Abstract
Breast tumors are heterogeneous and composed of different sub-population of cells, each with dynamic roles that can change with stage, site and microenvironment. Cellular heterogeneity is in part due to cancer stem-like cells (CSC) that share properties with stem cells and associated with treatment-resistance. CSCs rewire metabolism to meet energy demands of increased growth and biosynthesis. O-GlcNAc transferase enzyme (OGT) uses UDP-GlcNAc as a substrate for adding O-GlcNAc moieties to nuclear and cytoplasmic proteins. OGT/O-GlcNAc levels are elevated in multiple cancers and reducing OGT in cancer cells blocks tumor growth. Here, we report that breast CSCs enriched in mammosphere cultures contain elevated OGT/O-GlcNAcylation. Inhibition of OGT genetically or pharmacologically reduced mammosphere forming efficiency, the CD44H/CD24L, NANOG+ and ALDH+ CSC population in breast cancer cells. Conversely, breast cancer cells over-expressing OGT increased mammosphere formation, CSC populations in-vitro and also increased tumor initiation and CSC frequency in-vivo. Furthermore, OGT regulates expression of a number of epithelial to mesenchymal transition (EMT) and cancer stem-like cell markers including CD44, NANOG, and c-Myc. In addition, we identify KLF8 as a novel regulator of breast cancer mammosphere formation and a critical target of OGT in regulating cancer stem-like cells.
Keywords: O-GlcNAc, OGT, metabolism, cancer stem-like cell, mammospheres, CD44, NANOG, KLF8
Introduction
Breast cancer is the most common cancer in women and the leading cause of death in women worldwide (1, 2). As of 2018, the world incidence rate is 46 per 100,000 with more than 2 million cases diagnosed (2). Intratumoral cellular heterogeneity is considered a critical player of the malignant breast cancer state, persistent tumor growth, metastasis and therapeutic resistance (3). This heterogeneity is driven, in part, by a self-renewing population of cancer stem-like cells (CSCs) (4). This subpopulation of CSCs in a tumor can be isolated and are able to self-renew, differentiate and form the bulk of the tumor (5). Many cancers do not respond to traditional chemotherapy or radiotherapy, and those that initially respond, often relapse. Conventional chemotherapy preferentially targets proliferating cells, leaving behind a pool of resistant stem-like cells that can regenerate the heterogenous tumor and thus thought to drive recurrence (6). Understanding mechanisms that regulate cancer stem cell activity will lead to designing and developing novel effective therapeutics.
Mutations in various tumor suppressors and oncogenes deregulate metabolic pathways causing malignancy (7). Cancer cells heavily favor glycolysis for ATP production over oxidative phosphorylation even in oxygen proficient conditions. This phenomenon is termed “Warburg effect” (8) and its characteristic increased glucose uptake is used to fuel the synthesis of building blocks for the growth of cancer cells. Glycolytic environment also supports homeostasis of pluripotent cells and induction of pluripotency in differentiated somatic cell requires drastic remodeling of metabolic framework (9). Similarly, CSC populations also exhibit altered metabolic signatures (10).
The hexosamine biosynthetic pathway (HBP) takes up 2–5% of glucose that enters a cell and along with glutamine is used to produce UDP-GlcNAc (11). This amino sugar can serve as a substrate for the enzyme O-GlcNAc transferase (OGT) that post-translationally modifies proteins by adding O-linked β-N-acetylglucosamine moieties (O-GlcNAc) to serine and threonine residues of nuclear and cytoplasmic proteins (12). O-GlcNAcase (OGA) is the enzyme responsible for removing this post-translational modification (12). Functional regulation of proteins by O-GlcNAcylation is similar to phosphorylation and often competes for similar residues, modulating protein-protein interaction, protein stability, and other important functions (12). O-GlcNAc is highly elevated in many cancers (13) (13)including breast (14) and prostate (15) and contributing to various hallmarks of cancer such as proliferation, angiogenesis, invasion and survival (13). Targeting OGT can impair growth, invasion and metastasis of breast and prostate cancer in-vitro and in-vivo (14) (15). Importantly, inhibiting O-GlcNAcylation induces endoplasmic reticulum stress and apoptosis in breast cancer cells but not in normal mammary epithelial cells (16) (17). However, the possible role of OGT and O-GlcNAcylation in cancer stem cellregulation has not been examined.
Here we report that OGT/O-GlcNAc is required and sufficient for maintaining CSC phenotype in breast cancer cells in-vitro and play a critical role in tumor initiating potential in-vivo. In addition, we identify a key target of OGT, Krüppel-like factor 8 (KLF8) as a key regulator of breast cancer mammosphere formation.
Material and Methods
Tissue Culture
MDA-MB-231, MDA-MB-157 and MCF-7 cells were recently purchased from ATCC and cultured as previously described (17). SUM159 and HCI-10 cell line were received as a kind gift from Dr. Tiffany Seagroves (University of Tennessee) and cultured as previously described (18). Cells were cultured using Dulbecco’s Modified Eagle’s Medium (DMEM) culture medium supplemented with 10% fetal bovine serum (FBS), 5% 10,000 Units/mL Penicillin-10,000 μg/mL Streptomycin, and 5% 200 mM L-Glutamine. Cells stably overexpressing OGT (GeneCopoeia, EX-Z3428-M13–10- OGT plasmid) were generated through production of lentivirus. All cell lines were quarterly tested for mycoplasma (abmGood PCR Mycoplasma Detection Kit) and cell line verification was done using Genetica STR DNA profile. Following thawing, cells were only used for experiments for up to ten passages.
RNA interference
OGT knockdown was performed using two different lentiviral constructs. OGT shRNA constructs were acquired from Sigma and the sequences used for OGT-1: GCCCTAAGTTTGAGTCCAAATCTCGAGATTTGGACTCAAACTTAGGGC and for OGT-2: GCTGAGCAGTATTCCGAGAAACTCGAGTTTCTCGGAATACTGCTCAGC. KLF8 shRNA constructs were acquired from Sigma (TRCN0000015881, TRCN0000015878) and the sequences used for KLF8–1: CCGGCCTCAGTCAGTCTGCCAAATACTCGAGTATTTGGCA-GACTGACTGAGGTTTTT and for KLF8–2: CCGGCGCTGCTTTATTCTTTCCAATCTCGA-GATTGGAAAGAATAAAGCAGCGTTTTT. Control shRNA was acquired through Addgene (plasmid 1864), from D. Sabatini (Whitehead Institute for Biomedical Research, MIT) and the sequence used was: CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAAC-CTTAGG. pLKO-Puro vectors carrying control (scramble sequence) shRNA, and OGT/KLF8 shRNA sequences (OGT-1 or OGT-2, KLF8–1 or KLF8–2) shRNA were packaged into VSVG-pseudotyped lentiviruses, through co-transfection of HEK-293T packaging cells with 10 μg of vector DNA and appropriate packaging vectors. Stable cell lines for shRNA knockdowns were generated as previously described (14).
Mammosphere assay
Cells were cultured in low attachment polyhema coated (1.2% in 95% ethanol) plates, with DMEM/F12 (Gibco), 20ng/ml EGF (Sigma), 20ng/ml bFGF (Invitrogen: PHG0024), B27 50x (Invitrogen; 17504–044), and 1mg/ml Pen/Strep (Gibco). Cells were plated in 96-well plates or 6-well plates and mammospheres greater than 50um in diameter were counted after 7 days (5 days for HCI-10 cell line). For mammosphere forming efficiency (MFE) assay we plated 100 cells per well and calculated MFE using the formula: (number of mammospheres/number of cells plated) * 100 and as previously described (19).
