Abstract
Purpose:
To investigate the impact of rapid-turnaround exome sequencing in critically ill neonates using phenotype-based subject selection criteria.
Methods:
Intensive care unit babies aged <6 months with hypotonia, seizures, a complex metabolic phenotype, and/or multiple congenital malformations were prospectively enrolled for rapid (<7 day) trio-based exome sequencing. Genomic variants relevant to the presenting phenotype were returned to the medical team.
Results:
A genetic diagnosis was attained in 29 of 50 (58%) sequenced cases. Twenty-seven (54%) patients received a molecular diagnosis involving known disease genes; two additional cases (4%) were solved with pathogenic variants found in novel disease genes. In 24 of the solved cases, diagnosis had impact on patient management and/or family members. Management changes included shift to palliative care, medication changes, involvement of additional specialties, and the consideration of new experimental therapies.
Conclusion:
Phenotype-based patient selection is effective at identifying critically ill neonates with a high likelihood of receiving a molecular diagnosis via rapid-turnaround exome sequencing, leading to faster and more accurate diagnoses, reducing unnecessary testing and procedures, and informing medical care.
Keywords: exome sequencing, neonates, intensive care unit
INTRODUCTION
Many neonates requiring admission to intensive care units are eventually diagnosed with underlying genetic conditions. Their clinical presentations vary widely, from major anatomic malformations or striking biochemical abnormalities to more subtle signs and symptoms.1 Collectively, these conditions contribute significantly to morbidity and mortality, and the traditional path to diagnosis is often long, requiring extensive evaluations that may be invasive and/or costly.2
Increasingly, exome and genome sequencing (ES/GS) are being used to accelerate the diagnostic odyssey in a variety of clinical settings, including the neonatal intensive care unit (NICU). Several ES/GS studies in the NICU population have found diagnostic yields in the 21–60% range.3–13 Exactly which patients are most likely to benefit from testing remains incompletely defined, as enrollment criteria in these studies often rely upon expert opinion of highly specialized teams including medical and metabolic geneticists and neurologists. Meanwhile, a randomized study showed that without specific inclusion criteria, sequencing NICU patients yielded a diagnosis in only 1 of 29 cases.14 As the deployment of clinical ES/GS continues to expand, it is increasingly pressing to define which patients benefit most from these technologies, especially for institutions with limited access to clinical genetics expertise.
Motivated by this need, we developed and tested phenotype-based criteria for selecting neonates in the ICU for rapid-turnaround ES, seeking to maximize utility while allowing for easy implementation of this approach across (N) ICUs.
MATERIALS AND METHODS
Patient selection algorithm
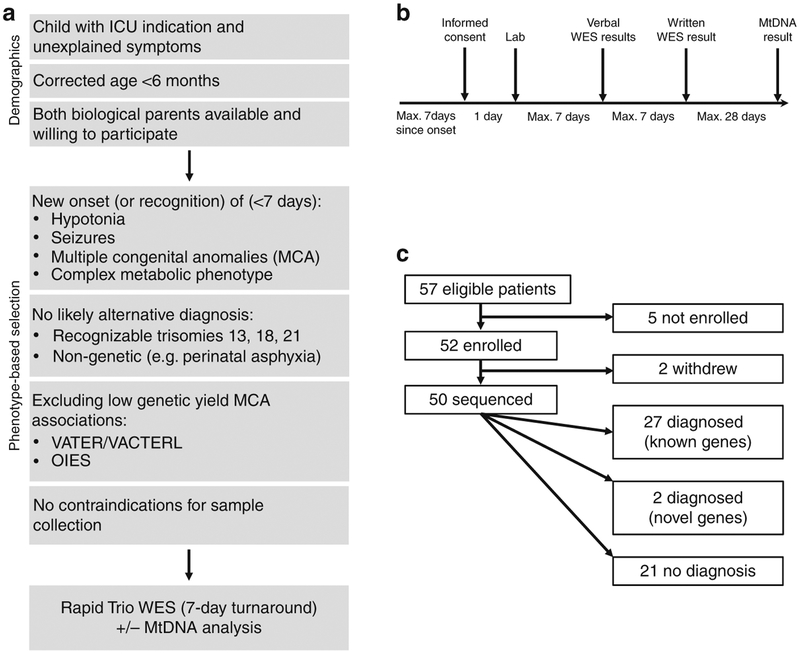
The patient selection algorithm is summarized in Fig. 1a. Enrollment criteria were developed by a multidisciplinary medical team (neonatology, genetics, and neurology) based on review of ten years of genetic testing results in the Boston Children’s Hospital (BCH) NICU population.2
Fig. 1. Details of patient enrollment.
(a) Phenotype-driven selection protocol. (b) Timeline for screening, enrollment, sendoff, and return of results. (c) Overview of patients. ES exome sequencing, ICU intensive care unit, mtDNA mitochondrial DNA.
Eligibility criteria included a corrected age <6 months, ICU admission (or awaiting ICU transfer), and recent presentation with one or more of the following unexplained clinical features: (1) seizures, (2) hypotonia, (3) multiple congenital anomalies (MCA), (4) complex metabolic phenotype (defined as the presence of clinical or biochemical features suspicious for a metabolic disorder, with more than one possible condition in the differential diagnosis, which could not be distinguished with one easily available laboratory test). These four criteria represented the core phenotypic eligibility criteria used for enrollment of the first 20 cases. After the first 20 cases, additional clinical phenotypes were added: (5) disorders of sex development (DSD), (6) interstitial lung disease, and (7) immunodeficiency. “Recent presentation” was defined as within 7 days of (a) onset of symptoms, (b) ordering of their first genetic test, (c) consulting genetics/metabolism, or (d) admission to the ICU.
