Abstract
Ticks are obligate hematophagous arthropods and must tolerate starvation during off-host periods. Macroautophagy (hereafter autophagy) is a well-conserved self-eating mechanism of cell survival and is essential for recycling cellular contents during periods of starvation, stress, and injury in organisms. Although the genome sequence of Ixodes scapularis (Say) is available, the characteristics and functions of autophagy-related gene families remain largely unknown. To advance our understanding of autophagy in I. scapularis, we used comprehensive genomic approaches to identify Atg genes. Homologues of 14 Atg genes were identified, and their protein motif compositions were predicted. Phylogenetic analysis indicated that ATGs in I. scapularis were evolutionarily closely related to their homologues in Haemaphysalis longicornis and Rhipicephalus microplus ticks. Expression patterns of Atg genes differed across tick developmental stages. Immunofluorescence results by monodansylcadaverine (MDC) staining indicated that autophagy was activated after amino acid starvation treatments in I. scapularis embryo-derived cell lines ISE6 and IDE8. Subsequently, the expression of key Atg genes involved in autophagy pathway in both cell lines were examined. In ISE6 cells, the expression levels of three Atg genes (Atg4B, Atg6 and Atg8A) increased significantly after amino acid starvation; similarly, four Atg genes (Atg4A, Atg4B, Atg6 and Atg8B) were upregulated in IDE8 cells in response to starvation. In parallel, the MDC and lysotracker staining results indicated that autophagy was triggered after amino acid starvation treatments in R. microplus embryo-derived cell line BME26. Our observations showed that Atg family genes are highly conserved in ticks and function in autophagy pathway induced by amino acid starvation. These results also provide valuable insight for further autophagy-related research as a new strategy for blocking the transmission of tick-borne pathogens.
Keywords: Ixodes scapularis, blacklegged tick, autophagy-related genes, functional identification, amino acid starvation, autophagosome
1. Introduction
Ticks are obligate hematophagous arthropods that undergo unique life cycles alternating between feeding periods and long non-feeding periods without a blood meal (Umemiya-Shirafuji et al., 2014). Unlike other blood-sucking vectors, most hard tick species can survive without a blood meal for several months to years (Needham and Teel, 1991), and this strategy is essential for persistence of tick-borne pathogens (TBPs). Starvation, induced by a shortage of nutrient supply, is the most frequent challenge to survival in nature. To overcome this threat, organisms can obtain amino acids by recycling cellular components for protein synthesis and survival (Ohsumi, 2014). Autophagy is an intracellular pathway for bulk protein degradation and beneficial to cell survival under adverse environmental conditions (Ravanan et al., 2017). Autophagy occurs at low levels under normal growth conditions, and is an important mechanism used by arthropods to control pathogen burdens (Malagoli et al., 2010; Caljon et al., 2016). However, starvation or low-energy reserves result in a dramatic activation of autophagy (Yorimitsu and Klionsky, 2005). The classical morphological feature of autophagy is mature, double-membrane vesicles, termed autophagosomes. During autophagy, phagophores, the autophagosome precursors, take up proteins, lipids, and damaged organelles from the cytoplasm (Delorme-Axford and Klionsky, 2018). In yeast or plant cells, the autophagosomes then fuse with vacuoles, while in metazoan cells, the autophagosomes fuse with lysosomes. Subsequently, the phagolysosomes deliver the autophagic cargo to the cellular degradation machinery (Gatica et al., 2018), where the break-down products are reused as building blocks for the synthesis of macromolecules, and to generate energy (Feng et al., 2015; He and Klionsky, 2009).
Autophagy is mediated by a set of evolutionarily conserved genes, namely the autophagy related genes (Atg) (Klionsky et al., 2003). Numerous Atg genes and their corresponding proteins have been identified by their functions in regulating the different types/stages of autophagy (Mizushima, 2018). In yeast, genetic screens for autophagy-defective mutants have revealed almost 40 Atgs, and many of these genes have orthologues in higher eukaryotes (Feng et al., 2015). The four conserved signaling modules encoded by yeast Atg genes which control key steps are well characterized, but in mammals, the autophagy system is more complex (Behrends et al., 2010). Systemic or tissue-specific deletion of Atg genes has led to a broad knowledge about the important roles of autophagy in adaptive responses to prolonged stress, homeostasis, and development (Jiang and Mizushima, 2014; Levine et al., 2011; Mizushima and Komatsu, 2011; Mizushima and Levine, 2010). The large phylum Arthropoda, which includes insects, ticks, spiders, crustaceans, etc., diverged from vertebrates as early as 500 million years ago (Hedges et al., 2004), but the molecular basis of some basic biological functions was evolutionarily conserved. Ground-breaking studies dating back to the 1960s (Gaudecker, 1963; Jenkins et al., 2013; Lőrincz et al., 2017) have revealed that the essential mechanisms of autophagy in Drosophila are quite similar to those in other eukaryotes. Therefore, it is not surprising that insects including Drosophila are classical models for autophagy research. Ticks are obligate blood-sucking parasites and among the most important vectors of pathogens affecting humans and animals world-wide. TBPs such as Rickettsia and Babesia spp., cause a heavy burden of deadly or disabling diseases that continue to damage livestock production or human health (de la Fuente et al., 2008; Gulia-Nuss et al., 2016; Jongejan and Uilenberg, 2004). Because autophagy plays an important role in the response of organisms to starvation, and ticks are highly tolerant to nutrient shortage, studies of the mechanism of autophagy in ticks will undoubtedly provide clues for developing tick control strategies. To date, most studies have focused on analyzing the transcription of a few Atg genes in the ticks Haemaphysalis longicornis, Rhipicephalus microplus and Amblyomma sculptum (Fernández et al., 2016; Flores Fernández et al., 2014; Kawano et al., 2011; Moura-Martiniano et al., 2017; Umemiya et al., 2008, 2007; Umemiya-Shirafuji et al., 2014), but the role of autophagy in ticks has not been well-studied, leaving many questions unanswered. As a vector of pathogens, Ixodes scapularis (Say) is responsible for Lyme disease, human granulocytic anaplasmosis, babesiosis and tick-borne encephalitis (Rodríguez et al., 2018). The recent publication of the I. scapularis genome provides invaluable reference to analyze Atg genes at a genome-wide scale (Gulia-Nuss et al., 2016; Kawano et al., 2011; Umemiya et al., 2008, 2007; Umemiya-Shirafuji et al., 2014). Here, we utilize the tick I. scapularis and cell lines derived from its embryos to demonstrate the function of the Atg gene family in response to amino acid starvation. In particular, we are interested in the following questions: 1) How large is the Atg gene family in I. scapularis ticks? 2) How conserved are the signaling modules in the tick autophagy system? 3) How does autophagy affect starvation, or how does starvation affect autophagy in ticks? 4) In addition, do TBPs trigger autophagy in ticks? 5) And if so, what are the autophagic molecular responses to pathogen infection in ticks? Unraveling these questions may reveal the core mechanisms of autophagy in ticks.