Anoikis assay
The anoikis assay was performed as described previously (20). In brief, 3×105 cells were cultured in suspension using low attachment polyhema coated plate (1.2% in 95% ethanol), or in adherence using 6-well plate in regular media. Cells cultured in both suspension and adherence were collected after 24h and 48h in lysis buffer for immunoblot analysis.
Inhibitors and treatments
The OGT inhibitor (Ac-5s-GlcNAc) and the O-GlcNAcase inhibitor (NButGT) were a kind gift from Dr. David Vocadlo (Simon Fraser University). O-GlcNAcase inhibitor Thiamet-G was obtained from Cayman Chemical. Cells were treated with inhibitor for 48hrs before being plated for the mammosphere assay.
Flow Cytometry
Cells were trypsinized (0.05% Trypsin), counted, washed twice with 1xPBS, and resuspended in 50ul 1%BSA in PBS. Cells were incubated with 5ul (anti-CD44) and 2ul (anti-CD24) antibody for 45 min on ice. Then washed with 1%BSA in PBS twice and resuspended in 1ml 1xPBS. Tubes were then read on Guava easyCyte flow cytometer. All data were collected and analyzed using a Guava EasyCyte Plus system and CytoSoft (version 5.3) software (Millipore). Data are gated and expressed relative to the appropriate unstained and single stained controls. Antibodies were purchased from; CD44-FITC (BD Biosciences; 555478), CD24-PE (Biolegend; 31106), CD24-PECy5 (Biolegend; 101812). For NANOG-GFP experiments, cells were stably transduced with NANOG-GFP (Addgene) and analyzed on GFP channel after respective treatments. The Aldeflour assay was performed using the ALDEFLOUR kit from STEMCELL Technologies as per manufacturer’s instructions. Unstained and single-stained controls were used for gating.
Western blotting
Western blotting procedures were carried out as previously described (15). Briefly, cells were collected in RIPA lysis buffer (150mM NaCl, 1% NP40, 0.5% DOC, 50mM Tris-HCl at pH 8, 0.1% SDS, 10% glycerol, 5mM EDTA, 20mM NaF and 1mM Na3VO4, 1 μg/ml each of pepstatin, leupeptin, and aprotinin, 200 μg/ml phenyl-methylsulfonyl-fluoride). Lysates were cleared by centrifugation at 14,000 x g for 20 minutes at 4 °C and analyzed by SDS-PAGE and autoradiography. Lysates from mammospheres were collected 7 days after culture in mammosphere media. Corresponding adherent controls were put on mammosphere media 24 hrs after seeding and collected the next day. Antibodies were purchased from the following companies; Anti-OGT (Santa Cruz, Cell-Signaling), Anti-O-GlcNAc (Santa Cruz, Sigma), Anti-c-Myc (Novus), Anti-Actin (Santa Cruz), Anti-CD44, Anti-Fibronectin, Anti-Vimentin, Anti-NANOG (Cell-Signaling), Anti-K8/18 (Thermo-Fisher), Anti-K14 (Thermo-Fisher), Anti-GAPDH (Genscript), Anti-KLF8 (Sigma). Densitometry was performed using Image J Software (National Institutes of Health, Bethesda, MA).
Quantitative RT-PCR (qRT-PCR)
Total RNA was isolated using RNeasy Mini kit (Qiagen) according to the manufacturer’s protocol for monolayer cell RNA extraction. Levels of expression were determined using the Applied Biosystems 7500, with Brilliant II qRT-PCR Master Mix Kit (Stratagene) according to the manufacturer’s protocol and as previously described (15). Taqman gene expression assay primer probes for OGT (Hs00914634_g1), CD44 (Hs01075861_m1), c-Myc (Hs00153408_m1), KLF8 (Hs05554122_s1) and Cyclophilin A (Hs99999904_m1) were purchased from Applied Biosystems (Foster City, CA, USA).
Animal studies
SUM159-Luc cells were generated by stable transfection with firefly luciferase in a pWZL-Hygro vector (kindly provided by Dr. Maureen Murphy, Wistar Institute), followed by stable transduction with either empty vector or Flag-tagged OGT lentivirus. Cells were injected into cleared inguinal mammary fat pads of 4–6 weeks old female NOD/SCID mice. Mice were weighed, palpated and imaged weekly for bioluminescence, after intraperitoneal injection with 200 ul of D-Luciferin (20mg/ml: Perkin Elmer) on the IVIS 200. Tumors were measured using digital calipers and tumor volume was calculated using the formula (LxW2)/2. Extreme limiting dilution analysis (ELDA) (21) was used to calculate tumor initiating cell (TIC) frequency. Mouse experiments were performed with the approval of Institutional Animal Care and Use Committee.
RNA-Seq
RNA was extracted using Trizol (Invitrogen) as per manufacturer’s instruction from SUM159 mammospheres expressing control or Fl-OGT (3 biological replicates per condition). RNA-Seq and analysis was performed by BGI Tech.
Survival analysis
Kaplan-Meier curves were generated using the online database Kaplan-Meier plotter (http://kmplot.com/analysis/) (22) to determine the relevance of KLF8 mRNA expression in Luminal A breast cancer patients (auto select best cutoff, overall survival, Intrinsic subtype: Luminal A), HER2+ breast cancer patients (auto select best cutoff, overall survival, Intrinsic subtype: HER2+), Mesenchymal breast cancer patients (auto select best cutoff, overall survival, Pietenpol subtype: mesenchymal), Basal-like 2 breast cancer patients (auto select best cutoff, overall survival, Pietenpol subtype: basal-like 2) to the overall survival (OS).
Statistical Analysis
Result from at least three independent experiments are presented as mean ± SEM. Statistical analysis was performed using Student’s t-test with 95% of confidence interval. * represents statistical significance with p-value ≤ 0.05.
Results
Increased levels of OGT & O-GlcNAc in breast CSC population
Cancer cells are highly dependent on OGT for growth in-vivo (14) (15), hence we hypothesized that OGT may regulate CSC populations and their growth. Since breast cancer stem-like cells have the ability to survive and form mammospheres on non-adherent substrates (23), we examined whether OGT and O-GlcNAc were enriched in these cell populations. Consistent with our hypothesis, we observed a significant increase of O-GlcNAc and OGT levels in mammosphere cultures from multiple human breast cancer cell lines including estrogen receptor positive (+) MCF7, triple-negative MDA-MB-231 and SUM159 as well as a triple-negative patient-derived xenograft cell line HCI-10 (Fig. 1A–C) compared to adherent cells cultured in mammosphere media suggesting an up-regulation of O-GlcNAcylation and OGT levels in conditions that enhance stem-like breast cancer cells. This increase in OGT and O-GlcNAc levels not due to lack of cell adhesion (or anoikis) in mammosphere culture as MCF7 (Fig. S1A) or MDA-MB-231 (Fig. S1B) cells did not increase OGT or O-GlcNAc levels when placed in suspension culture for 24 and 48 hours in regular media when compared to attached cells. Interestingly, OGT mRNA levels were also elevated in mammosphere cultures of MDA-MB-231 cells (30 fold) and SUM159 (6 fold) (Fig. 1D) compared to adherent cells. These data suggest that OGT mRNA and protein and total O-GlcNAcylation levels are elevated in conditions that enrich for breast CSCs.
