Abstract
Genomic rearrangements leading to the aberrant expression of ERG are the most common early events in prostate cancer and are significantly enriched for the concomitant loss of PTEN. Genetically engineered mouse models reveal that ERG overexpression alone is not sufficient to induce tumorigenesis, but combined loss of PTEN results in an aggressive invasive phenotype. Here we show that oncogenic ERG repressed PI3K signaling though direct transcriptional suppression of IRS2 leading to reduced RTK levels and activity. In accordance with this finding, ERG positive human prostate cancers had a repressed AKT gene signature and transcriptional down-regulation of IRS2. While overexpression of IRS2 activated PI3K signaling, promoting cell migration in a PI3K dependent manner, this did not fully recapitulate the phenotype seen with loss of PTEN as PI3K signaling is not as robust as observed in the setting of loss of PTEN. Importantly, deletions of the PTEN locus, which promotes active PI3K signaling, were among the most significant copy number alterations that co-occurred with ERG genomic rearrangements. This work provides insight on how initiating oncogenic events may directly influence the selection of secondary concomitant alterations to promote oncogenic signaling during tumor evolution.
Keywords: ERG genomic rearrangements, PTEN, INPP4B, PI3K, Tumorigenesis, Secondary genomic events
Introduction
Tumorigenesis is a multifaceted process involving the complex interplay of several biologic systems that are highly dependent on the activation of pro-proliferative, survival, and migration pathways. Genomic rearrangements of the ETS family transcription factor ERG are the most common early initiating events in prostate cancer, occurring in approximately 50% of primary prostate cancers (1–3). Mouse modeling studies have demonstrated that overexpression of ERG alone confers a phenotype of mild prostatic hyperplasia but is not sufficient to promote prostate cancer development (4,5). However, combined overexpression of ERG and loss of PTEN in pre-clinical models results in the development of a high grade invasive prostate cancer (4,5).
The PI3K pathway plays a dominant driver role in a variety of malignancies, most frequently activated through loss of the tumor suppressor PTEN. PI3K signaling originates at the cellular membrane through ligand and receptor binding of receptor tyrosine kinases, g-protein coupled receptors, and other membrane bound receptors (6). This results in the active recruitment of the PI3K complex which catalyzes the phosphorylation of down-stream substrates from the generation of PIP3. PTEN is a phospholipid and protein phosphatase that is responsible for dephosphorylating PIP3 to PIP2, and thus serves the repressive gate keeper for PI3K signaling. The PI3K pathway is responsible for the regulation of numerous cellular processes play a significant role in normal cellular physiology, and when aberrantly active contribute to tumorigenesis (6). Loss of the tumor suppressor PTEN, promoting aberrant PI3K signaling, occurs in approximately 20% of primary prostate cancer and nearly 50% of metastatic prostate cancer (1). Previous pre-clinical studies have shown that loss of PTEN resulting in activation of PI3K signaling is a driver of prostate cancer progression and occurs in a dose dependent fashion (7,8).
While activation of PI3K signaling is sufficient to initiate tumorigenesis in genetically engineered mouse (GEM) models, molecular profiling studies in prostate cancer suggest that loss of PTEN and activation of PI3K signaling are secondary progression events (1,9). In contrast to the sub-clonal PTEN genomic alterations in prostate cancer suggestive of a secondary event, genomic rearrangements of ERG are highly clonal, providing evidence that ERG is an early initiating event in prostate cancer that enriches for concomitant loss of PTEN. Given the significant biologic interaction of ERG and PTEN in prostate tumorigenesis we sought to further define the molecular interaction of these two dominant pathways in prostate cancer.
Methods
Isolation and culture of mouse prostate epithelial cells:
Murine prostates were digested with Collagenase/Hyaluronidase (STEMCELL; 07912) and subsequently with TrypLE (GIBCO). Cells were cultured in suspension for 5–10 days and transferred to collagen-coated plates as described previously (10,11). These cells were authenticated by PCR genotyping protocols established for the Ptenlox/lox and Rosa26-ERG Ptenlox/lox GEM models. The VCaP and 293FT were obtained from ATCC and validated by STR genotyping protocols. Normal human prostate organoids were generated by our group from a patient undergoing a radical prostatectomy after written and informed consent on our IRB approved protocols (IRB 06–107 and 12–001) and grown using standard organoid culture methodology. These two protocols are approved by MSKCC IRB for fresh tissue acquisition for the establishment of prostate organoids from patients and all experiments were conducted in accordance with recognized ethical guidelines (e.g., Declaration of Helsinki, CIOMS, Belmont Report, U.S. Common Rule). All cell lines and prostate organoids used in our studies have tested negative for mycoplasma using the MycoProbe Mycoplasma Detection Kit (R&D Systems). All cell lines and organoids were freshly thawed and only passaged to achieve the number of cells required for in vitro or in vivo experiments.
Immunoblot:
Cell lysates were prepared in RIPA buffer supplemented with proteinase and phosphatase inhibitors. Proteins were resolved on NuPAGE Novex 4–12% Bis-Tris Protein Gels (Thermo Fisher Scientific) and transferred electronically onto a PVDF 0.45 μM membrane (Millipore). All experiments were performed in triplicate and the representative blots are shown. Quantification was performed using Image J software.