Here, we utilize the tick as a model system to explore the functions of the autophagy-related gene family. We use comprehensive genomics approaches to identify Atg gene family members in the genome of I. scapularis. Homologues of 14 Atg genes were identified, and their protein motif compositions were predicted, and the expression patterns of the Atg genes were analyzed for all developmental stages. Autophagy was induced in two I. scapularis cell lines by amino acid starvation treatment and autophagic processes were visualized by monodansylcadaverine (MDC) and lysotracker staining. Our study aimed to provide valuable insight for further autophagy-related research towards a new strategy for blocking the transmission of tick-borne pathogens.
2. Materials and Methods
2.1. Identification of autophagy family genes in I. scapularis (IsAtg)
The complete genome sequence of I. scapularis is accessible at VectorBase (https://www.vectorbase.org/organisms/ixodes-scapularis). To identify the ATG proteins in I. scapularis, BlastP and tBlastN searches were performed against the genome of I. scapularis using the published ATG protein sequences from Homo sapiens, D. melanogaster, and Caenorhabditis elegans as queries with e-values ≤le–20. All significant hits were submitted to the NCBI CDD (https://www.ncbi.nlm.nih.gov/cdd) (Marchler-Bauer et al., 2017) and the Pfam database (https://pfam.xfam.org/) to confirm the presence of conserved domains of ATG proteins. Protein subcellular localization were predicted based on iLoc-Animal (Lin et al., 2013) and CELLO.V.2.5 (Yu et al., 2004).
2.2. Phylogenetic analysis of I. scapularis ATGs
All the ATG protein domains from I. scapularis, Anopheles gambiae, D. melanogaster, Apis mellifera, Bombyx mori, Daphnia pulex, Pediculus humanus, Strigamia maritima, Tetranychus urticae, Tribolium castaneum, C. elegans and H. sapiens were aligned by ClustalW v2.0 (Larkin et al., 2007; Marchler-Bauer et al., 2017) using the default parameters. The source information of all protein sequences was listed in Supplementary Table S1. The phylogenetic trees of all ATG proteins were constructed by maximum likelihood (ML) method using PhyML-3.0 software (Guindon et al., 2010) with 1000 bootstrap replicates. Different models were selected for each tree by Modelgenerator software (Keane et al., 2006) as listed in Supplementary Table S2. The neighbor-joining (NJ) phylogenetic trees of ATG proteins were constructed using MEGA7 (Guindon et al., 2010; Kumar et al., 2016) with 1000 bootstrap replicates.
2.3. Gene structure analysis and conserved motif detection
To investigate the genomic organizations of IsAtg genes, information about gene structure was extracted from the gff file of the I. scapularis genome. Then, Gene Structure Display Server (https://gsds.cbi.pku.edu.cn/index.php) (Guindon et al., 2010; Hu et al., 2015; Kumar et al., 2016) was utilized to graphically portray the numbers and positions of coding sequences (cds)/intron. To discover conserved motifs in the I. scapularis ATG proteins, the Multiple Em for Motif Elicitation (MEME v5.0.5) program was applied with the following parameters: motif sites to be distributed in sequences, zero or one per sequence; Minimum and maximum width of motif, 6 and 50, respectively (Bailey et al., 2006). Other options were left at default values and only motifs with an e-value of <le–20 were retained for further analysis.
2.4. Collection of tick samples
Engorged I. scapularis females (Tick Rearing Facility, Oklahoma State University) were transferred to a humidity chamber with 98% relative humidity, and maintained at 22 °C for oviposition. Eggs were collected for total RNA isolation on days 7-15 after initiation of oviposition. A proportion of eggs was kept until hatching, and unfed larvae (7-10 days post hatching) were collected for total RNA isolation. Remaining larvae were fed on hamsters, and engorged larvae were transferred to a humidity chamber (with 98% relative humidity) and kept at 22 °C for molting. Use of animals for this research was approved by the IACUC of the University of Minnesota under protocol Nr. 1905-37105 A. All standards of ethical animal use and care were strictly adhered to. Newly emerged nymphs (unfed, 7-10 days post molt) were collected for total RNA isolation, and remaining nymphs were kept for feeding. Engorged nymphs were transferred to a humidity chamber with 98% relative humidity and kept at 22 °C for molting. Unfed male (non-virgin), female (non-virgin) and partially engorged female ticks that had been membrane fed for 7 days (Oliver et al., 2016) were collected for total RNA isolation.
2.5. Cell culture
The I. scapularis cell lines ISE6 and IDE8 as well as the R. microplus cell line BME26 were derived from embryonated eggs in our laboratory (Munderloh et al., 1999, 1994, Munderloh and Kurtti, 1989; Esteves et al. 2008). The cell lines were cultured at 34 °C in L-15C300 medium (pH 7.2-7.5) with 5% fetal bovine serum (FBS) (RMBIO, FBS-BBT, Missoula, USA), 5% tryptose phosphate broth (Difco, Becton Dickinson, Bedford, MA), and 0.1% lipoprotein concentrate (MPBiomedical, Irvine, CA), hereafter referred to as complete medium.
2.6. RNA isolation and qPCR
Total RNA was extracted from above samples using TRI Reagent® (Sigma, T9424, St. Louis, MO, USA). RNA samples were purified using a RNA Clean & Concentrator kit (Zymo research, R1017, Irvine, USA) to remove genomic DNA. The quantity and quality of RNA were evaluated using a DS-11 Series Spectrophotometer / Fluorometer (DeNovix, Wilmington, USA). The cDNA was synthesized using the SYBR PrimeScript reverse transcription PCR (RT-PCR) kit II (Takara, RR037A, Otsu, Shiga, Japan). To decipher the expression profile of Atg genes in different developmental stages of I. scapularis, qPCR was employed to detect expression levels in eggs, larvae, nymphs, and adults (female and male). We used the Mx3005P Real-Time system (Stratagene, La Jolla, USA) with SYBR green detection (Agilent Technologies, 600830, Santa Clara, USA). All protocols were according to the manufacturer’s instructions. For qPCR, we used the glyceraldehyde 3-phosphate dehydrogenase and tubulin endogenous genes as controls. Primers used in this study are listed in Supplementary Table S3.
2.7. Amino acid starvation treatment
For amino acid starvation treatment, cells were washed three times with PBS and incubated with Hanks’ Balanced Salt Solution (HBSS; Sigma, 55037C, St. Louis, MO, USA) with no supplements at 34°C for different lengths of time (from 0 to 5 days). In control groups, cells were incubated in fresh complete medium and other conditions were the same as for treatment groups. Then, cells were collected for RNA isolation or immunofluorescence assay. All tests were biologically replicated three times.