Figure 1. Mammospheres are highly enriched for O-GlcNAc and OGT.
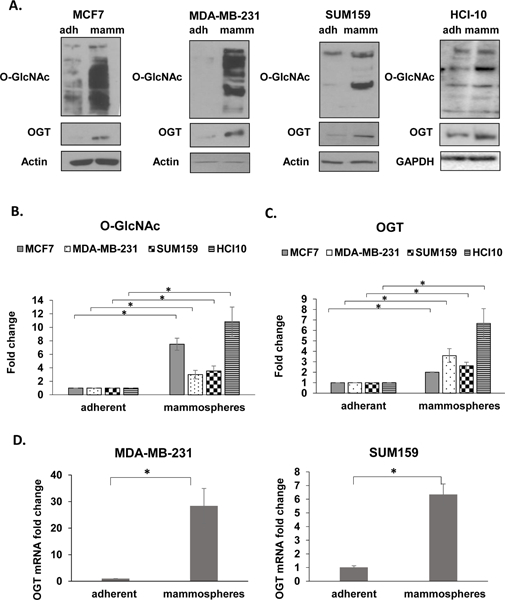
A. Immunoblotting analysis of MCF7, MDA-MB-231, SUM159 and HCI-10 cells grown in adherent (adh) and mammosphere (mamm) culture. Cell lysates from MCF7, MDA-MB-231, SUM159 and HCI-10 cells grown in adherent (adh) and mammosphere (mamm) culture were collected for immunoblot analysis with the indicated antibodies. B. Quantification of total O-GlcNAc from Fig 1A. C. Quantification of OGT from Fig 1A. D. Measurement of relative mRNA expression of OGT in MDA-MB-231 (left) or SUM159 cells (right) grown in adherent (adh) and mammosphere (mamm) culture conditions using qRT-PCR. All expression is normalized to internal control. Student’s t-test reported as mean ± SEM. * = p-value < 0.05.
Altering OGT and O-GlcNAc levels in breast cancer cells regulates mammosphere formation and CD44HCD24L CSC population
To test whether OGT and O-GlcNAcylation regulates stemness of breast cancer cells we used RNAi to stably knockdown OGT (Fig. 2A) in MDA-MB-231 cells and tested self-renewal ability through mammosphere forming efficiency (MFE). MFE was reduced more than ten-fold in MDA-MB-231 cells stably expressing OGT RNAi compared to controls (Fig. 2B). The few mammospheres that formed were smaller, suggesting that breast cancer cells require OGT to grow and form mammospheres (Fig 2B). Similar reduction in MFE was achieved in other breast cancer cells upon OGT knockdown including MCF7 (Fig. S2A), and SUM159 (Fig. S2C). CD44 and CD24 have been used extensively to isolate CSCs from breast tumors (5). Using CD44H/CD24L as a readout for breast cancer CSCs, we observed a significant reduction of CSC population in MDA-MB 231 cells with OGT knockdown compared to RNAi control cells (Fig. 2C). Similar inhibition of mammosphere formation and CD44H/CD24L levels associated with reduced OGT expression were observed in other breast cancer cells including MCF7 (Fig. S2A–B), SUM159 (Fig. S2C–D) and an additional triple-negative cell line MDA-MB-157 (Fig. S2E). To determine if cells were undergoing apoptosis and were therefore unable to form spheres, we measured early and late apoptosis through Annexin/PI staining in OGT RNAi expressing mammospheres but observed no significant induction of apoptosis (Fig. S2F). We also tested secondary mammosphere formation by dissociating primary mammospheres generated in the presence of OGT RNAi into single cells and placed in secondary mammosphere assays. OGT RNAi expressing cells (Fig. 2D) did not form secondary mammospheres as efficiently as cells from control mammospheres (Fig. 2E) suggesting OGT may play a role in self-renewal (24). To ensure OGTs contribution to mammosphere formation required its catalytic function, we also examined the effect on mammosphere formation upon treatment with a pharmacological inhibitor of O-GlcNAcylation, Ac-5S-GlcNAc (25). OGT inhibitor Ac-5s-GlcNAc reduced total O-GlcNAc levels in MDA-MB-231 (Fig. 2F), MCF7 (Fig. S3A), SUM159 (Fig. S3B) and HCI-10 treated cells (Fig. S3C). In agreement with RNAi data, mammosphere forming efficiency in the four breast cancer cells tested was also significantly impaired (Fig. 2F, S3A–C). These results suggest that OGT and O-GlcNAcylation are required for mammosphere formation in multiple breast cancer cell lines.
Figure 2. OGT inhibition attenuates mammosphere formation and CD44HCD24L TIC population.
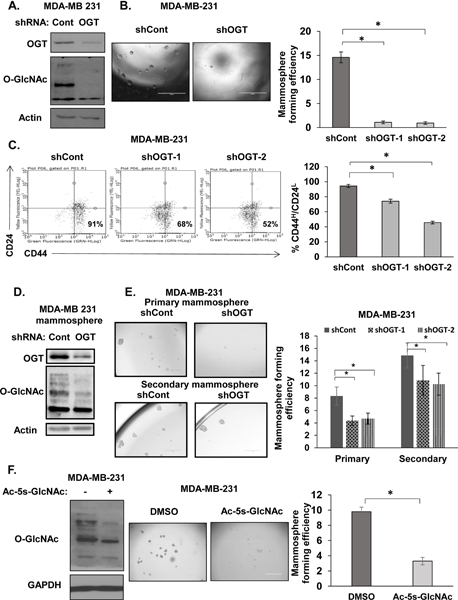
A. Cell lysates from MDA-MB-231 cells expressing control or OGT shRNA were collected for immunoblot analysis with the indicated antibodies. B. Representative images of mammospheres (left) and quantification of mammosphere forming efficiency (MFE) (right) in MDA-MB-231 cells stably expressing control and OGT shRNA C. Representative flow cytometry analysis of CD44HCD24L population in MDA-MB-231 cells stably expressing control, OGT-1, or OGT-2 shRNA and quantified (right). D. Cell lysates from MDA-MB-231 cells expressing control or OGT shRNA were collected for immunoblot analysis with the indicated antibodies following 7 days in mammosphere culture. E. Representative images of primary (top) or secondary (bottom) mammosphere culture from MDA-MB-231 cells expressing control or OGT shRNA and quantified MFE post 7 day in primary or secondary mammosphere culture (right). F. Cell lysates from MDA-MB-231 cells treated with DMSO and OGT inhibitor Ac-5s-GlcNAc (100 μM) (left) were collected for immunoblot analysis with the indicated antibodies. Representative mammosphere images (center) and quantified MFE post 7 day in mammosphere culture (right). Student’s t-test reported as mean ± SEM. * = p-value < 0.05.