Exclusion criteria included (1) presence of a likely nongenetic explanation for the phenotype (e.g., perinatal asphyxia explaining hypotonia); (2) clinical features pathognomonic for a recognizable chromosomal abnormality, including trisomy 13, 18, and 21; and (3) associations already known to have low genetic diagnostic yield, including VATER/VACTERL association and OEIS complex. An additional exclusion criterion for study purposes was (4) a contraindication for sample collection in the child or a parent.
Patient screening and informed consent
New ICU admissions were triaged daily by a research team member (C.S.G.), and clinical criteria for potential enrollees were then applied by C.S.G., P.B.A., and T.W.Y. For patients deemed eligible, the clinical team was asked to approach the family to assess parental interest in enrollment. If agreeable, the research team then set up an informational and consenting session with both parents. All patients were enrolled by either C. S.G., G.E.V., or J.A.M. While it was not necessary for enrollment to have an active genetics or metabolism inpatient consult, in practice all patients had the involvement of one or both teams. All clinical teams (including genetics, metabolism, neonatology, and neurology) were directed to continue standard of care treatment and diagnostic testing for all patients until the results of the ES were returned. There were no restrictions to or requirements of testing ordered by the clinical team. For example, chromosomal microarray testing was not required prior to enrollment.
Participating locations were Neonatal ICU, Cardiac ICU, Medical and Surgical ICU, and Intermediate Care Program (infants with an ICU indication who do not currently require advanced life support) at Boston Children’s Hospital; NICU at Brigham and Women’s Hospital (BWH); and NICU and Pediatric ICU at Massachusetts General Hospital (MGH). We have approval from the BCH institutional review board (IRB) under protocol number IRB-P00021883 with a reliance agreement from the Partners IRB, and the parents of each individual participant have signed informed consent. In one case, functional follow-up was needed, which was performed under IRB-P10–02–0053.
Rapid ES
Following informed consent, whole-blood samples were collected from probands and parents by venipuncture and shipped by courier to the sequencing provider for trio-based exome sequencing according to the timeline outlined in Fig. 1b. Patients with seizures, lethargy, biochemical abnormalities such as metabolic/lactic acidosis, cardiomyopathy, and/or ocular abnormalities such as cataracts, underwent additional mitochondrial DNA (mtDNA) sequencing including deletion/duplication testing.
Sequence analysis and interpretation
Rapid ES of DNA from the probands and both parents was performed using a CLIA-certified, commercially available clinical test (XomeDxXpress, GeneDx, Gaithersburg, MD). Exonic regions and flanking splice junctions of the genome were captured using the SureSelect Human All Exon V4 (50 Mb), the Clinical Research Exome kit (Agilent Technologies, Santa Clara, CA) or the IDT xGen Exome Research Panel v1.0. Massively parallel (next-generation) sequencing was done on an Illumina system with 100 bp or greater paired-end reads to a mean depth of 100–153× and ≥10× coverage of >90% of the targeted region. Reads were aligned to human genome build GRCh37/UCSC hg19 and variants were called using a custom-developed analysis tool, Xome-Analyzer (GeneDx). Additional sequencing and variant interpretation protocols have been previously described.15 The general assertion criteria for variant classification are publicly available on the GeneDx ClinVar submission page (https://www.ncbi.nlm.nih.gov/clinvar/submitters/26957/). All reportable variants were confirmed by an orthogonal method in the proband and both parents.
For mtDNA analysis, the entire mitochondrial genome was amplified and sequenced using next-generation sequencing. The sequence was assembled and analyzed in comparison with the revised Cambridge Reference Sequence (rCRS) and the reported variants and polymorphisms listed in the MITOMAP database (http://www.mitomap.org). A reference library of more than 6000 samples from different ethnic groups and online databases for mtDNA variations was used to evaluate variants of unknown clinical significance (VUS). The presence of a disease-associated sequence variant, if present, was confirmed by an orthogonal method in the proband and mother, if appropriate.
Return of results and data collection
Initial genetic testing results were returned verbally within ~7 days and written results were returned ~14 calendar days after sample receipt by the laboratory. Secondary findings (summarized in the American College of Medical Genetics and Genomics Secondary Findings [ACMG SF] v2.0 policy statement16) that were unrelated to the clinical presentation were not included in the analysis to focus only on results that pertained to the health of the sick infant.
Verbal and written results were relayed from the research team to the clinical team, who returned results to families. Members of the research team (C.S.G., G.E.V., or J.A.M.) participated in the result disclosure sessions to answer questions regarding the project and to observe how the ES result was used for clinical care (recording qualitative observations about how the result was shared and received, and whether genetic testing results led to any immediate clinical management decisions). Additional information about the clinical course following result disclosure was collected from the electronic medical record and the clinical teams.
Statistical analysis
STATA version 14.1 was used to calculate probability of diagnosis using Pearson chi-square test for presence of patient characteristics. A p value of 0.05 or less was deemed statistically significant.
RESULTS
Between March 2017 and November 2018, 57 neonates were identified as eligible for enrollment (Fig. 1c). Of 57 eligible families, all 57 agreed to participate in an information session with the research team, and 52 of 57 families (91%) subsequently enrolled. The BCH NICU served as the initial recruitment site beginning in March 2017, with expansion to other BCH ICUs in July 2017, and to BWH and MGH ICUs in May 2018.