Antibodies:
The following antibodies were used for Western blotting and ChIP: AR (Abcam; ab108341; 1:1,000 for Western blotting), beta-actin (Abcam; ab49900; 1:50,000 for Western blotting), ERG (Abcam; ab92513; 1:1,000 for Western blotting), histone H3 (acetyl K27) (Abcam; ab4729), IRS-1 (Cell Signaling; 2390S; 1:1,000 for Western blotting), IRS-2 (Cell Signaling; 4502S; 1:1,000 for Western blotting), phospho-AKT (Ser308) (Cell Signaling; 13038S; 1:1,000 for Western blotting), phospho-AKT (Ser473) (Cell Signaling; 4060L; 1:1,000 for Western blotting), PTEN (Cell Signaling; 9559L; 1:1,000 for Western blotting), phospho-IGF1R (Cell Signaling; 3024S; 1:1,000 for Western blotting), FKBP5(Cell Signaling; 12210S; 1:1,000 for Western blotting), phospho-Pras40 (Cell Signaling; 2997s; 1:1,000 for Western blotting), phospho-EGFR (Cell Signaling; 3777S; 1:1,000 for Western blotting), phosphor-GSK3B (Cell Signaling; 9336S; 1:1,000 for Western blotting).
Lentiviral CRISPR/Cas9-mediated knockout:
To knock out AR and ERG in mouse organoids, three pairs of single guide RNA (sgRNA) sequences were designed for human ERG and two pairs for murine AR using the design tool from the Feng Zhang Lab (MIT) and cloned into the LentiCRISPRv2 (Addgene, 52962). Murine prostate cells were infected with lentivirus for 48–72 hours and selected with puromycin (4 μg/mL) for 7–10 days. The target guides sequences are as follows: sgERG-1: F: CACCGACACCGTTGGGATGAACTA; sgERG-1: R: AAACTAGTTCATCCCAACGGTGTC; sgERG-2: F: CACCGTTCCTTCCCATCGATGTTC; sgERG-2: R: AAACGAACATCGATGGGAAGGAAC; sgERG-3: F: CACCGTACAGACCATGTGCGGCAG; sgERG-3: R: AAACCTGCCGCACATGGTCTGTAC; sgAr-1: F: CACCGGTGGAAAGTAATAGTCGAT; sgAr-1: R: AAACATCGACTATTACTTTCCACC; sgAr-2: F: CACCGGGTGGAAAGTAATAGTCGA; sgAr-2: R AAACTCGACTATTACTTTCCACCC
Lentiviral knockdown:
A hairpin sequence against Pten was cloned into Lenti (pRRL) (a gift from the Zuber lab) to make lentiviral particles. The sequence was as follows: Pten shRNA: GCCAGCTAAAGGTGAAGATATA
SiRNA:
To knockdown IRS-2, Rosa26-ERG Ptenlox/lox –CrisprERG cells were transfected with pre-designed siRNAs (25 nM) to knock down IRS2(Ambion; 4390771-s118458) or Silencer select negative control (Life Tech; 4390844).
Stable gene expression analysis:
Mouse IRS-2 (Origene; MR227167) was cloned into a Myc-DDK-tagged Lenti plasmid (Origene; PS100092).
Inducible gene expression by doxycycline:
pCW-CFP and pCW-CFP-ERG were transfected into 293FT cells to make lentiviruses. Wild-type normal mouse and human prostate organoids were infected with the viruses and treated with puromycin (4 ug/ml) for 7 days for cell selection. Cells were then treated with doxycycline (1 μg/ml) to induce ERG expression at different time points.
IRS2-promoter Luciferase assay:
A 1kb segment of the human IRS2 promoter was cloned into a firefly luciferase promoter expression construct (Switchgear genomics, Cat: S711020) and co-transfections were performed in 293T cells with MSCV-GFP or MSCV-ERG. A dual luciferase reporter assay was performed, and signals was quantified with normalization to Renilla luciferase to control for transfection efficiency. All experiments were performed in triplicates, and the mean signals were reported.
RNA sequencing:
QIAshredder (Qiagen; 79656) and RNeasy Mini kit (Qiagen; 74106) were used to isolate RNA from cell lines. RNA-seq was performed by New York genome center. RNA sequencing libraries were prepared using the TruSeq Stranded mRNA sample preparation kit in accordance with the manufacturer’s instructions. Final libraries were quantified using the KAPA Library Quantification Kit (KAPA Biosystems), and were sequenced on an Illumina HiSeq2500 sequencer (v4 chemistry) using 2 x 50bp cycles targeting 35M single-end reads per sample. RNAseq data deposited GEO GSE112469.
Chromatin Immunoprecipitation and Sequencing:
Following the protocol previously described (12), chromatin isolation from mouse organoid cell lines and immunoprecipitation using antibodies AR, ERG and H3K27ac was performed. Next generation sequencing was carried out on an Illumina HiSeq2000 platform with 50 bp or 100 bp single reads.
To identify the AR and Erg binding sites on chromosome, CHIP-Seq analyses were performed on the parental cells as well as AR or ERG Crispr knockout cell lines. Sequence reads were aligned to mm10 using bowtie program (13). MACS2 call peak program was used to call the peaks for each CHIP-Seq samples compared with input sequence using standard parameters (14). Peaks were annotated using Homer Annotate Peaks program with default parameters identifying promoter, gene body, and intergenic binding sites (15). ChIPseq data deposited GEO GSE112414.