Given that OGT depletion attenuates CSC potential, we examined whether OGT was sufficient to increase mammosphere formation. We stably over-expressed OGT in MCF7, SUM159 and MDA-MB 231 cells to determine whether increasing expression of OGT would enhance the CSC phenotype. MDA-MB-231 cells stably overexpressing OGT (Fig. 3A) showed nearly a 2-fold increase in MFE (Fig. 3B). Similarly, MFE of MCF7 significantly increased by 2-fold (Fig S4A) and SUM159 increased approximately 1.5-fold (Fig. S4C). Additionally, CD44H/CD24L population was significantly enriched in all breast cancer cell lines overexpressing OGT compared to controls (Fig. 3C, S4B, S4D). Although significant, the increase in CSCs in OGT overexpressing MDA-MB-231 and SUM159 cells is modest, as basal/triple negative breast cancer (TNBC) cells may contain higher cancer stem-like cell population compared to luminal breast cancer cells (26). We did not detect a significant increase in mammosphere size in OGT overexpressing cells (data not shown), however we cannot completely rule out OGT may contribute to mammosphere efficiency due to regulation of proliferation. To test whether elevating O-GlcNAc levels could also augment mammosphere formation, we indirectly increased O-GlcNAc levels utilizing the OGA inhibitor NButGT, that prevents the removal of O-GlcNAc and elevates total O-GlcNAcylation (27). After 48hr NButGT treatment, MFE of MDA-MB-231 cells increased by 3-fold (Fig. 3D) compared to control mammospheres. Similar results where seen in MDA-MB-231 (Fig. 3E) and MCF7 (Fig. S4E) cells treated with OGA inhibitor Thiamet-G. Overall, these data suggest that OGT and O-GlcNAcylation are required and sufficient in regulating mammosphere formation and may regulate CSC population.
Figure 3. Elevated OGT and O-GlcNAc increases mammosphere formation and CD44HCD24L TIC population.
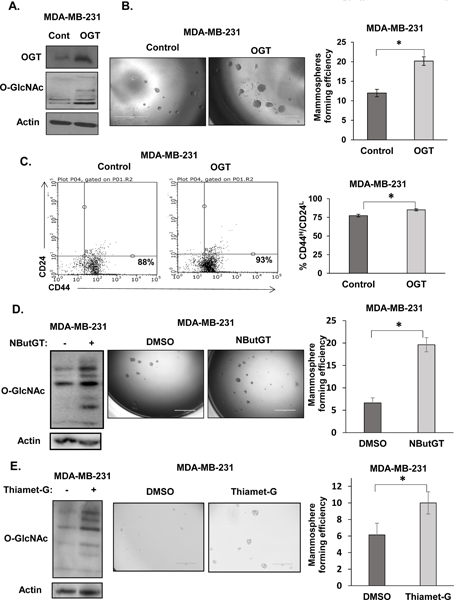
A. Cell lysates from MDA-MB-231 cells stably overexpressing control or OGT were collected for immunoblot analysis with indicated antibodies. B. Representative images of mammospheres formation of MDA-MB-231 cells stably overexpressing control or OGT (left) and quantification of MFE (right). C. Representative flow cytometry measuring CD44HCD24L population from MDA-MB-231 cells stably overexpressing control or OGT (left) and quantification (right). D. Cell lysates from MDA-MB-231 cells treated with DMSO and OGA inhibitor NButGT (100 μM) were collected for immunoblot analysis with indicated antibodies (left), representative mammospheres formation (center) and quantified MFE following 5–7day mammosphere culture (right). E. Cell lysates from MDA-MB-231 cells treated with DMSO and OGA inhibitor Thiamet-G (1 μM) were collected for immunoblot analysis with indicated antibodies (left), representative mammospheres formation (center) and quantified MFE following 7 day mammosphere culture (right). Student’s t-test reported as mean ± SEM. * = p-value < 0.05.
OGT and O-GlcNAc modulation affects NANOG GFP+ and ALDH+ populations
A tumor initiating cell detection assay was recently developed using NANOG-GFP reporter where the NANOG promoter drives GFP expression. Recent studies have shown that NANOG-GFP+ breast cancer cells are associated with elevated self-renewal and tumor initiation capacities (28). To test whether OGT also regulates NANOG-GFP+ breast CSCs we examined effects of altering OGT on this reporter system. Reducing OGT levels with stable RNAi expression (Fig. 4A) lead to reduction of NANOG-GFP+ population in MDA-MB-231 cells by 2-fold (Fig. 4B). Similar reduction of NANOG-GFP+ cells was detected in other TNBC/basal cell lines SUM159 (Fig S5A) and MDA-MB-157 (Fig. S5B) as well as luminal cell line MCF7 (Fig. S5C) expressing OGT RNAi. Conversely, under OGT overexpressing conditions (Fig. 4C), we detected a 3-fold increase in NANOG-GFP+ cells in MDA-MB-231 cells compared to control (Fig. 4D). Similar increase of NANOG-GFP+ cells was seen in other breast cancer cells overexpressing OGT including SUM159 (Fig. S5D) and MDA-MB-157 (Fig. S5E) cells compared to controls. Likewise, when we increased total O-GlcNAcylation by treating MDA-MB-231 cells with OGA inhibitor for 48hrs we detected a 4-fold increase in NANOG-GFP+ cells compared to control treated cells (Fig. S6A). Similar results were seen in SUM159 (Fig. S6B), MDA-MB-157 (Fig. S6C) and MCF7 (Fig. S6D) cells treated with NButGT compared to control cells.
Figure 4. OGT/O-GlcNAc modulation alters NANOG-GFP+ and ALDH+ TIC population.

A. Cell lysates from MDA-MB-231 cells expressing control, OGT-1 or OGT-2 shRNA were collected for immunoblot analysis with the indicated antibodies. B. Quantified flow cytometry graph showing NANOG-GFP+ population in MDA-MB-231 cells expressing control, OGT-1 or OGT-2 shRNA. C. Cell lysates from MDA-MB-231 cells stably overexpressing control or OGT were collected for immunoblot analysis with indicated antibodies. D. Quantified flow cytometry graph showing NANOG-GFP+ population in MDA-MB-231 cells stably overexpressing control or OGT. E. Cell lysates from SUM-159 cells expressing control, OGT-1 or OGT-2 shRNA were collected for immunoblot analysis with the indicated antibodies. F. Quantified flow cytometry graph showing ALDH+ population in SUM-159 cells expressing control, OGT-1 or OGT-2 shRNA. G. Cell lysates from SUM-159 cells stably overexpressing control or OGT were collected for immunoblot analysis with indicated antibodies. H. Quantified flow cytometry graph showing ALDH+ population in SUM-159 cells stably overexpressing control or OGT. Student’s t-test reported as mean ± SEM. * = p-value < 0.05.
In addition, we evaluated breast cancer cells lines with altered OGT levels using the Aldeflour assay that identifies Aldehyde dehydrogenase (ALDH) expressing cells, previously associated with increased tumor initiating and metastatic capacity (29). Consistent with other assays, SUM159 (Fig. 4E, 4F). MDA-MB-231 (Fig. S6E), MCF7 (Fig. S6F) and MDA-MB-157 (Fig. S6G) cells stably expressing OGT RNAi exhibited reduced ALDH activity compared to controls. Conversely, SUM159 cells stably overexpressing OGT (Fig. 4G) increased the ALDH+ cell fraction by 2-fold (Fig. 4H). Together, these data suggest that OGT and O-GlcNAcylation are required and sufficient to regulate in-vitro breast CSC phenotypes.