2.8. Labeling of autophagic vacuoles with monodansylcadaverine (MDC) and lysotracker
For immunofluorescence assay, the control and starvation treated cells were transferred to 35-mm dishes (MatTek, P35G-1.5-14-C, Ashland MA, USA) and incubated with 50 μM MDC at 37 °C for 10 minutes (Biederbick et al., 1995; Pattingre et al., 2004). After incubation, cells were washed 3 times with PBS. Subsequently, those cells were incubated with 1 μM LysoTracker Red DND-99 (Molecular Probes, L-7528, Eugene, USA) in PBS for 10 seconds at RT. After incubation, cells were washed 3 times with PBS and immediately imaged on a Nikon A1si Spectral Confocal Microscope with the same camera settings for control and treated groups. Images were processed using the program NIS-Elements View 4.50 (University Imaging Centers at the University of Minnesota, Twin Cities). All tests were performed in triplicate for each group.
2.9. Statistical analysis
The relative gene expression level of individual Atg genes was calculated by the comparative CT method (2ΔΔCT). Statistical analysis was carried out using the data obtained from three separate cDNA sets of three independent biological samples, and heatmaps for expression profiles were generated with Mev 4.0 software (Saeed, 2003). Student’s two tailed t-tests were used to compare the control and starvation treated group. Differences between treatments were considered significant when p < 0.05, and are indicated by different letters. All statistical analyses were conducted using SPSS 20.0.
3. Results
3.1. Identification of the Atg gene family in I. scapularis
We utilized ATG protein sequences from three model organisms (D. melanogaster, C. elegans and H. sapiens) to search against the I. scapularis genome database. After removing redundant sequences and alternatively spliced isoforms from the same gene, 4 candidate IsAtg genes were identified. Their domains were further confirmed with the Pfam database (http://pfam.xfam.org/). In parallel, the ATGs were also identified in eight other representative species: An. gambiae, A. mellifera, B. mori, D. pulex, P. humanus, S. maritima, T. urticae and T. castaneum (Table 1). The lengths of the deduced amino acid sequences, the predicted molecular weights, and the predicted isoelectric points (pI) differed greatly among the 14 IsATG proteins (Table 2). In I. scapularis, the numbers of amino acids encoded by identified Atg genes varied from 110 for IsATG8B to 702 for IsATG7, and their corresponding molecular weight ranged between 11.5 kDa for IsATG8B to 76.4 kDa for IsATG7. Protein subcellular localization may provide important clues to help determine its function. In this study, we found that most ATG proteins were predicted to be located in the cytoplasm, and only two ATGs (ATG13, ATG16) were predicted to be in the outer membrane.
Table 1:
Atg genes used in this study.
| Genome size (Mb) |
Atg subfamily | Total | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Order | Organism | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 16 | 17 | 18 | 24 | 101 | ||
| Diptera | Anopheles gambiae | 250.71 | 0 | 1 | 1 | 2 | 1 | 2 | 0 | 1 | 2 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 2 | 0 | 0 | 19 |
| Drosophila melanogaster | 137.68 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 2 | 0 | 1 | 20 | |
| Hymenoptera | Apis mellifera | 235.3 | 0 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 15 |
| Lepidoptera | Bombyx mori | 397.68 | 0 | 0 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 12 |
| Cladocera | Daphnia pulex | 197.2 | 0 | 1 | 1 | 2 | 1 | 1 | 0 | 2 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 15 |
| Phthiraptera | Pediculus humanus | 110.78 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 16 |
| Geophilomorpha | Strigamia maritima | 176.21 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 4 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 13 |
| Ixodida | Ixodes scapularis | 1765.38 | 0 | 1 | 1 | 2 | 1 | 1 | 1 | 3 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 14 |
| Trombidiformes | Tetranychus urticae | 90.82 | 0 | 1 | 1 | 3 | 1 | 2 | 1 | 4 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 19 |
| Coleoptera | Tribolium castaneum | 165.94 | 0 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 15 |
| Rhabditida | Caenorhabditis elegans | 101.16 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 2 | 0 | 1 | 0 | 0 | 16 |
| Primates | Homo sapiens | 2991.11 | 2 | 2 | 1 | 4 | 1 | 1 | 1 | 7 | 2 | 1 | 0 | 1 | 1 | 1 | 2 | 1 | 2 | 2 | 1 | 33 |
Table 2:
Characteristics of the putative Ixodes scapularis ATG genes.
| IsATGs | Transcript ID | Length(aa) | pI | MW(KD) | Subcellular location |
|---|---|---|---|---|---|
| ATG2 | ISCW008987-RA | 422 | 5.17 | 46.591 | Cytoplasmic |
| ATG3 | ISCW007083-RA | 323 | 4.1 | 19.846 | Cytoplasmic |
| ATG4A | ISCW004712-PA | 382 | 5.34 | 43.396 | Cytoplasmic |
| ATG4B | ISCW010488-PA | 433 | 7.80 | 49.791 | Cytoplasmic |
| ATG5 | ISCW013143-RA | 194 | 6.44 | 21.893 | Cytoplasmic |
| ATG6 | ISCW014200-RA | 443 | 4.7 | 51.351 | Cytoplasmic |
| ATG7 | ISCW008151-RA | 702 | 5.56 | 76.444 | Cytoplasmic |
| ATG8A | ISCW000710-RA | 110 | 9.72 | 13.001 | Cytoplasmic |
| ATG8B | ISCW004756-RA | 101 | 9.53 | 11.492 | Cytoplasmic |
| ATG8C | ISCW017654-RA | 117 | 6.91 | 13.982 | Cytoplasmic |
| ATG11 | ISCW006294-RA | 334 | 4.6 | 20.118 | Cytoplasmic |
| ATG13 | ISCW018154-RA | 454 | 8.5 | 49.762 | Outer membrane |
| ATG16 | ISCW000705-RA | 525 | 9.55 | 19.958 | Outer membrane |
| ATG101 | ISCW015657-RA | 124 | 6.28 | 14.313 | Cytoplasmic |
3.2. Phylogenetic analysis of I. scapularis ATGs compared with ATGs of other species
To assess the phylogenetic relationship of ATG proteins in I. scapularis and other species surveyed here, all ATG proteins were aligned to generate unrooted maximum likelihood trees (Fig.1). Unlike in H. sapiens, the size of ATG families in most arthropods was small. Phylogenetic analysis indicated that most IsATGs were phylogenetically closely related to their homologues in other species. The high bootstrap values of internal branches indicated pairs of possibly orthologous proteins sharing similar functions that had diverged from a common ancestor. Several IsATG subfamilies were most closely related to those in the centipede S. maritima which had a small ATG family comprising 13 members. In addition, the ATG subfamilies in I. scapularis were quite small, and most possessed one single gene (Atg2, Atg3, Atg5, Atg6, Atg7, Atg11, Atg13, Atg16 and Atg101), except that the ATG4 subfamily had two members (Atg4A, Atg4B) and the ATG8 subfamily had three members (Atg8A, Atg8B, Atg8C).