Cell proliferation and migration assay:
Cells per well were plated (1x103) in a collagen-coated 96-well plate. The number of viable cells were counted using Cell Titer-Glo Luminescent Cell Viability Assay kit (Promega; G7573). Cell migration was analyzed using a xCELLigence real-time cell analyzer linked Boyden chamber assay.
Mouse Xenograft procedure:
2.0 × 106 cells resuspended in 100 µL of 1:1 mix of growth media and Matrigel (Corning; 356237) were injected into 6–8 weeks old CB17-SCID male mice (Taconic). Tumor size was measured weekly by Peira TM900 (Peira Scientific Instruments). The volume of tumors was calculated using the formula: Volume = Length x Width x Height. A total of 10 tumors per group were used to assay tumor growth in vivo. All experiments were approved by our IACUC protocol 06-07-012.
Histology and immunohistochemistry:
Xenografts were fixed using 4% paraformaldehyde for 2–3 days and embedded using a Leica ASP6025 tissue processor (Leica Biosystems).
Results
ERG aberrant expression suppresses upstream PI3K signaling
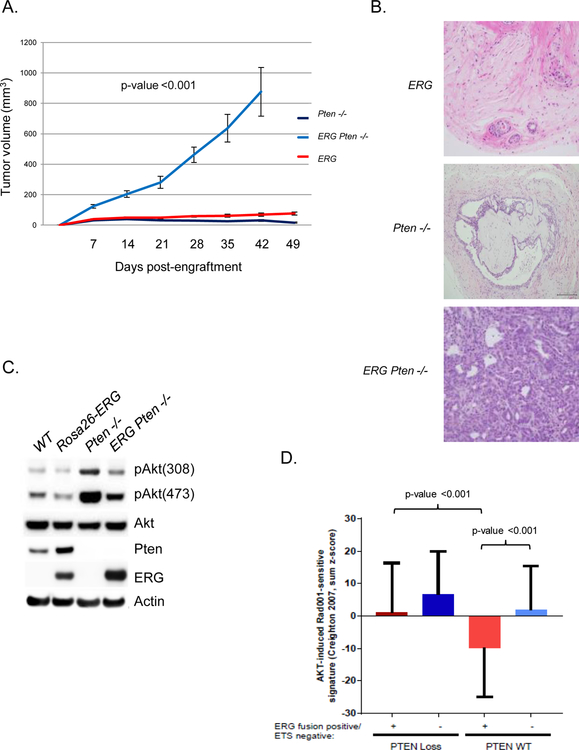
To directly study the molecular interaction of ERG and PTEN loss on PI3K signaling we took advantage of the prostate organoid culture methodology and generated prostate organoids from wild-type, Pb-Cre Rosa26-ERG, Pb-Cre Ptenlox/lox, and Pb-Cre Rosa26-ERG Ptenlox/lox GEM models. In concordance with previous GEM model data, ERG overexpression or Pten loss alone was not sufficient for tumorigenesis after injection of these organoids into the flanks of SCID mice whereas combined ERG overexpression and loss of Pten resulted in high grade invasive prostate cancer (Figure 1A, 1B). Surprisingly, we found that ERG expression was associated with reduced levels of pAkt both in the context of Pten wild-type and Pten loss prostate organoids (Figure 1C). Interestingly, ERG overexpression, in the context of wild-type Pten, was associated with an increase in Pten protein levels. To address whether ERG could be a transcriptional activator of Pten, we performed qRT-PCR and found that Pten mRNA levels did not differ between ERG positive and negative prostate organoids (Supplementary Figure 1A). Given that the repression of PI3K signaling in the setting of ERG overexpression occurred in both the presence and absence of Pten demonstrates that this phenotype was ERG and not Pten dependent. To further study this, we analyzed the TCGA primary prostate cancer genomic profiling dataset categorizing tumors based on ERG/ETS and PTEN status. Indeed, we found that ERG genomic rearrangements are correlated with repression of a PI3K/AKT RNA expression signature and PTEN loss significantly enhances this signature, p<0.001 (Figure 1D) (2,16).
Figure 1. ERG suppresses PI3K signaling constraining tumorigenesis.
A) Rosa26-ERG (n=10), Pten−/− (n=10) and Rosa26-ERG Pten−/− (n=10) prostate organoids were injected into the flanks of SCID mice and tumor volumes were measured weekly, error bars reporting standard deviation from the mean. B) Histology (200x) of in vivo specimens from the prostate organoids at the end of study. C) Prostate organoids were generated from Wild-type, Rosa26-ERG, Pb-Cre Ptenlox/lox and Pb-Cre Rosa26-ERG Ptenlox/lox mice and western blot analysis was performed demonstrating expression of ERG, loss of Pten, and repressed PI3K signaling in organoids over-expressing ERG (experiment run in triplicate). D) Analysis of the TCGA profiling data set demonstrating that ERG positive human primary prostate cancers are significantly correlated with a repressed AKT gene signature and loss of PTEN results in activation of this signature.