OGT regulates EMT and CSC factors
EMT is associated with cancer cell stemness and tumor initiating potential (30) (31). Therefore, we examined changes in EMT and cancer-associated stem-lke cell markers under conditions of altered O-GlcNAcylation. Consistent with the model that OGT regulates tumor initiation ability, in OGT knockdown luminal-like MCF7 cells, we found reduction of mesenchymal markers including fibronectin and vimentin, while luminal epithelial marker cytokeratin 8/18 increased (Fig. 5A). We saw analogous effect in basal-like MDA-MB-231 cells containing OGT stable knockdown with reduction in mesenchymal markers such as fibronectin, vimentin and increase in basal epithelial marker cytokeratin 14 (Fig. 5B). Conversely, overexpressing OGT increased mesenchymal markers fibronectin and vimentin in luminal MCF7 cells (Fig. 5C) as well as in basal MDA-MB-231 cells overexpressing OGT (Fig. 5D). Thus, OGT regulates EMT markers consistent with increased cancer stem cell potential. To further understand whether OGT was regulating key CSC-associated factors, we also probed for NANOG, c-Myc and CD44 all of which have been shown to regulate breast cancer tumor initiation (28) (32) (33). We detected a decrease in c-Myc and NANOG protein levels under OGT knockdown conditions in the luminal MCF7 cells (Fig. 5A, S7A) and increase in c-Myc and NANOG protein levels in MCF7 cells overexpressing OGT (Fig 5C, S7B). In the basal/TNBC cells MDA-MB-231, we found that OGT RNAi reduces c-Myc, NANOG and CD44 protein levels (Fig. 5B, S7C) while overexpression of OGT increases protein levels of Myc, NANOG and CD44 (Fig. 5D, S7D). Similar results were seen in SUM-159 cells (Fig. S7E, S7F). This data is consistent with the idea that NANOG-GFP+ reporter identifies NANOG-expressing cells, we found that breast cancer cells overexpressing OGT contained elevated NANOG protein expression compared to control cells (Fig. 5C, 5D, S7F). Furthermore, we also found that OGT regulates CD44 and c-Myc at the RNA level in MDA-MB-231 (Fig. S8A, S8B) and in SUM159 cells (Fig. S8C, S8D). Taken together, we conclude that OGT regulates induction of EMT program and expression of CSC factors that are associated with stemness and differentiation in breast cancer cells.
Figure 5. OGT regulates EMT and CSC markers.
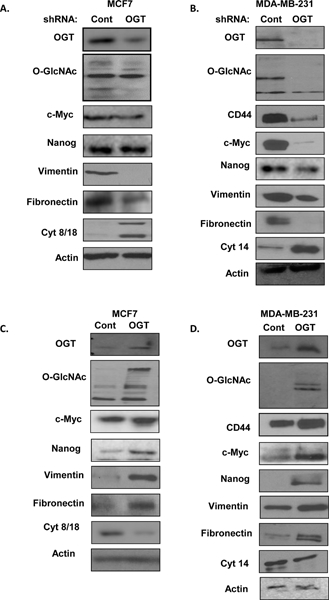
A. Cell lysates from MCF7 cells expressing control or OGT shRNA were collected for immunoblot analysis with the indicated antibodies. B. Cell lysates from MDA-MB-231 cells expressing control or OGT shRNA were collected for immunoblot analysis with the indicated antibodies. C. Cell lysates from MCF7 cells stably overexpressing control or OGT were collected for immunoblot analysis with indicated antibodies. D. Cell lysates from MDA-MB-231 cells stably overexpressing control or OGT were collected for immunoblot analysis with indicated antibodies.
OGT enhances tumor initiation in-vivo
To determine whether OGT enhances tumor initiation frequency in-vivo, we performed a limiting dilution experiment using four different dilutions of breast cancer cells (100, 500, 1000, 5000). SUM159 stably overexpressing OGT (Fig. 6A–B) or controls, that also express luciferase, were injected into cleared right inguinal fat pads of female mice. Mice were monitored and imaged for up to 8–10 weeks, and tumors were harvested for analysis. Tumor initiation was observed with as low as 100 cells in SUM159 cells over-expressing OGT whereas SUM159 control cells required at least 5000 cells to initiate a tumor (Fig. 6A–B). Limiting dilution analysis showed that OGT over-expressing SUM159 cells exhibited significantly higher frequency (1/153) of tumor initiation cells (TICs), compared to SUM159 control cells (1/4142) as determined using ELDA tool (21) (Fig. 6C). In contrast, control cells were deficient in tumor initiating ability at low densities (Fig 6A–C). Consistent with in-vitro results, xenograft tumors from OGT overexpressing SUM159 cells also contained elevated EMT markers and CSC marker levels compared to control tumors (Fig. 6D) including increased vimentin and fibronectin levels and higher levels of CD44, NANOG, and c-Myc respectively (Fig. 6D). Overall survival of mice with OGT over-expressing tumors was also significantly lower than mice with control tumors as represented using a Kaplan Meier survival curve (Fig. 6E). As OGT has been previously shown to regulate tumor growth in vivo (14), we cannot completely rule out that OGT may contribute to tumor initiation via regulation of proliferation. Thus, increased OGT expression in breast cancer cells was able to enhance tumor initiation frequency, capability and CSC markers in-vivo.
Figure 6. OGT enhances tumor initiation in-vivo.
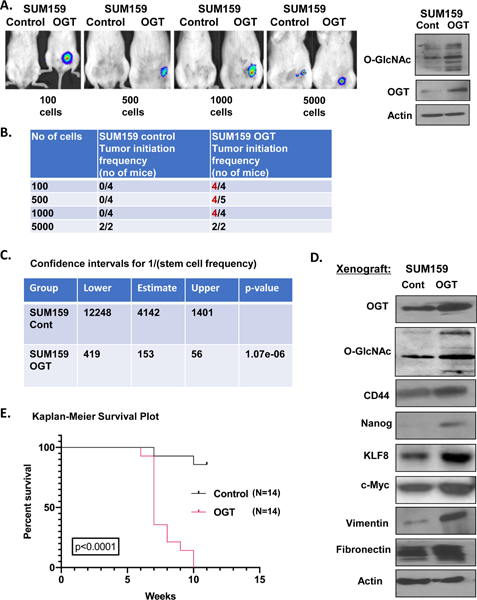
A. Representative bioluminescence imaging of mice that received orthotopic injection of increasing dilutions of SUM159-luciferase cells stably overexpressing control or OGT (left). Cell lysates from SUM159 ls stably overexpressing control or OGT were collected for immunoblot analysis with indicated antibodies (right). B. In-vivo limiting dilution assay of mice after mammary fat pad transplant of SUM159 cells overexpressing control or OGT. C. Confidence intervals for CSC frequency for SUM159 cells stably overexpressing control or OGT calculated using ELDA. D. Cell lysates from xenograft tumors of SUM159 cells stably overexpressing control or OGT were collected for immunoblot analysis with indicated antibodies. E. Kaplan-Meier overall survival curve of mice receiving SUM159 cells stably overexpressing control or OGT. Student’s t-test reported as mean ± SEM. * = p-value < 0.05.