Fig. 1.
Phylogenetic analysis of ATGs from I. scapularis, D. melanogaster, C. elegans, H. sapiens, A. gambiae, A. mellifera, B. mori, D. pulex, P. humanus, S. maritima, T. urticae and T. castaneum. Unrooted phylogenetic trees were constructed using the maximum likelihood method with 1000 bootstrap replications in PhyML-3.1 software.
3.3. IsATG gene structure and conserved motif analysis
To obtain further insight into the evolutionary relationships among IsATG family members, their exon-intron structures were predicted and compared (Fig. 2A). The exon-intron structures of IsAtg genes were analyzed by comparing their coding sequences (CDS) and corresponding genomic DNA sequences. IsAtg genes from different subfamilies varied structurally. For example, IsAtg13 consisted of a short sequence with only one exon, whereas IsAtg8B CDS were long, with 12 exons. Interestingly, members of Atg8 and Atg4 subfamilies that together formed one cluster showed different exon/intron structures and numbers (Fig. 2B). Further, the conserved motifs of IsATG proteins were identified using the MEME motif search tool (Fig. 2C). The top three motifs of IsATG8B and IsATG8C shared similar motif compositions, suggesting that the protein architecture was conserved within the subfamily and homologous groups. These were consistent with their phylogenetic relationships and may prove useful for the functional analysis of IsATGs.
Fig. 2.
IsATG gene structure and conserved motifs in the I. scapularis ATG family. (A) Phylogenetic analysis of I. scapularis ATGs. (B) Exon/intron structure of IsATG. (C) The sequence logos of conserved domains are based on full-length alignments of all IsATG proteins. The bit score indicates the information content for each position in the sequence.
3.4. Expression profiles of IsAtg genes across developmental stages
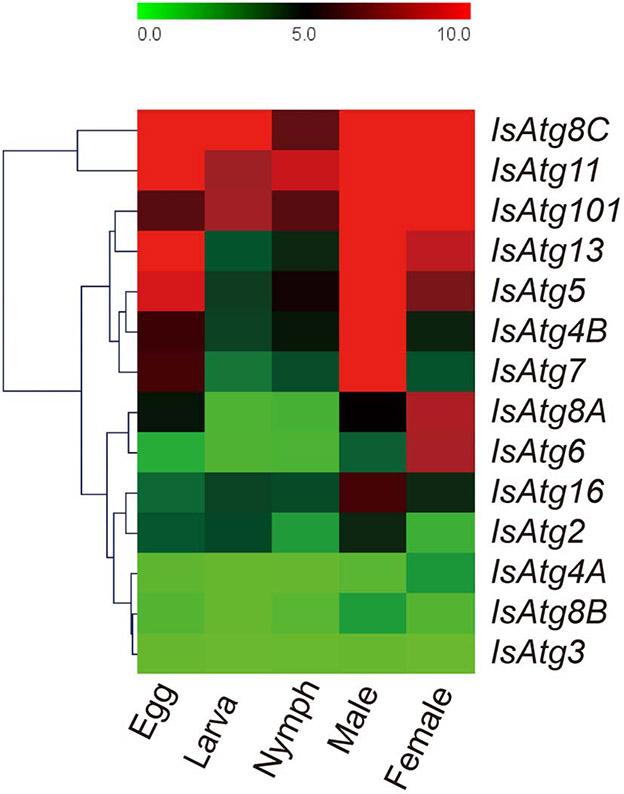
To investigate the function of Atg genes in I. scapularis in vivo, real-time PCR was performed to examine the expression patterns of IsAtgs in ticks at different developmental stages. Most IsAtg genes were expressed throughout the tick life cycle (Fig. 3), and IsAtg11 and IsAtg101 were strongly expressed in ticks from egg to adult. In contrast, IsAtg3, IsAtg8B, IsAtg8C, and others produced transcripts of low abundance, and IsAtg8A was only slightly expressed in female ticks. These results showed that the expression pattern of IsAtg8 subfamily members was variable during tick development. The expression profiles of IsAtg genes also varied with gender. IsAtg4B and IsAtg7 were strongly expressed in male ticks, but were barely expressed in females, whereas IsAtg8A and IsAtg6 were strongly expressed in female ticks, but only weakly in males. We also compared the expression pattern of IsAtg genes between unfed and partially fed female ticks (Fig. S2). In the unfed ticks group, the transcript abundance of most IsAtg genes was high. In contrast, the expression of most IsAtg genes decreased significantly after the ticks had taken a partial blood meal, which indicated most ATGs played a role during the unfed stages. These results suggested that different members of Atg gene families were functionally divergent in ticks.
Fig. 3.
Expression profiles of IsAtg genes in different developmental stages of I. scapularis. The clustering results, showing the gene expression patterns, are shown on the left. For heatmap, each row represents a sampling time point during the indicated development stage and each column represents Atg expression. The scale of relative expression levels is shown by the color bars, from green (low) to red (high).
3.5. Multiple sequence alignments of IsATG4 and IsATG8 subfamily
To analyze the sequence of IsATG4 and IsATG8 proteins, all the deduced proteins of ATG4 and ATG8 subfamilies members surveyed here were aligned. The alignment results showed that cysteine (Cys), aspartic acid (Asp), and histidine (His) residues of IsATG4 were widely conserved in all ATG4 homologues. The deduced amino acid sequence of IsATG4 contained a Peptidase C54 (IsATG4A, 37-320 aa; IsATG4B, 64-371 aa) domain (Fig. S3). We expected to identify the gamma-aminobutyric acid type A receptor-associated protein (GABARAP) binding site in all IsATG8 protein sequence, but interestingly, only IsATG8C possessed this putative active site (1-116 aa). The E1 ubiquitin-activating enzyme ATG7 performs a critical task in autophagy by activating two ubiquitin-like proteins, ATG8 and ATG12 (Geng and Klionsky, 2008; Noda et al., 2011). Additionally, all the I. scapularis ATG8 proteins contained the characteristic active sites, i.e., a catalytic cysteine for ATG7 cleavage (Fig. S4). All amino acid sequences compared contained the conserved C-terminal glycine residue. The conserved deduced amino acid composition indicated IsATG8C may play a role in the formation of autophagosomes.