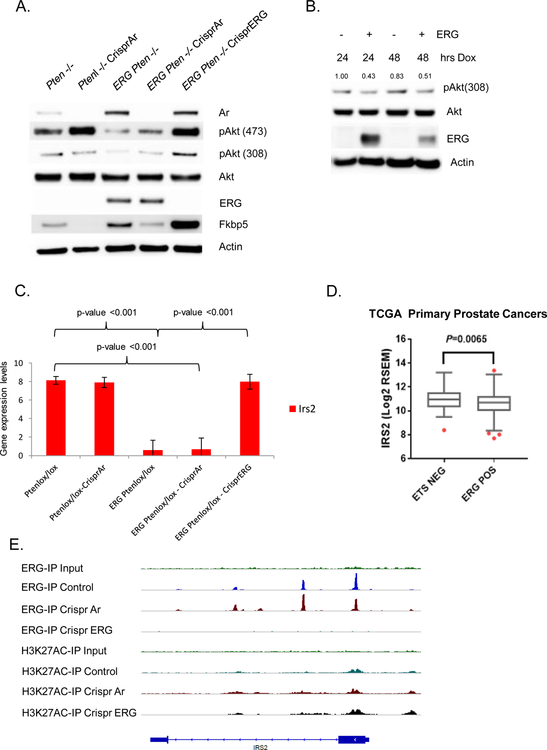
To determine if PI3K signaling was directly repressed in an ERG dependent manner, knock-out of ERG in the ERG Pten−/− organoids was performed using the CRISPR system. These cells displayed a profound increase in pAkt (Figure 2A). We then used a doxycycline inducible system in wild-type organoids and, demonstrated reduced levels of pAkt at T308, the direct PI3K-Pdk1 site, upon acute ERG expression (Figure 2B, Supplementary Figure 1B). We obtained similar results in the androgen regulated ERG-positive human prostate cancer cell line VCaP, where down-regulation of ERG promoted a dose dependent increase in PI3K signaling (Supplementary Figure 1C). Thus, both gain- and loss-of-function experiments in mouse and human models establish that ERG suppresses PI3K activity in prostate cells.
Figure 2. PI3K repression is ERG dependent and ERG suppresses IRS2 transcriptionally.
A) Using the Crispr/Cas9 technology, AR and ERG were knocked-out from our Pten−/− and Rosa26-ERG Pten−/− prostate organoids and protein lysates were analyzed by western blotting. As predicted, knock-out of AR in the setting of Pten loss robustly activated PI3K signaling. Surprisingly though knock-out of AR in the setting of ERG expression had minimal effect on PI3K signaling, while knock-out of ERG increased pAkt levels profoundly (experiment run in triplicate). B) Acute doxycycline induced overexpression of ERG in wild-type prostate organoids versus vector control demonstrated repression of PI3K signaling correlated with ERG expression levels (experiment run in triplicate, quantification of pAkt normalized to actin and Akt). C) mRNA expression of IRS2 was significantly repressed by ERG in our prostate organoids. Three independent lines for each genotype were analyzed with error bars reporting standard deviation from the mean. D) Analysis of IRS2 expression in the TCGA data set reveals that prostate cancers harboring ERG genomic rearrangements have a significantly repressed IRS2 expression. E) ChIP seq analysis demonstrates that ERG binds to the transcription start site of IRS2.
IRS2 is a direct target of ERG transcriptional repression
One potential mechanism for the suppressive effect of ERG on PI3K signaling could be through direct interplay with AR, as we have previously shown that AR inhibits PI3K through a reciprocal feedback mechanism and ERG alters the AR cistrome and promotes AR transcriptional activity (5) (17). To address this question, we generated a series of organoids using CRISPR technology to knock-out AR and ERG respectively. As expected, knock-out of AR results in up-regulation of PI3K signaling in the setting of Pten loss alone. However, knock-out of AR in ERG Pten−/− prostate organoids only marginally increased PI3K signaling (Figure 2A), indicating that PI3K suppression in the context of constitutive ERG overexpression is not primarily mediated by AR. While AR is not the direct dominant mechanism repressing PI3K signaling in the context of TMPRSS2:ERG rearrangements, it does indirectly influence PI3K signaling through regulation of ERG adding complexity to our original PI3K-AR reciprocal feedback model which was characterized in ERG negative model systems.
To further elucidate the mechanism responsible for PI3K repression, we performed RNAseq analysis across our panel of prostate organoids. We compared differential gene expression between ERG Pten−/− and ERG Pten−/− CRISPR ERG prostate cancer organoids and one of the top most significantly up-regulated genes following CRISPR ERG was IRS2 (Figure 2C, Supplementary Table 1). IRS2 is a cytoplasmic signaling molecule that mediates effects of insulin, insulin-like growth factor 1, and other cytokines by acting as a molecular adaptor between IGF1R/INSR and PI3K (18–20). In accordance with this data, we analyzed the expression of IRS2 in ERG positive prostate cancers profiled in TCGA and indeed, ERG positive tumors displayed a significant reduction in IRS2 levels (Figure 2D).