OGT regulates stem-cell transcription factor KLF8
OGT is sufficient to increase breast CSCs in-vitro and tumor initiation in-vivo. To identify critical factors involved in OGT-mediated CSC regulation, we performed RNA-sequencing to further analyze transcriptional changes in gene expression patterns in mammospheres overexpressing OGT. Transcriptome analysis of mammospheres of SUM159 cells overexpressing OGT (Fig. 7A) revealed changes in gene expression patterns compared to control mammospheres (Fig. 7B). We identified 329 differentially regulated genes, consisting of 161 up-regulated genes and 168 down-regulated genes using a cut-off value of fold change > 2 (and p-value <0.05) (Fig. 7B, Table S1). The pathway functional enrichment for up/down regulated genes shows that some pathways associated with stem cells or cancer stem-like cells are regulated including proteoglycans in cancer, ECM-receptor interactions, cell adhesion molecules, and hematopoietic cell lineage (Fig. S9A). Among the top 10 up-regulated genes (Fig. 7C) in OGT overexpressing mammospheres, KLF8 is of interest since recent evidence suggests it can promote EMT, invasion and metastasis (34). We confirmed KLF8 protein expression is increased in MCF7, MDA-MB-231 and SUM159 cells over-expressing OGT compared to controls (Fig. 7D) as well as in SUM159 xenograft tumors (Fig. 6D). In addition, we show that KLF8 protein is enriched in MCF7, MDA-MB-231 and HCI-10 mammosphere cultures compared to adherent cells (Fig. S9B). This data suggests that KLF8 may play an important role in mammosphere formation in breast cancer cells and may be downstream of OGT-signaling. Consistent with this hypothesis, stable knockdown of KLF8 in SUM159 cells reduced mammosphere forming ability compared to controls (Fig. 7E, S9C) and reduced expression of EMT/CSC markers fibronectin, c-Myc and NANOG (Fig. S9C). Similar results on mammosphere formation were seen in MDA-MB-231 with KLF8 knockdown (Fig. S9D). We tested whether KLF8 is required for O-GlcNAc-mediated mammosphere formation. Consistent with our idea of OGT regulating KLF8, treatment with OGA-inhibitor also increased KLF8 protein levels in breast cancer cells (Fig. S9E). Importantly, reducing KLF8 levels significantly reduced the OGA inhibitor-mediated increase in mammosphere formation (Fig. 7F–G) and protein expression of EMT/CSC markers vimentin, fibronectin, and c-Myc (Fig. S9E). Consistent with idea that OGT regulates KLF8 levels, we found that RNAi against OGT in breast cancer cells MDA-MB-231 (Fig. S10A), SUM-159 (Fig. 10B) and MCF7 (Fig. S10C) cells reduced KLF8 proteins levels and RNA levels in MDA-MB-231 cells compared to controls (Fig. S10D). Supporting the idea that KLF8 is a critical factor for breast cancer progression, Kaplan-Meier survival analysis of Luminal A breast cancer patients (Fig. S11A), HER2+ breast cancer patients (Fig. S11B) and TNBC subtypes (35) mesenchymal (Fig. S11C) and basal-like 2 (Fig. S11D) breast cancer patients when stratified by the level of KLF8 expression, indicated that overall survival was significantly lower in breast cancer patients with a high KLF8 expression compared to patients with low KLF8 expression. Thus, we identify KLF8 as a key regulator of breast cancer mammosphere formation, and importantly, show that KLF8 is regulated by OGT in mammospheres and contributes, in part, to O-GlcNAcylation-mediated regulation of mammosphere formation in breast cancer cells.
Figure 7. KLF8 is a critical regulator of mammosphere formation and required for OGT-mediated mammosphere growth.
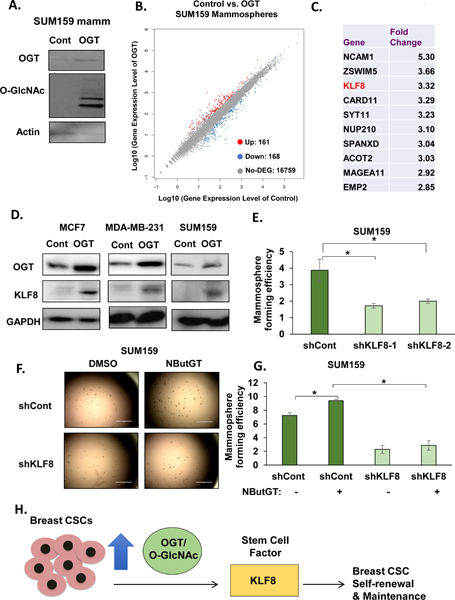
A. Cell lysates from SUM159 cells under mammosphere conditions stably overexpressing control or OGT were collected for immunoblot analysis with indicated antibodies. B. Scatter plot highlighting differentially expressed genes: upregulated (red) on top, downregulated on bottom (blue). C. Fold change increase of top 10 genes upregulated in OGT-overexpressing mammospheres compared to control. D. Cell lysates from SUM159, MCF7 and MDA-MB-231 cells stably overexpressing control or OGT were collected for immunoblot analysis with indicated antibodies. E. Quantified MFE in KLF8 knockdown in SUM159 cells using two shRNA constructs. F. Representative mammosphere images of KLF8 knockdown treated with DMSO or NButGT. G. Quantified MFE of SUM159 cells expressing shControl or shKLF8 after 48hr treatment with DMSO and NButGT (100 μM). H. Working model showing breast CSCs population increasing OGT and O-GlcNAc and OGT regulating CSC potential through, in part, stem cell factor KLF8 and responsible for maintaining CSC and tumor initiating potential of BC cells. Student’s t-test reported as mean ± SEM. * = p-value <= 0.05.
Discussion
CSCs are associated with tumor initiation, metastasis, relapse and resistance (36). CSCs are highly plastic and capable of adapting to new environment (26), therefore targeting single pathways may lead to CSC adaptation and treatment failure. This necessitates the discovery of novel signaling pathways and mechanisms that are involved in CSC maintenance. To this end, we identified OGT/O-GlcNAc as playing a critical role in maintenance and function of breast CSCs, identifying novel effectors like NANOG and CD44 and defining a new role for KLF8 in breast CSCs. Based on our observations, elevated levels of OGT/O-GlcNAcylation are required and sufficient for maintaining mammosphere formation in-vitro and plays a critical role in tumor initiating ability in-vivo. This ability of OGT to regulate TICs associates with self-renewal, increased EMT and regulation of multiple stem-cell factors like c-Myc, NANOG, and CD44. Importantly, we show that OGT-mediated mammosphere formation requires, in part, regulation of KLF8 (Fig. 7H).
Reprogrammed metabolic activities are well known to contribute to many aspects of cancer biology (37). The HBP and O-GlcNAcylation are key enablers of many of the “Hallmarks of Cancer” including growth, survival, invasion and metabolism (13). Our results indicate that OGT and O-GlcNAcylation can also regulate cancer cell self-renewal and tumor-initiating abilities. O-GlcNAcylation and OGT are known regulators of embryonic stem cells since direct O‐GlcNAcylation of stem cell factors including OCT4 are important to maintain embryonic stem cell self‐renewal (38). Interestingly, OGT and O-GlcNAc may play a similar role in tumor initiation as nearly all cancers examined to date contain increased O-GlcNAcylation either via increased HBP flux, and/or alterations in OGT or OGA expression (39). In line with our observations, recent studies have started to implicate O-GlcNAc’s role in regulating cancer stem-like cells. OGT knockdown reduced human colon CSC population in-vitro and self-renewal was associated with O-GlcNAc epigenetic regulation of transcription factor Myb-related protein B (MYBL1) (40). In lung and colon cancer cells, IL-8 is able to enhance sphere formation in-vitro and tumor initiation in-vivo by upregulating GFAT expression, glucose uptake, SOX2 expression and total O-GlcNAcylation in a GLUT-3 dependent manner (41). Our study is the first to show that increased OGT and O-GlcNAcylation is sufficient to enrich the cancer stem-like cell population in-vitro and promote tumor initiation in-vivo. It will be of interest to test whether OGT plays a similar role in tumor initiation ability in other cancers.
