Abstract
The present study was undertaken to confirm the genetic identity of Clostridium perfringens isolates from domestic livestock in Saudi Arabia and to characterize the genes encoding to alpha, beta, epsilon, and iota (α-, β-, ε-, and ι-) toxins. C. perfringens were confirmed in 104 out of 136 isolates on multiplex polymerase chain reaction using specific primers amplifying genes related to toxins produced by C. perfringens. Genes encoding α-toxins were detected in 104 samples. Of the isolates, 80.8% were diagnosed as type A, 15.4% as type D, 2.9% as type C, and 0.96% as type B. None of the isolates has genes encoding iota (ι-) toxin. All isolates investigated yielded enterotoxin (cpe) products and none yielded β2 (cpb2-toxin) or NetB products. PLC gene sequences encoding α-toxin showed >96.7% similarity. Isolates which had α-toxins as well as enterotoxin (cpe) are regarded as type F. Phylogenetic analysis using maximum likelihood analysis yielded two clades, and the majority of the isolates were in one group while only two isolates clustered on the second clade. Within the Kingdom of Saudi Arabia strains, 54 variable positions and 23 polymorphic amino acids were noticed. Isolates with ε- and β-toxins were variable and were found to be close to those published for C. perfringens. ETX gene sequences encoding ε-toxins were found to be related to CPE sequences.
1. Introduction
The anaerobe and spore-forming Clostridium perfringens is a Gram-positive rod-shaped bacterium detected in soil and water as well as among intestinal bacterial flora of animals and man [1]. It is categorized with the most widespread bacteria, which occurs in different environments such as soil, sewage, food, feces, and the normal intestinal flora of humans and animals [2]. The toxins produced by different species of Clostridium are more than those produced by other eubacteria [3]. All strains of C. perfringens possess the gene encoding for phospholipase C (plc) (also referred to as alpha toxin α) in combination with differential expression of 3 major toxin-encoding genes (beta β, epsilon ε, and iota ι) used to classify strains as toxinotypes A to E [4]. C. perfringens isolates from type A produce only α-toxin, those from type B produce α-, β-, and ε-toxins, those from type C secrete α- and β-toxins, those from type D produce α- and ε-toxins, and bacteria from type E produce α- and ι-toxins [5]. C. perfringens is a human and livestock pathogen, resulting in disease in different tissues as well as intestinal diseases in the form of enterotoxaemia. C. perfringens type A toxigenic group causes food poisoning in humans and enteritis in domestic and wildlife animal species, type B and C cause necrotizing enteritis and enterotoxaemia in cattle, sheep, and other animals, type D affects sheep, goats, and cattle mainly, and type E causes enteritis in rabbits, sheep, and cattle [6, 7]. Recent results by Rood and others suggested a modification for the toxinotyping of C. perfringens which required the inclusion of the enterotoxin (CPE) and NetB toxins produced by the bacterium [8]. They proposed that strains which produce α-toxin as well as CPE are assigned type F whereas those producing α-toxin and NetB toxins are assigned type G. In both cases, no other toxins such as β-, ε-, and ι-toxins are produced.
The prevalence of C. perfringens in Saudi Arabia was studied by some investigators [9, 10]. In 2006, during the annual Hajj pilgrimage, a group of Saudi soldiers were diagnosed for having gastroenteritis following a rice lunch contaminated with Bacillus cereus and C. perfringens [9]. Moussa and Hessan [10] detected C. perfringens in 20% of the calves, sheep, and poultry investigated. C. perfringens was detected in the dromedary camel in the eastern region of Saudi Arabia, where a prevalence of 23.4% was reported from the samples investigated [11].
Diagnosis of C. perfringens enterotoxaemia in various animal species is dependent on clinical signs as well as pathological findings; however, characterization of toxins in intestinal contents is of importance to confirm the clinical diagnosis.