Four of five families who declined cited logistical concerns (timing conflicts with clinical care), and one family cited privacy concerns. Further, two families withdrew from the study after enrollment: one because they were preparing for hospital discharge, and the other due to feeling overwhelmed with ongoing clinical concerns. A total of 50 families completed sequencing and received results per study protocol.
Characteristics of the enrolled patients are summarized in Table 1. Additional details of each case can be found in the Supplemental Materials. Forty-three of 50 probands were enrolled from neonatal intensive care units (33 BCH, 6 BWH, 4 MGH), 3 from the BCH cardiac ICU, 2 from the BCH medical or surgical ICUs, and two from the BCH intermediate care program (while awaiting transfer to an ICU). Median age of enrollment was 13 days of life (range: 3 days to 8 months [<6 months corrected for prematurity] of life). Patients were evenly divided between males and females (25 each).
Table 1.
Characteristics of sequenced patients and diagnostic yield
| Number of patients | Number of diagnoses | Diagnostic yield (%) | ||
|---|---|---|---|---|
| Overall | 50 | 29 | 58 | |
| Age at enrollment | ||||
| <7 days | 18 | 11 | 61 | |
| >7 days | 32 | 18 | 53 | |
| Sex | ||||
| Male (XY) | 25 | 14 | 56 | |
| Female (XX) | 25 | 15 | 60 | |
| Location | ||||
| NICU | 43 | 24 | 56 | |
| Other ICUs | 7 | 5 | 71 | |
| Clinical criteria | ||||
| Hypotonia | 20 | 16 | 80a | p = 0.01 |
| Seizures | 10 | 9 | 90a | p = 0.02 |
| MCA | 36 | 23 | 64 | |
| Metabolic | 16 | 8 | 50 | |
| DSD | 3 | 1 | 33 | |
| Immunodeficiency | 1 | 1 | 100 | |
| Combined criteria | ||||
| Isolated hypotonia | 1 | 1 | 100 | |
| Isolated seizures | 1 | 0 | 0 | |
| Isolated MCA | 11 | 5 | 45 | |
| Isolated metabolic | 7 | 3 | 43 | |
| Isolated DSD | 1 | 0 | 0 | |
| Hypotonia and seizures | 6 | 6 | 100a | p = 0.03 |
| Hypotonia and MCA | 15 | 12 | 80a | p = 0.04 |
| Hypotonia and metabolic | 4 | 3 | 75 | |
| Seizures and MCA | 6 | 6 | 100a | p = 0.03 |
| Seizures and metabolic | 2 | 2 | 100 | |
| MCA and metabolic | 6 | 4 | 67 | |
DSD disorders of sex development, ICU intensive care unit, MCA multiple congenital anomalies, NICU neonatal intensive care unit.
Statistically significant difference between patients with and those without characteristic on Pearson chi-square. p < 0.05 was considered significant.
The majority of enrolled patients (30/50) met more than one criterion (Table 1). Across all enrolled patients, the most common inclusion phenotype was MCA (n = 37), which most frequently included dysmorphic features, cardiac malformations, and/or brain abnormalities. The great majority of enrolled patients represented were recruited under the core phenotypic inclusion criteria (seizures, hypotonia, MCA, complex metabolic); only a single patient was enrolled solely with the expanded phenotypic eligibility criteria (case 30, a child with isolated DSD).
In 29 of 50 sequenced cases (58%), rapid genetic testing yielded a definitive and often unifying molecular diagnosis (Table 1, Supplemental Table). Of 29 diagnoses, 27 involved a previously published disease gene, and two additional patients were solved with pathogenic variants in novel disease genes that unified the clinical features observed (based on the identification of other, similar patients through GeneMatcher,17 or on existing animal model and biological functional data). The average time to verbal report was 4.9 days from sample receipt (range: 4–9 days), and the average time to written report was 10.2 days (range: 5–18 days).
Dominant de novo events were responsible for 10 of the 29 definitive diagnoses (34%), including the two novel disease genes (WDR3718 and PRKCE19). One case involved an X-linked de novo pathogenic variant (3%). Ten infants were found to have autosomal recessive conditions (34%), and four boys were hemizygous for a maternally inherited X-linked condition (14%). One, patient 19, had Prader–Willi syndrome due to maternal heterodisomy of chromosome 15 (diagnosed on ES due to missing paternal signal of chromosome 15). One child (patient 16) had a paternally inherited pathogenic variant in a maternally imprinted gene (MAGEL2). One child (patient 40) was identified to be a double carrier for an inborn error of metabolism (3-methylcrotonyl-CoA carboxylase deficiency, 3-MCC [OMIM 210200 and 210210]), with one pathogenic variant in MCCC1 and a likely pathogenic variant in MCCC2, explaining her biochemical abnormalities on neonatal screening. Finally, mtDNA analysis was performed in 16 cases, which led to a diagnosis in one patient (patient 1). Of note, all three cases found to have an underlying chromosomal abnormality (UPD15, deletion Xq26.3, deletion 3p21.2p14.2) were picked up by ES.