Metabolic plasticity may be a feature of cancer stem-like cells that allows them to adapt their metabolism to microenvironment nutritional state as there are studies showing that CSCs are mainly glycolytic or dependent on mitochondrial respiration (42). Similar to what has been found in multipotent stem cells (43), some studies have shown that CSCs are generally more glycolytic than their differentiated counterpart (44). Breast cancer cells made to increase CSC-like properties underwent metabolic reprograming from oxidative phosphorylation to glycolysis (45). Consistent with this idea, in cancer cells OGT and O-GlcNAc have been found to increase glucose uptake (16), increase de-novo lipid metabolism (46) and increase glycolytic flux (47) suggesting that OGT may contribute to the metabolism of proliferative cancer stem-like cells. More studies are needed to determine whether OGT and O-GlcNAcylation plays a direct role in CSC metabolism.
Our results also demonstrate that OGT regulates a number of established CSC factors including c-Myc, NANOG and CD44. OGT can directly modify c-Myc and regulate its protein stability (48). Here we show that OGT can also regulate c-Myc RNA levels (Fig. S8) in breast cancer cells. Our group has shown that one mechanism by which OGT and O-GlcNAcylation are elevated in cancer cells is via c-Myc regulation of OGT protein levels (17) and thus suggests a feed-forward mechanism that maintains OGT and c-Myc elevation in some cancer cells. Whether this mechanism is found in CSCs is not clear. We also show that OGT regulates protein levels of critical CSC factors NANOG and CD44 (Fig. 5, S7, S8). OGT regulation of NANOG and CD44 has not been previously reported. Our results show a positive regulation of NANOG and CD44 under increased O-GlcNAcylation and are the first to describe this regulation in cancer cells.
RNA-sequence analysis of mammospheres overexpressing OGT identified transcription factor KLF8 as a being novel effector of OGT. The Krüppel‐like factor (KLF) family of proteins includes 17 zinc‐finger transcription factors, 5 of which have been implicated in key stem cell functions, such as self‐renewal, pluripotency, as well as in cancer (34). KLF8 is elevated in cancer cell lines and primary tumors and can induce EMT and invasion in breast cancer cells (34). Mammary epithelial cell line MCF-10A cells overexpressing KLF8 gained CSC traits including increased mammosphere formation and increased expression of CD44High/CD24Low (49). Here, we show for the first time that breast cancer cell mammosphere cultures are enriched in KLF8 protein levels and KLF8 expression is required for breast cancer cell mammosphere formation. We also confirm that KLF8 is downstream effector of OGT in multiple breast cancer cells and in xenograft tumors in-vivo. Importantly, we show that OGT requires KLF8 function, in part, for mammosphere formation. Consistent with the role of KLF8 in breast CSCs, we also found that breast cancers containing high levels of KLF8 had poor overall survival.
Cancer stem-like cells are associated with resistance to chemotherapy and radiation therapy (6). Recent studies have linked increased O-GlcNAcylation to increased chemoresistance and radio-resistance in cancer cells. Importantly, suppression of O-GlcNAcylation reduced the resistance of both established and primary cancer cells to chemotherapy (39). CSCs also contain higher radio-resistance compared to the mass of tumor cells, supporting the use of CSC related biomarkers for prediction of radiotherapy outcome (36). Recent studies have also implicated the HBP pathway in protecting from radiotherapy by enhancing DNA repair pathways in-vitro and in-vivo as increasing protein O-GlcNAc modification protected tumor xenografts against radiation (50). Thus, OGT/O-GlcNAcylation may provide a promising target for chemo/radio-resistant cancers. Our results also suggest that OGT’s role in cancer stem-like cell may contribute to CSC-mediated chemo- and radio-resistance. However, a direct link between OGT/O-GlcNAcylation to CSC-chemoresistance will need to be further established.
In this study, we have described a novel role for OGT in tumor initiation and stemness in breast cancer. As OGT is required for tumor growth of many different cancers (13), it will be of interest to determine whether OGT itself is a general cancer stem-like cell factor and test whether OGT inhibitors may be viable therapeutic agents used to target CSCs.
Supplementary Material
Implications:
These findings demonstrate that OGT plays a key role in the regulation of breast CSCs in-vitro and tumor initiation in-vivo, in part, via regulation of KLF8 and thus inhibition of OGT may serve as a therapeutic strategy to regulate tumor-initiating activity.
Acknowledgements
We thank all members of Reginato Lab for helpful discussions. This research was partly supported by CA155413 (NIH/NCI) (to M.J. Reginato) and by CA192868 (NIH/NCI) (to V.L. Sodi).
Footnotes
Disclaimer
The content is soley the responsibility of the authors and does not necessarily represent the official views of the NIH.
No potential conflicts of interest were disclosed.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Fund WCR. Continuous Update Project. 2018; Available from: https://http://www.wcrf.org/sites/default/files/Breast-cancer-report.pdf.
- 2.Organization IAfRoC-WH. Cancer Today. 2018. [May 9, 2019]; Available from: http://gco.iarc.fr/today/home.
- 3.Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, et al. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. [Research Support, Non-U.S. Gov’t]. 2010. January 8;140(1):62–73. [DOI] [PubMed] [Google Scholar]
- 4.Hong D, Fritz AJ, Zaidi SK, van Wijnen AJ, Nickerson JA, Imbalzano AN, et al. Epithelial-to-mesenchymal transition and cancer stem cells contribute to breast cancer heterogeneity. J Cell Physiol. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Review]. 2018. December;233(12):9136–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003. April 1;100(7):3983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butti R, Gunasekaran VP, Kumar TVS, Banerjee P, Kundu GC. Breast cancer stem cells: Biology and therapeutic implications. Int J Biochem Cell Biol. 2019. February;107:38–52. [DOI] [PubMed] [Google Scholar]
- 7.Seyfried TN, Flores RE, Poff AM, D’Agostino DP. Cancer as a metabolic disease: implications for novel therapeutics. Carcinogenesis. 2014. March;35(3):515–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. J Gen Physiol. 1927. March 7;8(6):519–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, et al. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011. August 3;14(2):264–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng W, Gentles A, Nair RV, Huang M, Lin Y, Lee CY, et al. Targeting unique metabolic properties of breast tumor initiating cells. Stem Cells. 2014. July;32(7):1734–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marshall S, Bacote V, Traxinger RR. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem. 1991. March 15;266(8):4706–12. [PubMed] [Google Scholar]
- 12.Bond MR, Hanover JA. A little sugar goes a long way: the cell biology of O-GlcNAc. J Cell Biol. 2015. March 30;208(7):869–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrer CM, Sodi VL, Reginato MJ. O-GlcNAcylation in Cancer Biology: Linking Metabolism and Signaling. J Mol Biol. 2016. August 14;428(16):3282–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caldwell SA, Jackson SR, Shahriari KS, Lynch TP, Sethi G, Walker S, et al. Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S.]. 2010. May 13;29(19):2831–42. [DOI] [PubMed] [Google Scholar]