The widely used identification of the toxins is based on mice neutralization test, but this method is expensive, time-consuming, and subjecting laboratory animals to stress and unnecessary risk of being affected by disease [12]. Enzyme-linked immunosorbent assay (ELISA) kits have also been used for the detection of clostridial toxins [13, 14]. DNA-based techniques, such as the polymerase chain reaction (PCR) was developed for C. perfringens genotyping which is a reliable alternative method to replace testing in laboratory animals. Different PCR protocols were employed to genotype C. perfringens isolates with respect to the genes cpa, cpb, etx, iap, cpe, NetB, and cpb2, which encode the α-, β-, ε-, ι-toxins, CPE, NetB, and β2-toxins, respectively [4, 15].
In the present study, C. perfringens isolated from livestock in Saudi Arabia has been characterized and the sequence variation in α-, ε-, and β-toxins producing isolates has been investigated.
2. Materials and Methods
Isolates of C. perfringens were obtained from field investigations for the diagnosis of enterotoxaemia in domestic livestock in Saudi Arabia. Intestinal swabs and intestinal contents were taken to the laboratory of the Ministry of Environment, Water and Agriculture (MEWA), KSA. In the laboratory, samples were subjected to microscopic examination using Gram stain as well as to serological testing. Representative isolates were subjected to DNA extraction, polymerase chain reaction, and DNA sequencing.
2.1. Microscopic Examination of Gram-Stained Smears
Impression smears were prepared from parts of the small intestines and smears were also prepared from suspected bacterial isolates. Smears were air-dried, heat fixed, stained with Gram stain, and examined microscopically for detection of large-sized Gram-positive bacilli similar to clostridia.
2.2. Detection and Typing of C. perfringens by ELISA Assay
The Bio-X enterotoxaemia ELISA kit (Bio-X Diagnostics, Belgium) was used to detect α-, β-, and ε-toxins, cellular antigens of C. perfringens in clinical samples, and bacterial cultures according to the manufacturer's instructions. Typing of C. perfringens isolates into the different types A, B, C, or D was done by comparing the types of toxins detected in the clinical material or culture supernatants of pure colonies of C. perfringens isolates grown in the liquid Trypticase Glucose Yeast (TGY) extract medium. The TGY medium is composed of Tryptone, yeast extract, glucose, K2HPO4, agar, and water.
2.3. Bacterial DNA Extraction, Polymerase Chain Reaction (PCR), DNA Sequencing, and Phylogenetic Analyses
DNA was extracted from 136 supposedly C. perfringens isolates based on the microscopic examination, using the QuickExtract™ Bacterial DNA Kit from Epicentre® (an Illumina® Company). Polymerase chain reaction (PCR) and multiplex PCR were performed using the primers illustrated in Table 1. The plc gene was PCR amplified from extracted DNA or cDNA using the primer pair designed by Baums and others [15]. Amplifying NetB gene was performed using primers APK78-F and APK79-R as indicated by Keyburn and others [16].
Table 1.
Primers used for amplification of the genes responsible of producing specific toxins and the expected amplicon size together with the annealing temperature for the PCR.
| Gene | Primer sequence 5′ to 3′ | Amplicon size (bp) | Annealing temp (°C) |
|---|---|---|---|
| Plc, cpa (α-toxin) | CPA5L: AGTCTACGCTTGGGATGGAA CPA5R: TTTCCTGGGTTGTCCATTTC |
900 | 55 |
|
| |||
| cpb (β-toxin) | CPBL: TCCTTTCTTGAGGGAGGATAAA CPBR: TGAACCTCCTATTTTGTATCCCA |
612 | 55 |
|
| |||
| etx (ε-toxin) | CPETXL: GGGGAACCCTCAGTAGTTTCA CPETXR: ACCAGCTGGATTTGAGTTTAATG |
396 | 55 |
|
| |||
| ipa (ι-toxin) | CPIL: AAACGCATTAAAGCTCACACC CPIR: CTGCATACCCTGGAATGGCT |
293 | 55 |
|
| |||
| Cpe (CPE) | CPEF: GGAGATGGTTGGATATTAGG CPER: GGACCAGCAGTTGTAGATA |
233 | 56 |
|
| |||
| cpb2 (β2-toxin) | CPBL: CAAGCAATTGGGGGAGTTTA CPBR: GCAGAATCAGGATTTTGACCA |
200 | 55 |
|
| |||
| NetB toxin | AKP78: GCTGGTGCTGGAATAAATGC AKP79: TCGCCATTGAGTAGTTTCCC |
560 | 55-58 |
The plc (cpa), cpb, ext, ipa, cpe, NetB, and cpb2 genes were amplified in 25 μl volumes containing 5 μl PCR buffer, 1 pmol each specific primer, 200 mM each dNTP, 4 U MyTaq DNA polymerase (Bioline, London, UK), and 2 μl of DNA extract. Thermal cycling conditions were as follows: 2 min at 94°C, followed by 35 cycles of 94°C for 30 sec, 55°C (up to 58°C depending on the primer) for 30 sec, 72°C for 30 sec, and a final extension at 72°C for 5 min. The PCR products were visualized using stained ethidium bromide 1.5% agarose gel exposed to ultraviolet light, and digital images were taken from the PCR product through gel documentation system. Positive PCR products obtained were purified using the QIAquick kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions.