3.6. Autophagy was induced in ISE6 and IDE8 cells in response to amino acid starvation
We were interested in imaging cellular responses during autophagy, however, it is difficult to avoid nonspecific background and cross-reactivity resulting from the tick blood meal. Therefore, we used I. scapularis embryo-derived cell lines ISE6 and IDE8 as they offer an excellent alternative to monitor autophagy. We utilized HBSS to induce autophagy in tick cells and demonstrated that autophagy was induced in vitro. HBSS treated-ISE6 and IDE8 cells displayed intense, punctate lysosome signals (red) and punctate MDC-specific signals (green), while fewer autophagic and lysosome signals were detected in complete medium treated ISE6 (Fig. 4B) and IDE8 (Fig. S6B) cells. Interestingly, the co-localization results for autophagosomes and lysosomes in ISE6 were not consistent with those in IDE8. One reasonable explanation is that the two cell types have different functional characteristics and ontogenies (Oliver et al., 2015), which implies possible distinct biological properties. With the accumulation of lysosomes after amino acid starvation, the autophagosomes noticeably increased and peaked at 72 h and then decreased after 96 h (Fig. S5). The dynamics of autophagosomes and lysosomes at different time points in ISE6 cells indicated that autophagy was triggered after amino acid starvation treatments. We were particularly interested in Atg4, Atg6, and Atg8 not only because their products play an important role in the formation of the double-membrane autophagosomes but also these proteins were well conserved in organisms. Hence, the expression of those key Atgs involved in the autophagy pathway in both cell lines was examined. In ISE6 cells, the expression level of Atg4B, Atg6, and Atg8A increased significantly after a 24-hour treatment with HBSS (Fig. 4A); Similarly, expression of Atg4A, Atg4B, Atg6, and Atg8B was upregulated in IDE8 cells in response to starvation (Fig. S6A), while in ISE6 cells, the expression level of Atg4A and Atg8B did not change significantly after amino acid starvation. Meanwhile, expression of Atg8C in ISE6, Atg8A and Atg8C in IDE8 decreased in response to starvation. These results suggested that autophagy was activated after amino acid starvation treatments in I. scapularis embryo-derived cell lines ISE6 and IDE8.
Fig. 4.
Amino acid starvation induces autophagy in ISE6 cells. (A) Relative expression of key Atg genes measured in ISE6 cells after HBSS treatment for 1 day by qPCR with GAPDH expression as the internal control. Significant differences are indicated by different letters. (B) MDC staining (green) for autophagosomes and lysotracker staining (red) for lysosomes in ISE6 cells after HBSS treatment for 1 day. Bar: 20 μm.
3.7. Autophagy was induced in BME26 cells in response to amino acid starvation
To further assess the phylogenetic relationship of Atg genes in I. scapularis, H. longicornis and R. microplus ticks, the EST sequences of Atg genes in H. longicornis and R. microplus were downloaded from NCBI and subsequently aligned to generate an unrooted maximum likelihood tree (Fig. 5). Phylogenetic analysis indicated that Atgs in I. scapularis were evolutionarily closely related to their homologues in H. longicornis and R. microplus ticks. To rule out a possible confounding effect of starvation during generation of the R. microplus EST database, BME26 cells were labeled with MDC and lysotracker. We measured the dynamics of autophagosomes and lysosomes at different time points in BME26 cells using confocal microscopy and immunofluorescence. Lysosomes and autophagosomes could barely be detected in control cells cultured in complete medium. With the accumulation of lysosomes after amino acid starvation, the autophagosomes noticeably increased and peaked at 5 days (Fig. 6). The results showed that autophagy was triggered after amino acid starvation treatments in the R. microplus embryo cell line BME26, indicating this pathway was active in this species as expected.
Fig. 5.
Phylogenetic analysis of ATGs from I. scapularis, H. longicornis and R. microplus ticks. Unrooted phylogenetic trees were constructed using the maximum likelihood method with 1000 bootstrap replications in PhyML-3.1 software.
Fig. 6.
Amino acid starvation induces autophagy in BME26 cells. MDC staining (green) for autophagosomes and lysotracker staining (red) for lysosomes in BME26 cells after HBSS treatment for 0, 1, 2, 3, and 5 days. Bar: 20 μm.
4. Discussion
Ticks are highly tolerant to nutrient shortage, and can survive long-term starvation. This exceptional ability is obviously linked to the autophagy pathway which plays an important role in the response of organisms to starvation stress. To date, the Atg genes have been identified in many taxa including yeast, plants, and animals. For example, around 40 Atg genes have been cloned in yeast. Subsequently, Atg genes have been identified in H. sapiens, Drosophila, C. elegans, Arabidopsis thaliana, Oryza sativa and many others, which indicates that the core ATG mechanism is evolutionarily conserved across kingdoms (Kwon and Park, 2008; Meijer et al., 2007; Meléndez and Levine, 2009; Xia et al., 2011; Zirin and Perrimon, 2010). For arthropod lineages, the autophagy machinery is less well characterized (Malagoli et al., 2010). As a vector of many pathogens such as Borrelia burgdorferi, Anaplasma phagocytophilum, Babesia microti and the tick-borne encephalitis virus, I. scapularis is the one of the most important ticks that are responsible for diseases of animals and humans in North America. However, currently little is known about the role of autophagy in I. scapularis development and in response to starvation stress, as well as in other tick species.
Here, we identified 14 Atg genes belonging to 11 classes in the genome of I. scapularis via bioinformatics analysis, and provide the first genome wide analysis of the Atg gene family in ticks. In order to understand the phylogenetic relationships of Atg genes in I. scapularis, we also investigated the homologues in eight other arthropod taxa. Results showed that the size of all Atg gene family is species-specific, and not proportional to the genome size (Table 1). The unrooted phylogenetic trees (Fig. 1) suggested that IsATGs were clustered tightly with their counterparts in other species, which was consistent with the present understanding of tick evolutionary history (Chipman et al., 2014). In addition, as with most species surveyed here, the size among the Atg gene subfamilies was uneven in ticks. The Atg genes of Atg4 and Atg8 gene subfamilies in I. scapularis were encoded by multiple copies, implying possible subfunctionalization or functional redundancy of those duplicated Atg gene subfamilies. Only one single representative was found for each of the remaining IsAtg gene subfamilies, suggesting they could serve as potentially attractive targets for reverse genetic studies in ticks to elucidate transcription and control.
In the evolution of duplicate genes, there are three alternative outcomes of their function, i.e., nonfunctionalization, which means one copy becomes silenced; neofunctionalization, which means one copy acquires novel function while the other one retains the original function; subfunctionalization, which means both copies become partially compromised relative to the function of ancestral genes (Lynch and Conery, 2000). The gene structure may provide information about the biological function. Unlike the conserved exon-intron arrangements of Atg genes within the same subfamilies in plants (Li et al., 2016; Shangguan et al., 2018), the exon-intron organization of Atg genes identified in I. scapularis were highly divergent even within the same subfamilies (Fig. 2B). This suggests that IsAtg genes may have undergone potential change of their function or the acquisition of new functions during evolution (Xu et al., 2012).