Of the 21 undiagnosed cases, there was one case (Supplemental Table, patient 48) in which a variant was found for which the testing laboratory and clinical team did not agree on pathogenicity (and was therefore considered not solved). There were two patients (patients 12 and 33) with one VUS in a gene associated with an autosomal recessive disorder fitting the phenotype, but no second allele was found (one family was lost to follow-up before additional studies could be done, and in the other del/dup analysis was normal, and no second allele was identified). Finally, in one case (patient 7) a maternally inherited VUS provided a possible explanation for part of the child’s clinical features: a missense variant in SOS1, a Noonan syndrome–associated gene, might account for the patient’s complex cardiac malformation, but not his additional eye and brain malformations. This patient also had a sibling with a potentially similar cardiac phenotype; unfortunately, the family was lost to follow-up before additional familial segregation could be established.
Impact of diagnostic findings
The diagnostic ES results informed medical management changes in 24 of 29 patients as described in detail below. Exceptions included one child (patient 26) whose diagnosis of MESP2-related spondylocostal dysostosis (OMIM 608681) was suspected prior to ES and was treated as such, one child (patient 3) who died prior to return of results, the abovementioned patient who is a double carrier for 3-MCC (patient 40), and the two patients with novel disease genes. Twenty-one of 24 management changes had an acute impact on care, for instance, by prompting a switch to comfort care, or revealing the need for a specialist referral to rule out additional medical complications predictable by the diagnosis with potential immediate clinical impact. Interestingly, management changes were not restricted to those patients with positive findings: in 4 of the 21 patients with nondiagnostic exome results, the lack of diagnostic findings were sufficient to prompt a decision not to pursue further diagnostic testing, thus also influencing care.
Referral to additional specialists and imaging/screening
The most common management change, occurring in 20 patients (Supplemental Table), was referral to a specialist for additional evaluation, including ophthalmology, neurology, and endocrinology specialty teams either for integrative medical decision-making and radiological diagnostic procedures, or in anticipation of future support needs, for example by referral to early intervention.
For example, patient 11 was a 6-week-old female with hypotonia, laryngomalacia, atrial septal defect (ASD), ambiguous genitalia, and renal pyelectasis who was transferred to BCH for surgical management. Exome sequencing revealed a de novo frameshift pathogenic variant in KAT6B, diagnostic of KAT6B-related disorder20 (OMIM 603736 and 606170). Based on the prognostic information provided by this diagnosis, she was consequently evaluated for hypothyroidism, gastrointestinal malformations, hip dysplasia, and hearing loss, and enrolled in early intervention.
In another example, patients 38 and 39 were a fraternal twin pair who each presented with distinct patterns of congenital malformations (ASD, ventricular septal defect [VSD], patent foramen ovale [PFO], club foot, and hydronephrosis in patient 38, and dysmorphic features, omphalocele, ambiguous genitalia, and hydronephrosis in patient 39). Both were identified to be compound heterozygous for variants of unknown significance in IFT80, a gene recently associated with a ciliopathy phenotype.21,22 Guided by this diagnosis, further medical workup was performed including a skeletal survey, ophthalmological and audiological evaluation, brain magnetic resonance image (MRI), and abdominal ultrasound.
Medication changes and new therapeutic options
In four cases, early genetic diagnoses opened up new therapeutic options for the child. In patient 14, the diagnostic finding of a pathogenic de novo missense variant in SCN2A prompted a change in anticonvulsant therapy to include phenytoin (sodium channel blockers are first-line therapies for this condition23), with subsequent clinical improvement in seizure control.
Two unrelated patients with hypotonia (patients 6 and 23) were found to have pathogenic variants in MTM1 causing X-linked myotubular myopathy (OMIM 310400). These findings allowed the care team to appropriately counsel the families about prognosis and potential therapies including an ongoing gene therapy trial ( NCT03199469). One family, in light of the prognosis,24 chose to focus on comfort care, while the other decided to continue all treatment given the possibility of future therapeutic options.
Finally, compound heterozygous pathogenic variants in DGUOK were found in a 9-day-old female (patient 13) presenting with lethargy, hypoglycemia, abnormal plasma amino acid levels, and brain MRI abnormalities. Biallelic pathogenic DGUOK variants are associated with mitochondrial DNA depletion syndrome 3 (OMIM 251880) with variable organ involvement and/or age of presentation.25 On this basis, the clinical team prepared a physician request for an emergency investigational new drug (IND) application to start nucleoside replacement therapy on compassionate use basis. Unfortunately, before permission could be granted, the patient deteriorated clinically and expired.
Comfort care
For nine children, a diagnosis helped the medical team and family adjust or adapt the patient’s clinical goals. In six of these patients, the diagnosis prompted a switch to a palliative trajectory with the goal of comfort care. In another three cases, sequencing results arrived after a clinical decision to redirect care had already been made, and the diagnosis was felt to have reassured the families about their decision. In several infants, even despite their palliative trajectory, a genetic diagnosis also allowed additional supportive measures to be instituted to preempt potential problems.
For example, patient 21 was a 7-day-old male presenting with hypotonia, apnea, and dysmorphic features after a pregnancy complicated by decreased fetal movement. He was found to have a maternally inherited 1.3-Mb deletion of the X chromosome that included the entire HPRT1 gene, which is associated with Lesch–Nyhan syndrome (OMIM 300322). A biochemical diagnosis was confirmed by HPRT1 enzyme activity measurement. Informed by the associated prognosis, nine days after result disclosure the parents decided to adjust clinical goals to a palliative trajectory, and the child died 18 days later.