Purified PCR products were sequenced using the Macrogen facility, Seoul, South Korea. The plc gene sequences were manually aligned according to their nucleotides and the deduced amino acid sequences using MEGA X, and all alignments were refined by manual inspection.
Phylogenetic analyses were conducted using the computer program MEGA X [17]. The best-fit substitution model was TN93+G+I. The statistical reliability of internal branches was assessed from 1000 bootstrap pseudoreplicates. Phylogenetic trees were generated in MEGA X using the maximum likelihood (ML) approach.
The deduced amino acids from the coding regions were obtained using MEGA X. Then, they were compared with the existing sequences in the NCBI Protein database to identify homologous sequences through PSI-BLAST. Sequences from different strains were aligned with the ClustalW program using the MAFFT, a multiple sequence alignment program [18]. The secondary structure of plc (α-helix and β-sheet) was predicted using the PDBSAS server [19]. Relevant annotations were mapped using a consensus sequence recovered from the PDB server. Phylogenetic trees were generated using the maximum likelihood (ML) analysis in MEGA X.
2.4. Nucleotide Sequence Accession Numbers
The plc gene sequences (α-toxins) have been deposited in the GenBank database under accession numbers (48 sequences) MN646319 to MN646366. etx gene sequences (13 sequences) accession numbers are from MN649858 to MN649870, cpb gene sequences (4 sequences) accession numbers are from MN683524 to MN68352527, and cpe gene sequences (40 sequences) are from MN683528 to MN683567.
3. Results
Clinical samples collected for bacteriological identification yielded C. perfringens based on cultural, microscopic, and cellular antigenic characterizations.
Molecular characterization of 136 isolates showed that 104 isolates yielded positive PCR products. Of those 138 isolates, 84 (80.8%) had only the plc gene (type A), 16 (15.4%) isolates had the etx gene as well as the plc gene (type D), 3 (2.9%) isolates had the cpb gene as well as the plc gene (type C), and 1 (0.96%) isolate had the plc, cpb, and etx genes (type B). All isolates investigated in the present study had the cpe gene; however, none had iap, cpb2, or NetB genes.
Figure 1 shows representative isolates which showed positive PCR products in a multiplex PCR. The expected sizes of the PCR product from different primers are shown from representative samples.
Figure 1.
A multiplex PCR typing of selected Clostridium perfringens isolates. Lane 1: empty lane, all the 18 isolates showed the PCR product resulting from amplification of plc (cpa) gene showing a fragment size of 900 bp. Lanes 4-7 produced etx gene (a fragment size of (396 bp) in addition to plc gene. Lanes 12 and 14 produced cpb gene (a fragment size of 612 bp) in addition to the plc gene. Lanes 2, 3, 8-11, 13, 15-19 produced only the plc gene. Lane 20: Bioline EasyLadder 1.
3.1. Alpha (α-plc) Gene Characterization
A total of 48 sequences from alpha toxins were analyzed. Sequences of alpha toxins obtained in the present study were highly similar, and they showed a similarity of >96.7%. There was no specific sequence type associated with a known location. The sequences investigated were from 5 different cities including Riyadh, Hofuf, Jeddah, Taif, and Qassim. The dataset used in the analysis included 104 sequences. Of those, 48 were new sequences (present study), 22 were published from animals and humans in the USA [20], 32 strains were isolated from Danish broiler chickens grouped according to pulsed-field gel electrophoresis (PFGE) profiles [21], one unpublished sequence (MK599266) was from camel, and one was out-group Clostridium novyi (D32125).