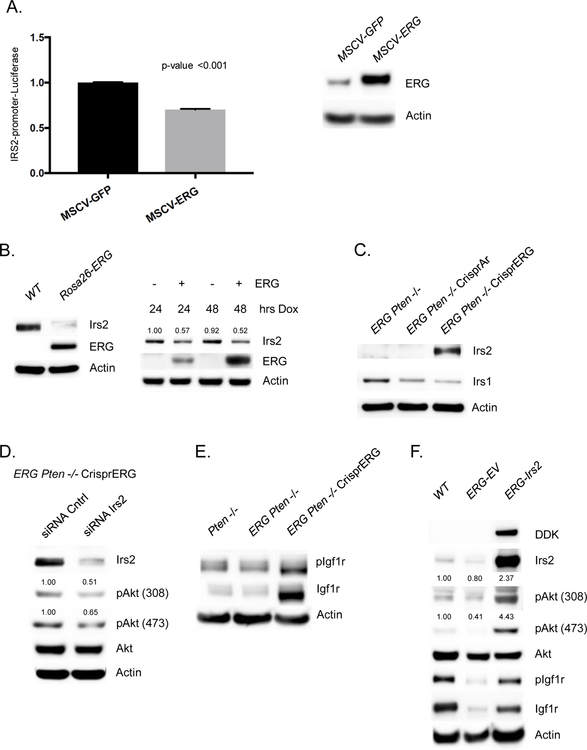
To determine if ERG is a direct regulator of IRS2, we performed ERG ChIP seq in our panel of ERG Pten−/− prostate cancer organoids and identified an ERG binding peak in the IRS2 promoter at an ETS consensus sequence, and this peak was absent after CRISPR deletion of ERG (Figure 2E, Supplementary figure 1D). To further investigate the role of ERG in directly repressing the IRS2 promoter, we transfected 293T cells with an IRS2-promoter-firefly luciferase construct and examined the impact of ERG expression on IRS2 promoter activity, controlling for transfection efficiency through normalization to a Renilla luciferase reporter. Indeed, we observed that ERG overexpression repressed the transcriptional activity of the IRS2 promoter (Figure 3A). Next, we examined the levels of ERG and IRS2 in wild-type and Rosa26-ERG prostate organoids following dox-inducible expression of ERG in wild-type prostate organoids. IRS2 levels were suppressed in the context of ERG expression (Figure 3B, Supplementary Figure 2A). Conversely, Crispr deletion of ERG in Rosa26-ERG Pten−/− organoids resulted in increased protein levels of IRS2 (Figure 3C). Furthermore, acute AR inhibition of in vivo VCaP tumors demonstrated reduced ERG levels leading to increased IRS2 levels and enhanced PI3K signaling (Supplementary Figure 2B). Importantly, siRNA knockdown of the increased IRS2 levels in the Rosa26-ERG Pten−/− organoids following ERG deletion reduced pAkt, consistent with a model whereby ERG impairs PI3K signaling by repression of IRS2 expression (Figure 3D, Supplementary Figure 2C).
Figure 3. ERG suppression of Down-regulation of IRS2 mediates IGF1R:PI3K signaling repression.
A) 293T cells transfected with IRS2-promoter-firefly luciferase construct with vector control or ERG co-expression demonstrates repressed promoter activity in the setting of ERG. Experiment performed in triplicate with error bars reporting standard deviation from the mean and normalized to Renilla luciferase. Western blot demonstrating ERG overexpression in the 293T cells. B) Acute doxycycline induced overexpression of ERG in wild-type prostate organoids versus vector control demonstrated a decrease in IRS2 levels correlated with ERG expression (experiment run in triplicate, quantification of Irs2 normalized to actin). C) IRS2 protein levels are repressed in Rosa-26 ERG Pten−/− organoids and knock-out of ERG is associated with a robust increase in IRS2 levels (experiment run in triplicate). D) siRNA knock-down of IRS2 following Crispr ERG in the Rosa26-ERG Pten−/− organoids reduced pAkt levels (experiment run in triplicate, quantification of pAkt normalized to actin and Akt). E) ERG Pten−/− organoids demonstrate reduced phosphorylation and total levels of Igf1R, that is increased following ERG knock-out (experiment run in triplicate). F) Prostate organoids derived from Rosa26-ERG mice demonstrate reduced IRS2 levels and repression of pIgf1r and PI3K signaling. Overexpression of IRS2 in Rosa26-ERG prostate organoids increases pIgf1r and PI3K signaling (experiment run in triplicate, quantification of pAkt normalized to actin and Akt).
ERG represses PI3K signaling in an RTK-IRS2 dependent manner
Given the role of IRS2 in mediating RTK signal transduction to PI3K, we took a focused look at the expression of a variety of RTK PI3K signaling mediators and did not observe any other differentially expressed genes (Supplementary Figure 2D). An exploratory phosphor-RTK array analysis was performed which demonstrated diminished pRTK signaling across several RTKs in the setting of ERG expression (Supplementary Figure 3A). We then explored the phosphorylation and total protein levels of Igf1r in our Pten−/−, ERG Pten−/−, and ERG Pten−/− Crispr ERG organoids. Interestingly, ERG overexpression was associated with reduced phosphorylation and total protein levels of Igf1r, and upon ERG knock-out, phosphorylation of Igf1R increase significantly with an associated increase in total Igf1r levels (Figure 3E). To demonstrate that the changes in RTK phosphorylation were IRS2 dependent we evaluated the levels of Igf1r in the setting of ERG aberrant expression. Indeed, we find that in the setting of ERG overexpression where IRS2 levels are suppressed Igf1r show reduced phosphorylation and total protein levels, and overexpression of IRS2 in this model system, increases the phosphorylation and total levels of Igf1r (Figure 3F, Supplementary Figure 3B).