Early supportive care
In three patients, genetic findings led to earlier institution of supportive care measures (tracheostomy and/or G-tube placement). For example, patient 15 was a 3-week-old female, transferred from a local hospital for workup and management of hypotonia and severe arthrogryposis. She had contractures of all four limbs and feeding difficulties. Rapid trio ES identified a de novo pathogenic variant in the BICD2 gene, associated with spinal muscular atrophy with lower extremity predominance (OMIM 618291). This diagnosis confirmed her need for tracheostomy and was pivotal in the care team’s decision-making process to place a G-tube. It also led to an adjustment of her clinical goals from home discharge to transition to a rehabilitation facility for longer-term care.
Identification of at-risk family members
In eight families, findings in the neonate had health-related implications for other family members, leading to additional genetic testing and/or clinical referrals.
In four families (patients 7, 16, 38/39 and 52) siblings had clinical features that might be related to the proband’s genetic finding. For example, patient 52 had an older brother with autism, prompting testing for the OPHN1 pathogenic variant found in the proband. In another case, patient 7 was found to have a VUS in SOS1 (p.N474H), which could contribute to the child’s complex cardiac malformation. Genetic testing on a sibling (who also had a cardiac malformation) and cardiology referral for the mother was arranged (results unfortunately not available as the family was lost to follow-up).
In three families (patients 1, 21 and 31), one or more apparently healthy parents and/or siblings underwent testing. Negative results in the healthy fraternal twin of patient 21 (Lesch–Nyhan syndrome) led to clarity and reassurance for the family. The parents of patient 31 (hypotonia, hydronephrosis, prune belly, dysmorphic features) underwent additional karyotyping to ensure neither carried the proband’s deletion-associated translocation, and the siblings of patient 1 (MT-ND3–associated Leigh syndrome) underwent mtDNA testing to assess their risk of disease.
Finally, in one case (patient 45), a proband’s genetic findings led to cardiology referral for other family members, but not genetic testing. Patient 45 (MCA including a complex cardiac malformation) had a maternally inherited nonsense variant in TNNT2 (p.W287X). While TNNT2 was not felt to be the cause of the child’s cardiac malformation, it is a cause of dilated cardiomyopathy (OMIM 601494), prompting cardiology referrals for the proband, mother, and grandparent.
Reproductive options
Finally, findings in all 17 cases involving inherited pathogenic variants prompted the clinical team to address future reproductive implications during result disclosure. Fourteen families were referred for reproductive counseling, of which at least three opted to explore preimplantation genetic diagnosis (PGD). For example, in the case of patient 3, who had 3-hydroxyisobutyryl-CoA hydrolase (HIBCH) deficiency but died before return of results, her parents elected to use PGD for their next pregnancy, and were able to have a healthy son.
DISCUSSION
In this study, we tested the utility of a phenotype-based decision algorithm to identify patients who benefit from rapid exome sequencing as a first-line test, using diagnostic yield and clinical management changes as outcome measures. A recent study suggests that sequencing of all infants admitted to the NICU without stratification may have a low diagnostic yield.14 Expert-based patient selection resulted in higher diagnostic yields;3–13 however, a widening gap between the increasing use of next-generation sequencing in clinical practice and declining numbers of clinical genetics trainees26 underscores the importance of developing and validating objective criteria for governing the use of genetic testing. These criteria may assist both specialists and nonspecialists and may decrease pressures for 24-hour consulting in these settings.
Our overall diagnostic yield (58%) was comparable with or higher than that seen in studies using expert-driven patient selection (Table 2). The greater the number of phenotypic criteria met prospectively, the more likely it was that ES would be diagnostic (Table 1). Breaking down yield by indication, the highest diagnostic yield was seen in patients with neurological manifestations. Diagnostic yields for patients presenting with either hypotonia or seizures were 80% (p = 0.01) and 90% (p = 0.02), respectively (and the diagnostic rate was 100% for six children enrolled with both symptoms [p = 0.03]) (Table 1). MCA had a diagnostic yield of 64%, and patients with complex metabolic phenotypes had a yield of 50% (both not statistically significantly different from the overall yield). The relatively lower diagnostic rate in this last category may be in part due to the fact that our definition of complex metabolic phenotype specifically excluded children with easily recognizable metabolic phenotypes and/or for whom a readily available diagnostic test was accessible. For example, a child who was admitted for workup after state-issued newborn screening was flagged for likely phenylketonuria (PKU) was deemed not eligible, and a baby boy admitted under suspicion of ornithine transcarbamylase (OTC) deficiency due to high levels of orotic acid also did not meet inclusion criteria. With respect to the phenotypic indications added after study launch (DSD, immunodeficiency, and interstitial lung disease), too few patients were enrolled to make definitive conclusions about their respective diagnostic yields (Table 1). Our diagnostic yield per indication was not dissimilar to numbers found by Willig et al.8 (using expert-based patient selection): 5 of their 9 infants with MCA received a diagnosis, 4/7 with neurological symptoms, and 2/4 with metabolic findings, and overall 20/35 (57%) received a diagnosis.
Table 2.