The ML topology showed two major clades with high bootstrap values, the first is divided into 10 subclades including 46 of the isolates. The most divergent isolates, 1 and 7, were localized in the second clade (Figure 2). The strain MK599266 grouped with strains from Hofuf and Riyadh.
Figure 2.
Maximum likelihood phylogenetic tree showing the relationship of the plc gene from 48 isolates reported in the present study with sequences from the GenBank. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Only significant bootstrap values are shown. This analysis involved 102 nucleotide sequences. There was a total of 842 positions in the final dataset. Evolutionary analyses were performed in MEGA X.
The plc gene was reported to be highly conserved in C. perfringens strains. We observed 275 and 47 variable positions, respectively, out of the 860 nucleotides and 272 deduced amino acids. The studied protein-coding region displayed moderate-level nucleotide diversity (Pi = 0.019). The dataset generated 74 haplotypes characterized by a high haplotype diversity (Hd = 0.987). Within KSA samples, we observed 54 variable positions and 23 polymorphic amino acids (Figure 3).
Figure 3.
Variable amino acid positions of the defined alpha toxin sequence types from USA, Denmark, and KSA (this study). Identical amino acids sites are shaded.
α-Gene sequences obtained in the present study were compared to similar sequences obtained from the GenBank. The sequences from the Kingdom of Saudi Arabia were scattered and clustered with sequences obtained from Denmark and USA (Figure 2). Fourteen sequences were clustered in one group with some other sequences from the GenBank while other sequences were scattered between other sequences. Protein sequences differed in 22 positions between isolates from Saudi Arabia, and they showed 31 different types. They showed differences in 41 amino acid position when compared with isolates from USA and Denmark; however, some of them have shown similarities (Figure 3). The sequence variation was not related to the pathogenicity of the strain as all organisms investigated in this study were obtained from pathological cases.
The secondary structure prediction revealed that plc amino acid sequences of α-toxin mainly consists of alpha-helices (Figure 4). The α-toxin enzyme contains an α-helical N-terminal domain (residues 1–246) encompassing the active site and a β-sandwich C-terminal domain (residues 247–255). The β-sandwich domain includes a calcium-binding pocket (Figure 4). The Protein Data Bank (PDB) multiple alignment confirmed the structural composition and showed 9 proteins with identity percentage varying between 95.4% and 98.9%, all α-toxin belonging to C. perfringens. Clostridium absonum α-toxin showed only 62% identity compared to those obtained from C. perfringens.
Figure 4.
Alpha toxin sequence annotation by structure SAS (sequence annotated by structure, EMBL-EBI) against all the proteins of known 3D structure in the Protein Data Bank (PDB code of the protein presenting the highest % of identity: 1qmd, 1qm6, 1sb4, and 1ca1).
3.2. Epsilon (ε-) Gene Characterization
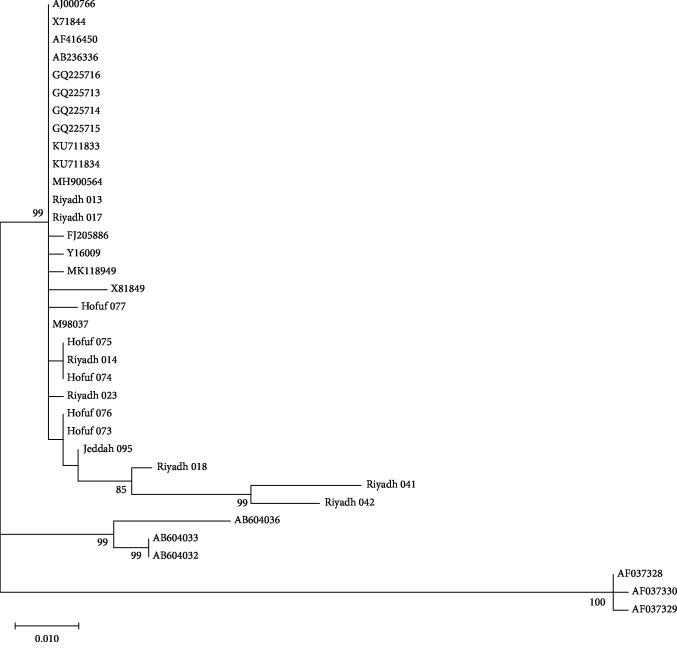
The dataset included 33 sequences of ε-gene, 13 new sequences (KSA) and 20 published previously (retail meat products, Japan; calves, lamb, human, and food, USA; and foal UK). We observed 69 variable sites and 48 were parsimony informative sites. We observed 32 variable positions out of the 146 deduced amino acids. The maximum likelihood (ML) tree generated revealed that ε-gene sequences from Saudi Arabia grouped with sequences obtained from cpe genes from different animal species and from humans (Figure 5).