Gene expression profiles can provide important clues for functional analysis. Overall, the developmental stage-specific expression profiles of IsAtg family genes in I. scapularis varied widely, indicating that IsAtg genes may fulfill versatile functions in tick development (Fig. 3). The different patterns of IsAtg gene between unfed and partially fed ticks indicated that most ATGs played a role in the unfed stages in I. scapularis. This phenomenon is consistent with our findings in ISE6 cells, i.e., most ATG genes were up-regulated by amino acid starvation treatment in the ISE6 cells. IsAtg8C, IsAtg11, and IsAtg101 were most highly expressed across all developmental stages in I. scapularis, whereas IsAtg2, IsAtg4B, IsAtg8B, and IsAtg3 displayed low transcript levels under all conditions. In addition, the expression patterns of IsAtg genes from the same subfamily (mainly Atg4 and Atg8) also differed. This is consistent with previous studies in H. longicornis ticks where the expression of HlAtg genes varied during the entire life cycle (Umemiya-Shirafuji et al., 2014). Interestingly, we found the expression profiles of IsAtg family genes were sex dependent, i.e., more IsAtg genes were highly expressed in males than in females. Overall, this indicated that the constitutive autophagy in ticks has development specific bias as well as sex-specific bias, which was consistent with findings in mice (Oliván et al., 2014).
The autophagic process is regulated by dozens of ATG components. In our study, Atg3, Atg4, Atg7, and Atg8 encoding core ATG proteins in ATG8 mediated ubiquitin-like pathways (Geng and Klionsky, 2008; Noda et al., 2011) were identified in I. scapularis. Not all homologues involved in the ATG12 ubiquitin-like pathways (Atg5, Atg7, Atg10, Atg12, and Atg16) were found. This is not surprising as only certain ATG proteins may be engaged in specific circumstances. For instance, ATG18 can compensate for ATG21 in the recruitment of ATG components in yeast (Nair et al., 2010). In B. mori, ATG10 can be replaced by ATG3 due to the highly conserved C-terminal catalytic cysteine in conjugation with the ATG12 ubiquitin system (Zhang et al., 2009). What’s more, even some ATG proteins considered essential in autophagy were lost in some model insects, such as honeybees, in which ATG10 and ATG12 were absent (Zhang et al., 2009). The ubiquitin-like ATG8 proteins participate prominently in the break-down and removal or recycling of cellular components such as proteins, carbohydrates, lipids, and DNA, or even organelles through autophagy (Nakatogawa et al., 2007), whereas ATG4, a unique protease, functions in processing the nascent ATG8 to expose a glycine residue and cleaving phosphatidylethanolamine (PE) which is referred to as deconjugating of ATG8. In this study, all the deduced protein sequences of IsATG8 subfamily members contained the conserved C-terminal glycine residue which was likely cleaved by cysteine protease ATG4 as suggested by using the InterproScan programme (Mitchell et al., 2019). However, the putative active GABARAP binding site only was found in IsATG8C (Fig. S3). Indeed, the IsAtg8C transcript levels were the most abundant across all tick developmental stages and sexes (Fig. 3). However, the expression level of IsAtg8C were decreased in both I. scapularis cell lines ISE6 (Fig. 4) and IDE8 (Fig. S6) in response to amino acid starvation treatment while IsAtg8A and IsAtg8B were induced, respectively. These results implied that the duplicated Atg8 genes had acquired neofunctionality in ticks to support responses to starvation stress. Furthermore, in R. microplus BME26 cells, autophagy was also induced by amino acid starvation treatment which was consistent with previous research (Esteves et al., 2008), indicating that autophagy was a conserved process in the response of ticks to starvation stress. It will be important to investigate further how IsAtg8 genes participate in the autophagy pathway in I. scapularis to gain additional knowledge beyond our phylogeny-based function prediction and expression profiles analysis.
Conclusion
To our knowledge, this is the first analysis of the Atg family at the whole-genome wide level in ticks. Our observations showed that the Atg gene superfamily is highly conserved in ticks and functions in autophagy pathways induced by amino acid starvation. Such basic knowledge will contribute to generating more specific and detailed information about tick starvation responses, and pave the way for innovative strategies to control TBPs.
Supplementary Material
Acknowledgments
We thank Guillermo Marqués and John Oja from the University image centers at the University of Minnesota, Twin Cities, for insightful suggestion for improvement of the images. The study was financially supported by a grant to UGM from the NIH (2R01AI049424).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- Bailey TL, Williams N, Misleh C, Li WW, 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW, 2010. Network organization of the human autophagy system. Nature 466, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederbick A, Kern HF, Elsässer HP, 1995. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur. J. Cell Biol 66, 3–14. [PubMed] [Google Scholar]
- Chipman AD, Ferrier DEK, Brena C, Qu J, Hughes DST, Schröder R, Torres-Oliva M, Znassi N, Jiang H, Almeida FC, Alonso CR, Apostolou Z, Aqrawi P, Arthur W, Barna JCJ, Blankenburg KP, Brites D, Capella-Gutiérrez S, Coyle M, Dearden PK, Du Pasquier L, Duncan EJ, Ebert D, Eibner C, Erikson G, Evans PD, Extavour CG, Francisco L, Gabaldón T, Gillis WJ, Goodwin-Horn EA, Green JE, Griffiths-Jones S, Grimmelikhuijzen CJP, Gubbala S, Guigó R, Han Y, Hauser F, Havlak P, Hayden L, Helbing S, Holder M, Hui JHL, Hunn JP, Hunnekuhl VS, Jackson L, Javaid M, Jhangiani SN, Jiggins FM, Jones TE, Kaiser TS, Kalra D, Kenny NJ, Korchina V, Kovar CL, Kraus FB, Lapraz F, Lee SL, Lv J, Mandapat C, Manning G, Mariotti M, Mata R, Mathew T, Neumann T, Newsham I, Ngo DN, Ninova M, Okwuonu G, Ongeri F, Palmer WJ, Patil S, Patraquim P, Pham C, Pu L-L, Putman NH, Rabouille C, Ramos OM, Rhodes AC, Robertson HE, Robertson HM, Ronshaugen M, Rozas J, Saada N, Sánchez-Gracia A, Scherer SE, Schurko AM, Siggens KW, Simmons D, Stief A, Stolle E, Telford MJ, Tessmar-Raible K, Thornton R, van der Zee M, von Haeseler A, Williams JM, Willis JH, Wu Y, Zou X, Lawson D, Muzny DM, Worley KC, Gibbs RA, Akam M, Richards S, 2014. The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 12, e1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente J, Estrada- Peña A, Venzal JM, Kocan KM, Sonenshine DE, 2008. Overview: Ticks as vectors of pathogens that cause disease in humans and animals. Front. Biosci 13, 6938–6946. [DOI] [PubMed] [Google Scholar]