Rapid exome/genome sequencing studies in ICU infants
| Patients | Selection | Type of sequencing | Diagnostic yield | |
|---|---|---|---|---|
| Willig et al.8 | Child <4 months, acute illness with suspected genetic cause | By research team | Trio rapid GS | 20/35 (57%) |
| Meng et al.7 | Child <100 days, referred for exome sequencing on clinical basis | By clinical provider (retrospective analysis) | ES, trio ES, rapid trio ES | 102/278 (36.7%); rapid trio ES: 32/63 (50.8%) |
| van Diemen et al.9 | Critically ill children <1 year old in NICU or PICU with MCA or neurological symptoms | Multidisciplinary working group | Rapid GS | 7/23 (30%) |
| Petrilkin et al.11 | Level IV NICU and PICU infants aged <4 months with suspected genetic disease defined as genetic test or consult ordered, one major or three minor congenital anomalies, abnormal lab test suggesting genetic disease, or abnormal response to therapy | Screening of NICU census | Rapid GS | 22/63 (32%) |
| Farnaes et al.10 | Acutely ill infants <1 year old, nominated by clinical provider | By clinical provider (retrospective analysis) | Trio rapid GS | 18/42 (43%) |
| Mestek-Boukhibar et al.13 | PICU and CICU patients | By multidisciplinary research team | Trio rapid GS with phased analysis | 10/24 (42%) |
| French et al.5 | NICU and PICU patients with possible single-gene disorder with congenital anomalies, neurological symptoms, suspected metabolic disease, surgical necrotizing enterocolitis, extreme IUGR or failure to thrive, unexplained critical illness, or at clinician’s request | Clinician/research team | Trio rapid GS | 40/195 (21%) |
| Eliott et al.12 | NICU neonates with unexplained seizures, metabolic disturbances, neurological abnormalities or depressed level of consciousness, MCA or significant physiological disturbance in keeping with genetic disorder for which diagnosis would likely change clinical manaqement of qenetic counselinq | Geneticist and research team | Trio rapid ES | 15/25 (60%) |
CICU cardiac intensive care unit, ES exome sequencing, GS genome sequencing IUGR, intrauterine growth restriction, MCA multiple congenital anomalies, NICU neonatal intensive care unit, PICU pediatric intensive care unit.
In some cases, even nondiagnostic exome sequencing results influenced clinical management, especially in patients undergoing workup for complex metabolic phenotypes. Clinicians often prioritized collecting blood samples for rapid ES (requiring 1 mL of blood) over sample collection for more extensive metabolic testing (requiring up to 15 mL, a large amount especially for premature babies). In some cases, negative ES results were used to spare neonates from metabolic tests that were deemed too unlikely. While out of scope for this report, this observation deserves further follow-up since typical metabolic workup algorithms (blood draws, muscle biopsies, gene panels) are often associated with significant time and cost.
The uptake of this study among families approached for enrollment was very high (52/57, 91%) and the dropout rate was very low (2/52, 4%), consistent with strong parental interest in diagnostic genetic testing in NICU patients. This is in contrast to a study randomizing all NICU patients to conventional exome sequencing (i.e., without rapid turnaround), where only 45 of 432 approached families in ICU settings elected to enroll.27 We interpret this to reflect evolving attitudes toward genetic testing, lack of randomization in our study, and expectations that rapid identification of a genetic diagnosis may aid in the child’s care. Over the course of the study, we also anecdotally observed a shift in the attitude of physicians and other caregivers towards rapid ES, from some initial skepticism regarding utility and concerns about potential risks, to acceptance and strong interest after several diagnoses were made.
While our study focused on the utility of rapid ES as a simple first-line test, it may be reasonable to ask what proportion of diagnoses achieved could have been arrived at by other methods. To address this, we analyzed our 16 hypotonia patients for whom rapid ES arrived at a diagnosis, and compared their findings with genes covered on the October 2019 version of GeneDx’s Congenital Hypotonia Xpanded panel. This is an extensive panel covering over 1400 genes (i.e., 30% of the ~4600 known disease-associated genes). Despite that, 2–4 (12.5–25%) of our patients would not have been picked up by panel testing alone. The first two (patient 21 and patient 31) had a copy-number variant that was picked up on exome analysis, but may not have necessarily been detected on the panel. The third patient had a variant in a mtDNA gene, which would not have been part of the panel. The fourth patient had a variant in a novel gene (PRKCE). Future studies will be needed to evaluate cost-effectiveness.
Unsurprisingly given the neonatal setting, many of the diagnoses achieved via this study represent cases that are among the youngest reported in the literature (Table 2). Diagnosed patients also tended to have more severe phenotypes than typically reported, especially for metabolic and neuromuscular disorders. Consistent with the severity of these presentations, the mortality rate in our cohort was 26% (13 of 50 sequenced patients), including 10 of the 30 patients who received a diagnosis (33%). For several genes, diagnosed cases also represented expansions of known phenotypes (Table 3). For example, patient 3 (HIBCH deficiency) presented with apneic episodes and seizures early in life, compared with more subtle initial presentations (e.g., failure to thrive at 3 months or older) in other reported cases.28
Table 3.