Validation of ERG – IRS2 – RTK pathway in human model systems
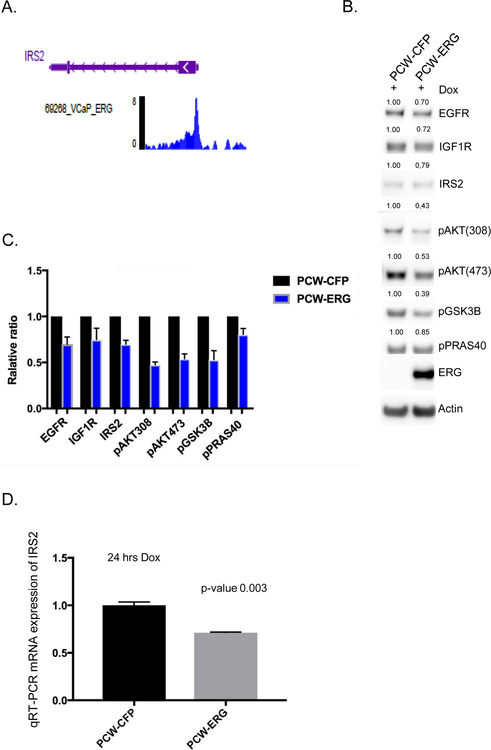
To further validate that ERG is a transcriptional repressor of IRS2, we performed an in silico analysis of publicly available ChIP-seq datasets using the ChIP-Atlas (chip-atlas.org). Indeed, we found that across various transcription factor ChIP experiments in the VCaP cell line, ERG demonstrated significant enrichment of peak reads at the IRS2 promoter region, similar to our GEM model prostate organoid experiments (Table 1, Figure 4A, Supplementary Figure 3C). Additionally, normal human prostate organoids were established, and ERG expression was performed in a dox-inducible fashion. The expression of ERG resulted in repression of IRS2 protein and mRNA expression, reduced total RTK levels (EGFR, IGF1R), and suppressed PI3K signaling across various nodes of the pathway (Figure 4B, 4C, 4D).
Table 1.
In silico analysis of publicly available VCaP transcription factor ChIP-Seq data sets demonstrating significant fold-enrichment for ERG binding in the region of the transcription start site for IRS2. Data analysis performed using ChIP-Atlas.
| Experiment ID | Antigen | Cell Line | No. Peaks | Overlaps/Control | Log p-value | Fold Enrichment |
|---|---|---|---|---|---|---|
| SRX1629251 | ERG | VCaP | 963 | 212/18438 | −1.9 | 86.97 |
| SRX471871 | BRD3 | VCaP | 1883 | 232/18438 | −1.9 | 79.47 |
| SRX1629253 | ERG | VCaP | 756 | 307/18438 | −1.8 | 60.06 |
| SRX1490930 | RB1 | VCaP | 499 | 334/18438 | −1.7 | 55.2 |
| SRX595673 | FOXP1 | VCaP | 8264 | 755/18438 | −1.4 | 24.42 |
| SRX2642375 | HOXB13 | VCaP | 9544 | 758/18438 | −1.4 | 24.32 |
Figure 4. Validation of ERG repression of IRS2-RTK signaling in human model systems.
A) In silico ChIP seq analyses in the prostate cancer ERG positive cell line VCaP demonstrates significant enrichment for ERG binding at the IRS2 promoter region. Representative read peaks of ERG binding in the IRS2 transcription start site region. B) Dox-inducible overexpression of ERG (24 hrs) in human normal prostate organoids demonstrates reduced levels of IRS2, total EGFR, total IGF1R, and downstream PI3K phosphorylation of AKT, GSK3B, PRAS40. Experiment performed in triplicate, representative western blot shown, and quantification of proteins normalized to actin. C) Bar graph representing mean and standard deviation for protein quantification across three independent experiments of ERG over-expression in human normal prostate organoids, normalized to control. D) IRS2 mRNA levels are significantly repressed following acute overexpression of ERG (24 hrs) in human normal prostate, experiment performed in triplicate, and mean and standard deviation reported.