Figure 5.
Maximum likelihood phylogenetic tree showing the relationship of the epsilon toxin-producing genes from 13 sequences reported from C. perfringens isolates in the present study with related sequences from the GenBank. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Only significant bootstrap values are shown. This analysis involved 35 nucleotide sequences. There was a total of 438 positions in the final dataset. Evolutionary analyses were performed in MEGA X.
3.3. Beta (β-) Gene Characterization
The dataset included 16 sequences comprising 4 new and 12 reference sequences published previously (HQ111476-82 and KU836730 from Iran, L13198 from the United Kingdom, and X83275 from Iceland). Annotation of the obtained sequence revealed that the region comprising between 63 and 68 corresponds to a regulatory class=“ribosome binding site.” The fragment from the position 76 to 560 corresponds to a partial product of the β-toxin. Multiple alignments of cpb gene sequences yielded 21 variable sites, and only three were in the coding region.
Homology of β-toxin with other proteins: the deduced partial β-toxin amino acid sequence was screened against the Swiss-Prot. A significant homology was found with the deduced amino acid sequence of Staphylococcus aureus α-toxin (sequence ID: P09616, 26% identities, 47% positives substitutions), ϒ-toxin component B (sequence ID: Q6GE12, 25% identities, 44% positives substitutions), and leukocidin-F (sequence ID: P31715, 25% identities, 44% positives substitutions). The sizes of these proteins vary between 319 and 325 amino acids.
3.4. Enterotoxin (cpe) Production
All isolates showed positive results for the C. perfringens enterotoxin (cpe). Different haplotypes were reported; however, the majority of the haplotypes were identical to those reported for C. perfringens available in the GenBank.
4. Discussion
The identity of Clostridium perfringens isolated from domestic livestock in Saudi Arabia has been confirmed using molecular techniques. Furthermore, different genes responsible of excreting each toxin have been identified and characterized. C. perfringens type A has been reported in 80.8% of the bacterial isolates, type D was reported in 15.4%, type C was isolated from 2.9%, and type B was reported in 0.98%. This finding was similar to a previous work which recorded the four types of C. perfringens reported in this study but with variable rates of prevalence [11]. They reported C. perfringens type A in 33.3%, type B in 5.3%, and types C and D both in 5.3% of the camel investigated. On an earlier study, C. perfringens types A in 81.5%, B in 3.7%, C in 3.7%, and D in 11.1% of sheep, goats, and chicken investigated in Riyadh region were reported [10]. In both studies, the neutralization test in mice were used and they did not include either cpe or NetB gene in their studies. Organisms which were recognized, in the present study, as type A may be designated as type F rather than type A as all produced CPE toxin together with α-toxin according to the suggestion made by Rood and others [8]. We were able to demonstrate the presence of C. perfringens type F for the first time in Saudi Arabia. Results of the present study indicate that C. perfringens type A (or type F) is the most prevalent type, and these results were also similar to those found in previous studies [22–27]. The gene coding to α-toxin in Saudi Arabia was found to show variability, and it is interesting that some of the strains are related to strains isolated from Japan and USA [20, 21]. Furthermore, strains which were reported from Riyadh clustered with those obtained from Taif, Jeddah, and Qassim. Such finding can be attributed to the fact that animal movement in Saudi Arabia is inevitable and Riyadh is a big market for livestock; therefore, animals are transported between different cities, and hence, bacterial infection can easily be found in various places.