Unusual phenotypes identified in the course of this study
| Phenotypic expansions | ||||||
|---|---|---|---|---|---|---|
| Patient | Gender | Age at enrollment | Test result | Diagnosis | Additional features | Literature |
| 3 | F | 3 weeks | HIBCH: c.852delS (p.L284 FfsX10)/c.488G>T (p.C163F) | HIBCH deficiency | Severe clinical course with recurring apnea | Loupatty et al.28 |
| 14 | M | 6 weeks | SCN2A: c.4919T>A (p.I1640N) | SCN2A-related disorder | Polymicrogyria | - |
| 28 | F | 2 months | POMGNT2: homozygous c.1234G>A (p.G412R) | Walker-Warburg syndrome | Cardiac defect, bilateral cataracts | Manzini et al.34 |
| 40 | F | 3 weeks | MCCC1: c.1155A>C (p.R385A)IMCCC2: (c.463C>T (p.R155W) | 3-methylcrotonyl-CoA carboxylase deficiency | Double carrier 3-MCC with biochemical phenotype | - |
| 47 | F | 6 months | AHI1: homozygous c.2755G>A (p.D919N) | AHI1-related disorder (ciliopathy) | No molar tooth, cardiac defect | Valente et al.35 |
| Earliest onset | ||||||
| Patient | Gender | Age at enrollment | Test result | Diagnosis | Typical age at presentation | Literature |
| 3 | F | 3 weeks | HIBCH: c.852delS (p.L284 FfsX10)/c.488G>T (p.C163F) | HIBCH deficiency | 3–7 months | Loupatty et al.28 |
| 9 | F | 7 days | PACS1: c.607C>T (p.R203W) | PACS1 related disorder | Late infancy | Schuurs-Hoeijmakers et al.36 |
| 21 | M | 5 days | Deletion Xq26.3(133605263_134910134) | Lesch-Nyhan syndrome | 3–6 months | Liu et al.37 |
| 38 | F | 5 days | IFT80: C.869A >G (p.N290S)/c.1936G >C (p.V646L) | Short-rib thoracic dysplasia 2/ciliopathy | Embryonic lethal in severe form, early childhood in ciliopathy phenotype | Moran et al.21 Bizaoui et al.22 |
| 39 | M | 5 days | IFT80: C.869A >G (p.N290S)/c.1936G>C (p.V646L) | |||
Diagnoses were elusive in some cases despite striking phenotypes. Patient 42, for example, had persistent metabolic acidosis, hypertrophic cardiomyopathy, and consanguineous parentage, yet no definitive diagnosis could be found. These patients may benefit from GS,8,29–31 RNA sequencing,32 and/or future exome reanalysis.33 These techniques offer interesting avenues for future research and clinical use.
In conclusion, we find our phenotype-driven protocol for rapid ES in the neonatal ICU to be associated with a high diagnostic yield (58%), and enabling of tangible management changes in patients and family members. Dissemination of such protocols in the NICU may allow for faster and more accurate diagnoses, reduction of unnecessary testing and procedures, and improvements in medical care.
Supplementary Material
ACKNOWLEDGEMENTS
The research team thanks all clinical teams involved for their help, as well as the patients and their families. This work was supported in part by a sponsored research agreement between GeneDx and Boston Children’s Hospital; the National Institutes of Health under award numbers U19HD077671, U54HD090255, R01AR068429, and R01HD075802; the William F. Milton Fund; and the resources of The Manton Center for Orphan Disease Research Gene Discovery Core. C.S.G. was partly supported by a William Randolph Hearst Fellowship. M.H.W. was supported by T32 GM 7748-40.
Footnotes
SUPPLEMENTARY INFORMATION
The online version of this article (https://doi.org/10.1038/s41436-019-0708-6) contains supplementary material, which is available to authorized users.
DISCLOSURE
Rapid exome sequencing for this study was performed by GeneDx, a commercial genetic diagnostic company. Authors D.C., S.Y., and J.J. are employed by GeneDx. The other authors declare no conflicts of interest.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
REFERENCES
- 1.Yoon PW, Olney RS, Khoury MJ, Sappenfield WM, Chavez GF, Taylor D. Contribution of birth defects and genetic diseases to pediatric hospitalizations. Arch Pediatr Adolesc Med. 1997;151:1096. [DOI] [PubMed] [Google Scholar]
- 2.Wojcik MH, Schwartz TS, Yamin I, et al. Genetic disorders and mortality in infancy and early childhood: delayed diagnoses and missed opportunities. Genet Med. 2018;20:1396–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iglesias A, Anyane-Yeboa K, Wynn J, et al. The usefulness of whole-exome sequencing in routine clinical practice. Genet Med. 2014;16:922–931. [DOI] [PubMed] [Google Scholar]
- 4.Daoud H, Luco SM, Li R, et al. Next-generation sequencing for diagnosis of rare diseases in the neonatal intensive care unit. Can Med Assoc J. 2016;188:E254–E260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.French CE, Delon I, Dolling H, et al. Whole genome sequencing reveals that genetic conditions are frequent in intensively ill children. Intensive Care Med. 2019;45:627–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stark Z, Schofield D, Alam K, et al. Prospective comparison of the cost-effectiveness of clinical whole-exome sequencing with that of usual care overwhelmingly supports early use and reimbursement. Genet Med. 2017;19:867–874. [DOI] [PubMed] [Google Scholar]
- 7.Meng L, Pammi M, Saronwala A, et al. Use of exome sequencing for infants in intensive care units. JAMA Pediatr. 2017;171:e173438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willig LK, Petrikin JE, Smith LD, et al. Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: a retrospective analysis of diagnostic and clinical findings. Lancet Respir Med. 2015;3:377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Diemen CC, Kerstjens-Frederikse WS, Bergman KA, et al. Rapid targeted genomics in critically ill newborns. Pediatrics. 2017;140: e20162854. [DOI] [PubMed] [Google Scholar]
