.1117/1.JEI.21.3.033010 [DOI] [Google Scholar]
- 31.Chakraborty J, Rangayyan RM, Banik S, et al. (2012) Detection of architectural distortion in prior mammograms using statistical measures of orientation of texture In: van Ginneken B, Novak CL (eds) Medical Imaging 2012: Computer-Aided Diagnosis; SPIE, p 831521 [Google Scholar]
- 32.Chakraborty J, Midya A, Mukhopadhyay S, Sadhu A (2013) Automatic characterization of masses in mammograms 2013 6th International Conference on Biomedical Engineering and Informatics; IEEE, pp 111–115 [Google Scholar]
- 33.Dice LR (1945) Measures of the Amount of Ecologic Association Between Species. Ecology 26:297. doi: 10.2307/1932409 [DOI] [Google Scholar]
- 34.ISO 5725–2:1994 - Accuracy (trueness and precision) of measurement methods and results -- Part 2: Basic method for the determination of repeatability and reproducibility of a standard measurement method. https://www.iso.org/standard/11834.html. Accessed 14 Dec 2018
- 35.Carrasco JL, Jover L (2003) Estimating the generalized concordance correlation coefficient through variance components. Biometrics 59:849–858. doi: 10.1111/j.0006-341X.2003.00099.x [DOI] [PubMed] [Google Scholar]
- 36.Balagurunathan Y, Gu Y, Wang H, et al. (2014) Reproducibility and Prognosis of Quantitative Features Extracted from CT Images. Transl Oncol 7:72–87. doi: 10.1593/tlo.13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balagurunathan Y, Kumar V, Gu Y, et al. (2014) Test-retest reproducibility analysis of lung CT image features. J Digit Imaging 27:805–823. doi: 10.1007/s10278-014-9716-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Armato SG, Meyer CR, Mcnitt-Gray MF, et al. (2008) The Reference Image Database to Evaluate Response to therapy in lung cancer (RIDER) project: a resource for the development of change-analysis software. Clin Pharmacol Ther 84:448–456. doi: 10.1038/clpt.2008.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zwanenburg A, Leger S, Vallières M, et al. (2016) Image biomarker standardisation initiative. arXiv [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