IRS2 expression promotes cell migration in a PI3K dependent manner but is not sufficient for tumorigenesis
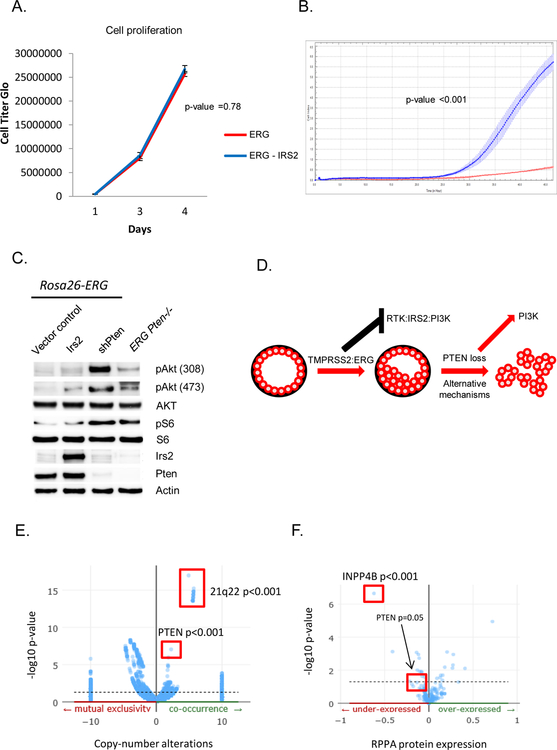
Our studies of mouse and human prostate cancer models establish that ERG impairs PI3K activation by direct repression of IRS2. We therefore postulated that restoration of IRS2 expression might promote ERG tumorigenesis in a similar manner as loss of Pten. Although in vitro cell proliferation was not augmented by IRS2 expression, IRS2 significantly enhanced cell migration (Figure 5A, 5B). Using p110a and p110b isoform selective inhibitors we found that the increase in cell migration occurred in a PI3K dependent fashion (Supplementary Figure 4A, 4B, 4C). Similarly, loss of PTEN promoted cell migration in a PI3K dependent fashion (Supplementary Figure 4D, 4E). Despite this enhanced migration, expression of IRS2 did not promote in vivo tumor growth in the setting of ERG expression as we observe with loss of Pten (Supplementary Figure 5, Figure 1A). While this may be secondary to non-PI3K related functions of PTEN, we find that PI3K signaling is significantly higher in the setting of Pten loss compared to IRS2 expression (Pten wild-type) and we believe this explains these results as previous studies have demonstrated that robust AKT activation through expression of myristylated AKT cooperates with ERG to promote tumorigenesis (Figure 5C) (21). Collectively our data provides a conceptual framework where an early oncogenic event (ERG aberrant expression in prostate) inhibits a mitogenic cellular signaling network (PI3K pathway) and may select for subsequent genetic events to activate signaling (loss of PTEN) resulting in transformation and progression (Figure 5D). To address this selection of concomitant genetic events, we analyzed the TCGA data to identify gene alterations that were significantly enriched in prostate cancers harboring ERG genomic rearrangements. The most significantly associated copy number alterations in ERG positive prostate cancers involved deletion of chromosome 21q22 which is the site of the TMPRSS2:ERG genomic rearrangement. Supporting our model, loss of PTEN (10q23) was the next most significantly enriched copy number alteration in ERG positive prostate cancers (p-value 1.4x10–7, q-value 2.4x10–5, Figure 5E). Surprisingly, analysis of the protein expression data in TCGA revealed that the most significantly under-expressed protein in ERG rearranged prostate cancers was INPP4B, which is the PIP2 phosphatase (Figure 5F). Thus, in prostate cancer ERG genomic rearrangements significantly co-occur with molecular alterations that promote active PI3K signaling, although a subset of ERG positive prostate cancers will evolve with alterations of other oncogenic pathways.
Figure 5. IRS2 expression promotes cell migration in a PI3K dependent manner but is not sufficient for tumorigenesis.
A) Overexpression of IRS2 in Rosa26-ERG organoids is not associated with increased cell proliferation. Experiment performed in triplicate with error bars reporting standard deviation from the mean. B) Overexpression of IRS2 promotes cell migration in a Boyden chamber assay. Experiment performed in triplicate with measuring cell density with error bars reporting standard deviation from the mean. C) Western blot comparison of IRS2 overexpression compared to shPten demonstrates a greater increase in pAkt signaling following knock-down of Pten (experiment run in triplicate). D) Model of the selective evolution of prostate cancer progression in tumor harboring ERG genomic rearrangements. E) Genomic data from TCGA profiled primary prostate cancers demonstrate a significant enrichment for PTEN loss in ERG positive prostate cancers. F) RPPA data from TCGA profiled primary prostate cancers demonstrated a significant reduction in protein expression of INPP4B and PTEN in ERG positive tumors.
Discussion
Tumorigenesis is a multi-step process of selection for pathway alterations that promote cell proliferation, survival, migration, and invasion. Here we show that ERG, an established oncogene, represses PI3K signaling through direct transcriptional suppression of IRS2. This repression of the PI3K pathway in turn may select for concomitant alterations, such as PTEN loss, that activate PI3K signaling to promote tumorigenesis during the evolution of prostate cancer. Given that ERG is an attractive therapeutic target in prostate cancer and there are inhibitors in pre-clinical development, our model demonstrates that inhibition of ERG may result in hyperactive PI3K signaling potentially impacting response to therapy and necessitating combined inhibition of PI3K. This finding is similar to the interaction we have discovered between PI3K and AR, and trials of combined PI3K and AR inhibitors are advancing in the clinic (17) (22). This work has broad implications with regards to furthering our understanding of the evolutionary biology of cancer and potential impact of cancer therapeutics.
IRS2 is a cytoplasmic signaling molecule that mediates effects of insulin, insulin-like growth factor 1, and other cytokines by acting as a molecular adaptor between diverse receptor tyrosine kinases and downstream effectors (18,20). IRS2 interacts with RTKs and upon tyrosine phosphorylation, binds the p85 regulatory subunit of the PI3K complex promoting activation and downstream signaling. Previous studies have demonstrated that IRS2 overexpression can promote active PI3K signaling and knock-out mice demonstrate a diabetic phenotype associate with resistance to insulin signaling (23). We have shown that the transcription of IRS2 is directly suppressed by ERG through promoter binding. While IRS2 expression can restore PI3K signaling levels in ERG positive Pten wild-type cells, this is still not sufficient for tumorigenesis, which explains why PTEN loss, and not IRS2 amplification or overexpression is more commonly selected for in the evolution of prostate cancer.