Epsilon-coding gene sequences were found to be related to cpe genes produced by type C and D isolates from different animal species in the study conducted by some investigators [28, 29]. CPE loci were found to be divergent in C. perfringens isolates. All cpe-positive type D isolates studied previously were found to carry both cpe and etx genes on the same plasmid [28, 30]. It is likely the case of having etx grouped with cpe in the current study. Such observations may indicate that a similar genetic element found in the plasmid has mobilized the conserved cpe locus from a progenitor cpe-carrying plasmid found in type D isolates into other plasmids in the same isolate. This finding requires further investigations to clarify it.
In the present study, enterotoxin gene (cpe) was reported in all bacterial isolates while the cbp2 gene was not detected in any of the isolates. Unlike what we found in our investigation, where β2-toxin was reported from all isolates, they were obtained from the dromedary camel [11]. β2-Toxin toxin-producing C. perfringens type A has been associated with disease in several animal species, such as piglets suffering from hemorrhagic enteritis in 1997, and it has been reported to possess a cytological effect on certain cell lines and is lethal to mice [31]. The β2-toxin is being associated mainly with type A but has also been identified in association with types C and E in piglets and calves, respectively [32]. Similar findings by other investigators have also indicated that none of the isolates studied was positive to specific sequences of the gene coding the enterotoxin production (cpe) [33, 34]. The enterotoxin (cpe) plays an important role in the development of intestinal disease in many animal species including man [7, 35]. Tight junction components between the intestinal cells which regulate the permeability of ions and macromolecules are included in the smaller and larger protein complexes which cause paracellular permeability changes [36, 37]. The mechanism of action of enterotoxin is binding to the intestinal epithelial cells forming a series of protein complexes in cell membranes, which ultimately results in altering membrane permeability and cell lysis. β2-Toxin was found to be present among cpe-positive C. perfringens type A isolates from humans with antibiotic-associated diarrhea. Of 35 cpb2-harbouring enterotoxigenic C. perfringens type A strains, production of β2-toxin was produced in 34 isolates in vitro [38]. The presence of cpb2 was less common among C. perfringens strains isolated from humans suffering from food poisoning [38].
Genotypic differences exist between cpe type A-positive isolates causing food poisoning and those causing nonfoodborne gastrointestinal disease [39]. It has been demonstrated that most, if not all, C. perfringens type A food poisoning isolates carry their cpe gene on the chromosome while most, if not all, CPE-associated nonfoodborne human gastrointestinal diseases carry a plasmid cpe gene [40, 41]. CPE sequences reported in the present study showed high similarity to those described from the GenBank, and they are of plasmid origin. This suggested that the cpe sequences reported from all α-toxin-producing strains are probably of plasmid origin and not of chromosomal origin.
Overall, the mutations that were found in the amino acid sequences of α-toxin did not significantly alter the predicted structure of the proteins. The studied isolates of plc showed high sequence identity (>95%) to PDB Clostridium strain. None of the detected amino acid mutations occurred in the active site or in the partial C-terminal domain (Figure 2). These regions have been shown to play key roles in protein-membrane interactions prior to phospholipid cleavage [42].
5. Conclusion
Clostridium perfringens types A, B, C, and D have been reported from the Kingdom of Saudi Arabia. C. perfringens type A has been reported in 80.8% of the bacterial isolates, type D was reported in 15.4%, type C was isolated from 2.9%, and type B was reported in 0.98%. There was some genetic variation in the conserved plc gene which is responsible of the production of alpha toxins. In the present study, 275 and 47 variable positions, respectively, out of the 860 nucleotides and 272 deduced amino acids, were observed. All isolates studied produced CPEs. When considering vaccine production, field isolates which comprise all the strains must be considered.
Acknowledgments
This project was funded by the National Plan for Science, Technology and Innovation (MAARIFAH) and King Abdulaziz City for Science and Technology, Kingdom of Saudi Arabia, studies on enterotoxaemia in livestock in the Kingdom of Saudi Arabia, Award Number 11-BIO-1855-02.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflict of interest regarding the publication of this paper.