- 10.Farnaes L, Hildreth A, Sweeney NM, et al. Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genomic Med. 2018;3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrikin JE, Willig LK, Smith LD, Kingsmore SF. Rapid whole genome sequencing and precision neonatology. Semin Perinatol. 2015;39:623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott AM, du Souich C, Lehman A, et al. RAPIDOMICS: rapid genome-wide sequencing in a neonatal intensive care unit—successes and challenges. Eur J Pediatr. 2019;178:1207–1218. [DOI] [PubMed] [Google Scholar]
- 13.Mestek-Boukhibar L, Clement E, Jones WD, et al. Rapid Paediatric Sequencing (RaPS): comprehensive real-life workflow for rapid diagnosis of critically ill children. J Med Genet. 2018;55:721–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ceyhan-Birsoy O, Ceyhan-Birsoy O, Murry JB, et al. Interpretation of genomic sequencing results in healthy and ill newborns: results from the BabySeq Project. Am J Hum Genet. 2019;104:76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Retterer K, Juusola J, Cho MT, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2016;18:696–704. [DOI] [PubMed] [Google Scholar]
- 16.Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:249–255. [DOI] [PubMed] [Google Scholar]
- 17.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanca O, Andrews JC, Lee P, et al. De novo variants in WDR37 are associated with epilepsy, colobomas, dysmorphism, developmental delay, intellectual disability, and cerebellar hypoplasia. Am J Hum Genet. 2019;105:413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alcantara D, Elmslie F, Tetreault M, et al. SHORT syndrome due to a novel de novo mutation in PRKCE (Protein Kinase Cɛ) impairing TORC2-dependent AKT activation. Hum Mol Genet. 2017;26:3713–3721. [DOI] [PubMed] [Google Scholar]
- 20.Marangi G, Di Giacomo MC, Lattante S, et al. A novel truncating variant within exon 7 of KAT6B associated with features of both Say-Barber-Biesecker-Young-Simpson syndrome and genitopatellar syndrome: further evidence of a continuum in the clinical spectrum of KAT6B-related disorders. Am J Med Genet Part A. 2018;176:455–459. [DOI] [PubMed] [Google Scholar]
- 21.Moran J,G Sanderson K, Maynes J, et al. IFT80 mutations cause a novel complex ciliopathy phenotype with retinal degeneration. Clin Genet. 2018;94:368–372. [DOI] [PubMed] [Google Scholar]
- 22.Bizaoui V, Huber C, Kohaut E, et al. Mutations in IFT80 cause SRPS type IV. Report of two families and review. Am J Med Genet Part A. 2019;179:639–644. [DOI] [PubMed] [Google Scholar]
- 23.Dilena R, Striano P, Gennaro E, et al. Efficacy of sodium channel blockers in SCN2A early infantile epileptic encephalopathy. Brain Dev. 2017;39:345–348. [DOI] [PubMed] [Google Scholar]
- 24.Beggs AH, Byrne BJ, De Chastonay S, et al. A multicenter, retrospective medical record review of X‐linked myotubular myopathy: the recensus study. Muscle Nerve. 2018;57:550–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Hattab AW, Craigen WJ, Scaglia F. Mitochondrial DNA maintenance defects. Biochim Biophys Acta Mol Basis Dis. 2017;1863:1539–1555. [DOI] [PubMed] [Google Scholar]
- 26.Maiese DR, Keehn A, Lyon M, Flannery D, Watson M. Current conditions in medical genetics practice. Genet Med. 2019;21:1874–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genetti CA, Schwartz TS, Robinson JO, et al. Parental interest in genomic sequencing of newborns: enrollment experience from the BabySeq Project. Genet Med. 2019;21:622–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loupatty FJ, Clayton PT, Ruiter JPN, et al. Mutations in the gene encoding 3-hydroxyisobutyryl-CoA hydrolase results in progressive infantile neurodegeneration. Am J Hum Genet. 2007;80:195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller NA, Farrow EG, Gibson M, et al. A 26-hour system of highly sensitive whole genome sequencing for emergency management of genetic diseases. Genome Med. 2015;7:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petrikin JE, Cakici JA, Clark MM, et al. The NSIGHT1-randomized controlled trial: rapid whole-genome sequencing for accelerated etiologic diagnosis in critically ill infants. NPJ Genomic Med. 2018;3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saunders CJ, Miller NA, Soden SE, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012;4:154ra135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonorazky HD, Naumenko S, Ramani AK, et al. Expanding the boundaries of RNA sequencing as a diagnostic tool for rare Mendelian disease. Am J Hum Genet. 2019;104:466–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitz-Abe K, Li Q, Rosen SM, et al. Unique bioinformatic approach and comprehensive reanalysis improve diagnostic yield of clinical exomes. Eur J Hum Genet. 2019. April 12; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manzini MC, Tambunan DE, Hill RS, et al. Exome sequencing and functional validation in zebrafish identify GTDC2 mutations as a cause of Walker-Warburg syndrome. Am J Hum Genet. 2012;91:541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valente EM, Brancati F, Silhavy JL, et al. AHI1 gene mutations cause specific forms of Joubert syndrome–related disorders. Ann Neurol. 2006;59:1527–34. [DOI] [PubMed] [Google Scholar]
- 36.Schuurs-Hoeijmakers JHM, Landsverk ML, Foulds N, et al. Clinical delineation of the PACS1 -related syndrome—report on 19 patients. Am J Med Genet Part A. 2016;170:670–675. [DOI] [PubMed] [Google Scholar]
- 37.Liu N, Zhuo Z-H, Wang H-L, et al. Prenatal diagnosis based on HPRT1 gene mutation in a Lesch–Nyhan family. J Obstet Gynaecol. 2015;35:490–493. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.