In accordance with these findings concomitant loss of PTEN resulting in hyperactive PI3K signaling promotes the development and progression of an invasive prostate cancer. These findings are well established in pre-clinical models and corroborated by the genomic profiling studies in prostate cancer revealing a significant enrichment for loss of PTEN in ERG positive tumors. Additionally, reduced protein levels of the PIP2 phosphatase INPP4B was also significantly enriched in human ERG positive prostate cancers. Prior studies have shown that the loss of INPP4B activates downstream PI3K signaling and cooperate with loss of PTEN to promote tumorigenesis but no studies have evaluated the cooperative impact of INPP4B loss and ERG overexpression in prostate cancer (24). While alterations in the major regulators of PI3K signaling are enriched in ERG positive prostate cancers, there still remains a subset of ERG positive cancers that evolve in a PI3K independent manner.
Conclusion
Genomic rearrangements leading to the aberrant expression of ERG represses PI3K signaling in an IRS2 dependent manner. This finding explains in part, the enrichment for secondary alterations, such as loss of PTEN, that activate PI3K signaling and promote the progression of ERG positive prostate cancers. Our work provides insight on how initiating oncogenic events may directly influence the selection of secondary concomitant alterations to promote oncogenic signaling during tumor evolution.
Supplementary Material
Significance.
This work provides insight on how initiating oncogenic events may directly influence the selection of secondary concomitant alterations to promote tumorigenesis.
Acknowledgements
A special thanks to members of the Chen, Sawyers, and Rosen labs for providing informative discussion.
Funding
This work was funded in part through: NIH/NCI Prostate SPORE P50-CA092629-14, NIH/NCI R01-CA182503-01A1 (B.S.C.), PCF Challenge Award (B.S.C.) and the MSKCC NIH/NCI Cancer Center Support Grant P30 CA008748. C. L. S. is a Howard Hughes Medical Institute Investigator. Funding through the STARR Cancer Consortium (Y.C., BSC.) allowed for establishment of a prostate organoid core to assist in our experiments.
Footnotes
Competing Interests: Dr. Sawyers serves on the Board of Directors of Novartis, is a co-founder of ORIC Pharm and co-inventor of enzalutamide and apalutamide. He is a science advisor to Agios, Beigene, Blueprint, Column Group, Foghorn, Housey Pharma, Nextech, KSQ, Petra and PMV.
References
- 1.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18:11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research N. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015;163:1011–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morais CL, Guedes LB, Hicks J, Baras AS, De Marzo AM, Lotan TL. ERG and PTEN status of isolated high-grade PIN occurring in cystoprostatectomy specimens without invasive prostatic adenocarcinoma. Hum Pathol 2016;55:117–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet 2009;41:619–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med 2013;19:1023–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantley LC. The phosphoinositide 3-kinase pathway. Science 2002;296:1655–7 [DOI] [PubMed] [Google Scholar]
- 7.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 2005;436:725–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trotman LC, Niki M, Dotan ZA, Koutcher JA, Di Cristofano A, Xiao A, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol 2003;1:E59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A, et al. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol 2009;22:1083–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 2014;159:163–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drost J, Karthaus WR, Gao D, Driehuis E, Sawyers CL, Chen Y, et al. Organoid culture systems for prostate epithelial and cancer tissue. Nat Protoc 2016;11:347–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi P, Chen Y, Zhang L, Guo X, Wongvipat J, Shamu T, et al. ETV1 is a lineage survival factor that cooperates with KIT in gastrointestinal stromal tumours. Nature 2010;467:849–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu T Use model-based Analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol Biol 2014;1150:81–95 [DOI] [PubMed] [Google Scholar]
- 15.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Creighton CJ. A gene transcription signature of the Akt/mTOR pathway in clinical breast tumors. Oncogene 2007;26:4648–55 [DOI] [PubMed] [Google Scholar]
- 17.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 2011;19:575–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaw LM. Identification of insulin receptor substrate 1 (IRS-1) and IRS-2 as signaling intermediates in the alpha6beta4 integrin-dependent activation of phosphoinositide 3-OH kinase and promotion of invasion. Mol Cell Biol 2001;21:5082–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szabolcs M, Keniry M, Simpson L, Reid LJ, Koujak S, Schiff SC, et al. Irs2 inactivation suppresses tumor progression in Pten+/− mice. Am J Pathol 2009;174:276–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khamzina L, Gruppuso PA, Wands JR. Insulin signaling through insulin receptor substrate 1 and 2 in normal liver development. Gastroenterology 2003;125:572–85 [DOI] [PubMed] [Google Scholar]
- 21.Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON. Identification of a cell of origin for human prostate cancer. Science 2010;329:568–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Bono JS, De Giorgi U, Nava Rodrigues D, Massard C, Bracarda S, Font A, et al. Randomized Phase II Study of Akt Blockade With or Without Ipatasertib in Abiraterone-Treated Patients With Metastatic Prostate Cancer With and Without PTEN Loss. Clin Cancer Res 2018 [DOI] [PubMed]
- 23.Sadagurski M, Weingarten G, Rhodes CJ, White MF, Wertheimer E. Insulin receptor substrate 2 plays diverse cell-specific roles in the regulation of glucose transport. J Biol Chem 2005;280:14536–44 [DOI] [PubMed] [Google Scholar]
- 24.Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, Bowtell D, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell 2009;16:115–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.