Authors' Contributions
Sawsan A. Omer and Osama B. Mohammed contributed equally to this work.
References
- 1.Florence L. C. H., Hakim S. L., Kamaluddin M. A., Thong K. L. Determination of toxinotypes of environmental Clostridium perfringens by polymerase chain reaction. Tropical Biomedicine. 2011;28:171–174. [PubMed] [Google Scholar]
- 2.McClane B. A., Robertson S. L., Li J. Clostridium perfringens. In: Buchanan R. L., Doyle M. P., editors. Food Microbiology: Fundamentals and Frontiers. 4th. Washington, DC, USA: American Society for Microbiology; 2013. pp. 465–489. [Google Scholar]
- 3.Johnson E. A. Clostridial toxins as therapeutic agents: benefits of nature’s most toxic proteins. Annual Review of Microbiology. 1999;53:551–575. doi: 10.1146/annurev.micro.53.1.551. [DOI] [PubMed] [Google Scholar]
- 4.Petit L., Gibert M., Popoff M. R. Clostridium perfringens: toxinotype and genotype. Trends in Microbiology. 1999;7(3):104–110. doi: 10.1016/s0966-842x(98)01430-9. [DOI] [PubMed] [Google Scholar]
- 5.McDonel J. L. Clostridium perfringens toxins (type A, B, C, D, E) Pharmacology & Therapeutics. 1980;10(3):617–655. doi: 10.1016/0163-7258(80)90031-5. [DOI] [PubMed] [Google Scholar]
- 6.Li J., Adams V., Bannam T. L., et al. Toxin plasmids of Clostridium perfringens. Microbiology and Molecular Biology Reviews. 2013;77(2):208–233. doi: 10.1128/MMBR.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Songer J. G. Clostridial enteric diseases of domestic animals. Clinical Microbiology Reviews. 1996;9(2):216–234. doi: 10.1128/cmr.9.2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rood J. I., Adams V., Lacey J., et al. Expansion of the Clostridium perfringens toxin-based typing scheme. Anaerobe. 2018;53:5–10. doi: 10.1016/j.anaerobe.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Joudi A. S. An outbreak of foodborne diarrheal illness among soldiers in Mina during Hajj: the role of consumer food handling behaviors. Journal of Family & Community Medicine. 2007;14(1):29–33. [PMC free article] [PubMed] [Google Scholar]
- 10.Moussa I. M., Hessan A. M. Molecular typing of Clostridium perfringens toxins recovered from central Saudi Arabia. Saudi Medical Journal. 2011;32(7):669–674. [PubMed] [Google Scholar]
- 11.Fayez M. M., Suleiman M. B., Al Marzoog A., Al Taweel H. H. Clostridium perfringens enterotoxaemia in camel (Camelus dromedarius) calves. International Journal of Advanced Research. 2013;1:239–247. [Google Scholar]
- 12.Yoo H. S., Lee S. U., Park K. Y., Park Y. H. Molecular typing and epidemiological survey of prevalence of Clostridium perfringens types by multiplex PCR. Journal of Clinical Microbiology. 1997;35(1):228–232. doi: 10.1128/JCM.35.1.228-232.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Idrissi A. H., Ward G. E. Evaluation of enzyme-linked immunosorbent assay for diagnosis of Clostridium perfringens enterotoxemias. Veterinary Microbiology. 1992;31(4):389–396. doi: 10.1016/0378-1135(92)90131-c. [DOI] [PubMed] [Google Scholar]
- 14.Naylor R. D., Martin P. K., Barker L. T. Detection of Clostridium perfringens toxin by enzyme-linked immunosorbent assay. Research in Veterinary Science. 1997;63(1):101–102. doi: 10.1016/s0034-5288(97)90168-5. [DOI] [PubMed] [Google Scholar]
- 15.Baums C. G., Schotte U., Amtsberg G., Goethe R. Diagnostic multiplex PCR for toxin genotyping of _Clostridium perfringens_ isolates. Veterinary Microbiology. 2004;100(1-2):11–16. doi: 10.1016/S0378-1135(03)00126-3. [DOI] [PubMed] [Google Scholar]