- 15.Lynch TP, Ferrer CM, Jackson SR, Shahriari KS, Vosseller K, Reginato MJ. Critical role of O-Linked beta-N-acetylglucosamine transferase in prostate cancer invasion, angiogenesis, and metastasis. J Biol Chem. 2012. March 30;287(14):11070–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrer CM, Lynch TP, Sodi VL, Falcone JN, Schwab LP, Peacock DL, et al. O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol Cell. 2014. June 5;54(5):820–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sodi VL, Khaku S, Krutilina R, Schwab LP, Vocadlo DJ, Seagroves TN, et al. mTOR/MYC Axis Regulates O-GlcNAc Transferase Expression and O-GlcNAcylation in Breast Cancer. Mol Cancer Res. 2015. May;13(5):923–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fatima I, El-Ayachi I, Taotao L, Lillo MA, Krutilina RI, Seagroves TN, et al. The natural compound Jatrophone interferes with Wnt/beta-catenin signaling and inhibits proliferation and EMT in human triple-negative breast cancer. PLoS One. 2017;12(12):e0189864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lombardo Y, de Giorgio A, Coombes CR, Stebbing J, Castellano L. Mammosphere formation assay from human breast cancer tissues and cell lines. J Vis Exp. 2015. March 22(97). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haenssen KK, Caldwell SA, Shahriari KS, Jackson SR, Whelan KA, Klein-Szanto AJ, et al. ErbB2 requires integrin alpha5 for anoikis resistance via Src regulation of receptor activity in human mammary epithelial cells. J Cell Sci. [Research Support, U.S. Gov’t, Non-P.H.S.]. 2010. April 15;123(Pt 8):1373–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009. August 15;347(1–2):70–8. [DOI] [PubMed] [Google Scholar]
- 22.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010. October;123(3):725–31. [DOI] [PubMed] [Google Scholar]
- 23.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003. May 15;17(10):1253–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. [Research Support, U.S. Gov’t, P.H.S.]. 2003. May 15;17(10):1253–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gloster TM, Zandberg WF, Heinonen JE, Shen DL, Deng L, Vocadlo DJ. Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat Chem Biol. 2011. March;7(3):174–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei W, Lewis MT. Identifying and targeting tumor-initiating cells in the treatment of breast cancer. Endocr Relat Cancer. 2015. June;22(3):R135–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macauley MS, Whitworth GE, Debowski AW, Chin D, Vocadlo DJ. O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J Biol Chem. 2005. July 8;280(27):25313–22. [DOI] [PubMed] [Google Scholar]
- 28.Thiagarajan PS, Hitomi M, Hale JS, Alvarado AG, Otvos B, Sinyuk M, et al. Development of a Fluorescent Reporter System to Delineate Cancer Stem Cells in Triple-Negative Breast Cancer. Stem Cells. 2015. July;33(7):2114–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007. November;1(5):555–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008. May 16;133(4):704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Reports. 2014. January 14;2(1):78–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee KM, Giltnane JM, Balko JM, Schwarz LJ, Guerrero-Zotano AL, Hutchinson KE, et al. MYC and MCL1 Cooperatively Promote Chemotherapy-Resistant Breast Cancer Stem Cells via Regulation of Mitochondrial Oxidative Phosphorylation. Cell Metab. 2017. October 3;26(4):633–47 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Louderbough JM, Schroeder JA. Understanding the dual nature of CD44 in breast cancer progression. Mol Cancer Res. 2011. December;9(12):1573–86. [DOI] [PubMed] [Google Scholar]
- 34.Tetreault MP, Yang Y, Katz JP. Kruppel-like factors in cancer. Nat Rev Cancer. [Research Support, N.I.H., Extramural Review]. 2013. October;13(10):701–13. [DOI] [PubMed] [Google Scholar]
- 35.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. 2011. July;121(7):2750–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saygin C, Matei D, Majeti R, Reizes O, Lathia JD. Targeting Cancer Stemness in the Clinic: From Hype to Hope. Cell Stem Cell. 2019. January 3;24(1):25–40. [DOI] [PubMed] [Google Scholar]
- 37.Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between Metabolism and Cancer Biology. Cell. 2017. February 9;168(4):657–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jang H, Kim TW, Yoon S, Choi SY, Kang TW, Kim SY, et al. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell. 2012. July 6;11(1):62–74. [DOI] [PubMed] [Google Scholar]
- 39.Akella NM, Ciraku L, Reginato MJ. Fueling the fire: emerging role of the hexosamine biosynthetic pathway in cancer. BMC Biol. [Research Support, Non-U.S. Gov’t Review]. 2019. July 4;17(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo H, Zhang B, Nairn AV, Nagy T, Moremen KW, Buckhaults P, et al. O-Linked N-Acetylglucosamine (O-GlcNAc) Expression Levels Epigenetically Regulate Colon Cancer Tumorigenesis by Affecting the Cancer Stem Cell Compartment via Modulating Expression of Transcriptional Factor MYBL1. J Biol Chem. 2017. March 10;292(10):4123–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimizu M, Tanaka N. IL-8-induced O-GlcNAc modification via GLUT3 and GFAT regulates cancer stem cell-like properties in colon and lung cancer cells. Oncogene. 2019. February;38(9):1520–33. [DOI] [PubMed] [Google Scholar]
- 42.Peiris-Pages M, Martinez-Outschoorn UE, Pestell RG, Sotgia F, Lisanti MP. Cancer stem cell metabolism. Breast Cancer Res. 2016. May 24;18(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Folmes CD, Dzeja PP, Nelson TJ, Terzic A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell. 2012. November 2;11(5):596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mancini R, Noto A, Pisanu ME, De Vitis C, Maugeri-Sacca M, Ciliberto G. Metabolic features of cancer stem cells: the emerging role of lipid metabolism. Oncogene. 2018. May;37(18):2367–78. [DOI] [PubMed] [Google Scholar]
- 45.Dong C, Yuan T, Wu Y, Wang Y, Fan TW, Miriyala S, et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013. March 18;23(3):316–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sodi VL, Bacigalupa ZA, Ferrer CM, Lee JV, Gocal WA, Mukhopadhyay D, et al. Nutrient sensor O-GlcNAc transferase controls cancer lipid metabolism via SREBP-1 regulation. Oncogene. [Research Support, N.I.H., Extramural]. 2018. February 15;37(7):924–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang B, Zhou P, Li X, Shi Q, Li D, Ju X. Bitterness in sugar: O-GlcNAcylation aggravates pre-B acute lymphocytic leukemia through glycolysis via the PI3K/Akt/c-Myc pathway. Am J Cancer Res. 2017;7(6):1337–49. [PMC free article] [PubMed] [Google Scholar]
- 48.Itkonen HM, Minner S, Guldvik IJ, Sandmann MJ, Tsourlakis MC, Berge V, et al. O-GlcNAc transferase integrates metabolic pathways to regulate the stability of c-MYC in human prostate cancer cells. Cancer Res. 2013. August 15;73(16):5277–87. [DOI] [PubMed] [Google Scholar]
- 49.Wang X, Lu H, Li T, Yu L, Liu G, Peng X, et al. Kruppel-like factor 8 promotes tumorigenic mammary stem cell induction by targeting miR-146a. Am J Cancer Res. 2013;3(4):356–73. [PMC free article] [PubMed] [Google Scholar]
- 50.Efimova EV, Appelbe OK, Ricco N, Lee SS, Liu Y, Wolfgeher DJ, et al. O-GlcNAcylation Enhances Double-Strand Break Repair, Promotes Cancer Cell Proliferation, and Prevents Therapy-Induced Senescence in Irradiated Tumors. Mol Cancer Res. 2019. March 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