- Delorme-Axford E, Klionsky DJ, 2018. Transcriptional and post-transcriptional regulation of autophagy in the yeast. J. Biol. Chem 293, 5396–5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves E, Lara FA, Lorenzini DM, Costa GH, Fukuzawa AH, Pressinotti LN, Silva JRM, Ferro JA, Kurtti TJ, Munderloh UG and Daffre S, 2008. Cellular and molecular characterization of an embryonic cell line (BME26) from the tick Rhipicephalus (Boophilus) microplus. Insect Biochem Molec. 38, 568–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Yao Z, Klionsky DJ, 2015. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol. 25, 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández JMF, Álvarez CPB, Hernández CVS, Camberos EP, Castillo CG, Sahagún DO, Velázquez MM, 2016. Molecular characterization and expression analysis of three novel autophagy-related genes from the cattle tick Rhipicephalus (Boophilus) microplus (Acari: Ixodidae). Parasitology. 10.1017/s0031182016001542 [DOI] [PubMed] [Google Scholar]
- Flores Fernández JM, Gutiérrez Ortega A, Rosario Cruz R, Padilla Camberos E, Alvarez AH, Martínez Velázquez M, 2014. Molecular cloning and characterization of two novel autophagy-related genes belonging to the ATG8 family from the cattle tick Rhipicephalus (Boophilus) microplus (Acari: Ixodidae). Exp. Appl. Acarol 64, 533–542. [DOI] [PubMed] [Google Scholar]
- Gatica D, Lahiri V, Klionsky DJ, 2018. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol 20, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudecker B, 1963. Über den Formwechsel einiger Zellorganelle bei der Bildung der Reservestoffe im Fettkörper von Drosophila-Larven. Zeitschrift für Zellforschung und Mikroskopische Anatomie [PubMed] [Google Scholar]
- Geng J, Klionsky DJ, 2008. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. “Protein modifications: beyond the usual suspects” review series. EMBO Rep. 9, 859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O, 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol 59, 307–321. [DOI] [PubMed] [Google Scholar]
- Gulia-Nuss M, Nuss AB, Meyer JM, Sonenshine DE, Roe RM, Waterhouse RM, Sattelle DB, de la Fuente J, Ribeiro JM, Megy K, Thimmapuram J, Miller JR, Walenz BP, Koren S, Hostetler JB, Thiagarajan M, Joardar VS, Hannick LI, Bidwell S, Hammond MP, Young S, Zeng Q, Abrudan JL, Almeida FC, Ayllón N, Bhide K, Bissinger BW, Bonzon-Kulichenko E, Buckingham SD, Caffrey DR, Caimano MJ, Croset V, Driscoll T, Gilbert D, Gillespie JJ, Giraldo-Calderón GI, Grabowski JM, Jiang D, Khalil SMS, Kim D, Kocan KM, Koči J, Kuhn RJ, Kurtti TJ, Lees K, Lang EG, Kennedy RC, Kwon H, Perera R, Qi Y, Radolf JD, Sakamoto JM, Sánchez-Gracia A, Severo MS, Silverman N, Šimo L, Tojo M, Tornador C, Van Zee JP, Vázquez J, Vieira FG, Villar M, Wespiser AR, Yang Y, Zhu J, Arensburger P, Pietrantonio PV, Barker SC, Shao R, Zdobnov EM, Hauser F, Grimmelikhuijzen CJP, Park Y, Rozas J, Benton R, Pedra JHF, Nelson DR, Unger MF, Tubio JMC, Tu Z, Robertson HM, Shumway M, Sutton G, Wortman JR, Lawson D, Wikel SK, Nene VM, Fraser CM, Collins FH, Birren B, Nelson KE, Caler E, Hill CA, 2016. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat. Commun 7, 10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Klionsky DJ, 2009. Regulation Mechanisms and Signaling Pathways of Autophagy. Annual Review of Genetics. 10.1146/annurev-genet-102808-114910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Blair JE, Venturi ML, Shoe JL, 2004. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol. Biol 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G, 2015. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins VK, Timmons AK, McCall K, 2013. Diversity of cell death pathways: insight from the fly ovary. Trends Cell Biol. 23, 567–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Mizushima N, 2014. Autophagy and human diseases. Cell Research. 10.1038/cr.2013.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongejan F, Uilenberg G, 2004. The global importance of ticks. Parasitology 129 Suppl, S3–14. [DOI] [PubMed] [Google Scholar]
- Kawano S, Umemiya-Shirafuji R, Boldbaatar D, Matsuoka K, Tanaka T, Fujisaki K, 2011. Cloning and characterization of the autophagy-related gene 6 from the hard tick, Haemaphysalis longicornis. Parasitol. Res 109, 1341–1349. [DOI] [PubMed] [Google Scholar]
- Keane TM, Creevey CJ, Pentony MM, Naughton TJ, Mclnerney JO, 2006. BMC Evolutionary Biology, 10.1186/1471-2148-6-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cregg JM, Dunn WA Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y, 2003. A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5, 539–545. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K, 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SI, Park OK, 2008. Autophagy in plants. J. Plant Biol 51, 313–320. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG, 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW, 2011. Autophagy in immunity and inflammation. Nature 469, 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W-Z, Fang J-A, Xiao X, Chou K-C, 2013. iLoc-Animal: a multi-label learning classifier for predicting subcellular localization of animal proteins. Mol. Biosyst 9, 634–644. [DOI] [PubMed] [Google Scholar]
- Li W, Chen M, Wang E, Hu L, Hawkesford MJ, Zhong L, Chen Z, Xu Z, Li L, Zhou Y, Others, 2016. Genome-wide analysis of autophagy-associated genes in foxtail millet (Setaria italica L.) and characterization of the function of SiATG8a in conferring tolerance to nitrogen starvation in rice. BMC Genomics 17, 797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lőrincz P, Mauvezin C, Juhász G, 2017. Exploring Autophagy in Drosophila. Cells 6 10.3390/cells6030022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS, 2000. The evolutionary fate and consequences of duplicate genes. Science. 290, 1151–1155. [DOI] [PubMed] [Google Scholar]
- Malagoli D, Abdalla FC, Cao Y, Feng Q, Fujisaki K, Gregorc A, Matsuo T, Nezis IP, Papassideri IS, Sass M, Silva-Zacarin ECM, Tettamanti G, Umemiya-Shirafuji R, 2010. Autophagy and its physiological relevance in arthropods: current knowledge and perspectives. Autophagy 6, 575–588. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH, 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Research. 10.1093/nar/gkw1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer WH, van der Klei IJ, Veenhuis M, Kiel JAKW, 2007. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy 3, 106–116. [DOI] [PubMed] [Google Scholar]
- Meléndez A, Levine B, 2009. Autophagy in C. elegans. WormBook; 1–26. [DOI] [PubMed] [Google Scholar]
- Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang H-Y, El-Gebali S, Fraser MI, Gough J, Haft DR, Huang H, Letunic I, Lopez R, Luciani A, Madeira F, Marchler-Bauer A, Mi H, Natale DA, Necci M, Nuka G, Orengo C, Pandurangan AP, Paysan-Lafosse T, Pesseat S, Potter SC, Qureshi MA, Rawlings ND, Redaschi N, Richardson LJ, Rivoire C, Salazar GA, Sangrador-Vegas A, Sigrist CJA, Sillitoe I, Sutton GG, Thanki N, Thomas PD, Tosatto SCE, Yong S-Y, Finn RD, 2019. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, 2018. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol 20, 521–527. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M, 2011. Autophagy: renovation of cells and tissues. Cell 147, 728–741. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, 2010. Autophagy in mammalian development and differentiation. Nat. Cell Biol 12, 823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura-Martiniano NO, Machado-Ferreira E, Gazêta GS, Soares CAG, 2017. Relative transcription of autophagy-related genes in Amblyomma sculptum and Rhipicephalus microplus ticks. Exp. Appl. Acarol 73, 401–428. [DOI] [PubMed] [Google Scholar]
- Munderloh UG, Kurtti TJ, 1989. Formulation of medium for tick cell culture. Exp Appl Acarol. 7, 219–229. [DOI] [PubMed] [Google Scholar]
- Munderloh UG, Jauron SD, Fingerle V, Leitritz L, Hayes SF, Hautman JM, Nelson CM, Huberty BW, Kurtti TJ, Ahlstrand GG, Greig B, Mellencamp MA, Goodman JL, 1999. Invasion and intracellular development of the human granulocytic ehrlichiosis agent in tick cell culture. J. Clin. Microbiol 37, 2518–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munderloh UG, Liu Y, Wang M, Chen C, Kurtti TJ, 1994. Establishment, maintenance and description of cell lines from the tick Ixodes scapularis. J. Parasitol 80, 533–543. [PubMed] [Google Scholar]
- Nair U, Cao Y, Xie Z, Klionsky DJ, 2010. Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy. J. Biol. Chem 285, 11476–11488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Ichimura Y, Ohsumi Y, 2007. Atg8, a Ubiquitin-like Protein Required for Autophagosome Formation, Mediates Membrane Tethering and Hemifusion. Cell. 10.1016/j.cell.2007.05.021 [DOI] [PubMed] [Google Scholar]
- Needham GR, Teel PD, 1991. Off-host physiological ecology of ixodid ticks. Annu. Rev. Entomol 36, 659–681. [DOI] [PubMed] [Google Scholar]
- Noda NN, Satoo K, Fujioka Y, Kumeta H, Ogura K, Nakatogawa H, Ohsumi Y, Inagaki F, 2011. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol. Cell 44, 462–475. [DOI] [PubMed] [Google Scholar]
- Oliver JD, Chávez ASO, Felsheim RF, Kurtti TJ, & Munderloh UG 2015. An Ixodes scapularis cell line with a predominantly neuron-like phenotype. Exp. Appl. Acarol 66, 427–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver JD, Lynn GE, Burkhardt NY, Proce LD, Nelson CM, Kurtti TJ, Munderloh UG 2015. Infection of immature Ixodes scapularis (Acari: Ixodidae) by membrane feeding. J. Med. Entomol 53, 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y, 2014. Historical landmarks of autophagy research. Cell Res. 24, 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliván S, Calvo AC, Manzano R, Zaragoza P, Osta R, 2014. Sex differences in constitutive autophagy. Biomed Res. Int 2014, 652817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Petiot A and Codogno P, 2004. Analyses of Gα-interacting protein and activator of G-protein-signaling-3 functions in macroautophagy In Methods in enzymology (Vol. 390, pp. 17–31). Academic Press. [DOI] [PubMed] [Google Scholar]
- Ravanan P, Srikumar IF, Talwar P, 2017. Autophagy: The spotlight for cellular stress responses. Life Sci. 188, 53–67. [DOI] [PubMed] [Google Scholar]
- Rodríguez Y, Rojas M, Gershwin ME, Anaya J-M, 2018. Tick-borne diseases and autoimmunity: A comprehensive review. J. Autoimmun 88, 21–42. [DOI] [PubMed] [Google Scholar]
- Saeed A, 2003. A Short Total Synthesis of rac-Peniolactol. ChemInform. 10.1002/chin.200328221 [DOI] [Google Scholar]
- Shangguan L, Fang X, Chen L, Cui L, Fang J, 2018. Genome-wide analysis of autophagy-related genes (ARGs) in grapevine and plant tolerance to copper stress. Planta 247, 1449–1463. [DOI] [PubMed] [Google Scholar]
- Umemiya R, Matsuo T, Hatta T, Sakakibara S-I, Boldbaatar D, Fujisaki K, 2008. Autophagy-related genes from a tick, Haemaphysalis longicornis. Autophagy 4, 79–81. [DOI] [PubMed] [Google Scholar]
- Umemiya R, Matsuo T, Hatta T, Sakakibara S-I, Boldbaatar D, Fujisaki K, 2007. Cloning and characterization of an autophagy-related gene, ATG12, from the three-host tick Haemaphysalis longicornis. Insect Biochem. Mol. Biol 37, 975–984. [DOI] [PubMed] [Google Scholar]
- Umemiya-Shirafuji R, Galay RL, Maeda H, Kawano S, Tanaka T, Fukumoto S, Suzuki H, Tsuji N, Fujisaki K, 2014. Expression analysis of autophagy-related genes in the hard tick Haemaphysalis longicornis. Vet. Parasitol 201, 169–175. [DOI] [PubMed] [Google Scholar]
- Xia K, Liu T, Ouyang J, Wang R, Fan T, Zhang M, 2011. Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 18, 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Guo C, Shan H, Kong H, 2012. Divergence of duplicate genes in exon-intron structure. Proceedings of the National Academy of Sciences. 10.1073/pnas.1109047109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ, 2005. Autophagy: molecular machinery for self-eating. Cell Death Differ. 12 Suppl 2, 1542–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C-S, Lin C-J, Hwang J-K, 2004. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 13, 1402–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Hu Z-Y, Li W-F, Li Q-R, Deng X-J, Yang W-Y, Cao Y, Zhou C-Z, 2009. Systematic cloning and analysis of autophagy-related genes from the silkworm Bombyx mori. BMC Mol. Biol 10, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirin J, Perrimon N, 2010. Drosophila as a model system to study autophagy. Semin. Immunopathol 32, 363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.