- 16.Keyburn A. L., Boyce J. D., Vaz P., et al. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 2008;4(2):p. e26. doi: 10.1371/journal.ppat.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Troshin P. V., Procter J. B., Barton G. J. Java bioinformatics analysis web services for multiple sequence alignment—JABAWS: MSA. Bioinformatics. 2011;27(14):2001–2002. doi: 10.1093/bioinformatics/btr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milburn D., Laskowski R. A., Thornton J. M. Sequences annotated by structure: a tool to facilitate the use of structural information in sequence analysis. Protein Engineering. 1998;11(10):855–859. doi: 10.1093/protein/11.10.855. [DOI] [PubMed] [Google Scholar]
- 20.Rooney A. P., Swezey J. L., Friedman R., Hecht D. W., Maddox C. W. Analysis of core housekeeping and virulence genes reveals cryptic lineages of Clostridium perfringens that are associated with distinct disease presentations. Genetics. 2006;172(4):2081–2092. doi: 10.1534/genetics.105.054601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abildgaard L., Schramm A., Rudi K., Højberg O. Dynamics of plc gene transcription and α-toxin production during growth of Clostridium perfringens strains with contrasting α-toxin production. Veterinary Microbiology. 2009;139(1-2):202–206. doi: 10.1016/j.vetmic.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 22.Grant K. A., Kenyon S., Nwafor I., et al. The identification and characterization of Clostridium perfringens by real-time PCR, location of enterotoxin gene, and heat resistance. Foodborne Pathogens and Disease. 2008;5(5):629–639. doi: 10.1089/fpd.2007.0066. [DOI] [PubMed] [Google Scholar]
- 23.Ozcan C., Gurcay M. Enterotoxaemia incidence in small ruminants in Elazig and surrounding provinces in 1994-1998. Turkish Journal of Veterinary and Animal Sciences. 2000;24(3):283–286. [Google Scholar]
- 24.Miwa N., Masuda T., Terai K., Kawamura A., Otani K., Miyamoto H. Bacteriological investigation of an outbreak of Clostridium perfringens food poisoning caused by Japanese food without animal protein. International Journal of Food Microbiology. 1999;49(1-2):103–106. doi: 10.1016/s0168-1605(99)00059-8. [DOI] [PubMed] [Google Scholar]
- 25.Sting R. Detection of beta2 and major toxin genes by PCR in Clostridium perfringens field isolates of domestic animals suffering from enteritis or enterotoxaemia. Berliner und Münchener Tierärztliche Wochenschrift. 2009;122:341–347. [PubMed] [Google Scholar]
- 26.Gurjar A. A., Hegde N. V., Love B. C., Jayarao B. M. Real-time multiplex PCR assay for rapid detection and toxintyping of Clostridium perfringens toxin producing strains in feces of dairy cattle. Molecular and Cellular Probes. 2008;22(2):90–95. doi: 10.1016/j.mcp.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Sipos W., Fischer L., Schindler M., Schmoll F. Genotyping of Clostridium perfringens isolated from domestic and exotic ruminants and swine. Journal of Veterinary Medicine Series B. 2003;50(7):360–362. doi: 10.1046/j.1439-0450.2003.00690.x. [DOI] [PubMed] [Google Scholar]
- 28.Li J., Miyamoto K., Sayeed S., McClane B. A. Organization of the cpe locus in CPE-positive Clostridium perfringens type C and D isolates. PLoS One. 2010;5(6, e10932) doi: 10.1371/journal.pone.0010932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyamoto K., Li J., Sayeed S., Akimoto S., McClane B. A. Sequencing and diversity analyses reveal extensive similarities between some epsilon-toxin-encoding plasmids and the pCPF5603 Clostridium perfringens enterotoxin plasmid. Journal of Bacteriology. 2008;190(21):7178–7188. doi: 10.1128/JB.00939-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sayeed S., Li J., McClane B. A. Virulence plasmid diversity in Clostridium perfringens type D isolates. Infection and Immunity. 2007;75(5):2391–2398. doi: 10.1128/IAI.02014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibert M., Jolivet-Reynaud C., Popoff M. R. Beta2 toxin, a novel toxin produced by Clostridium perfringens. Gene. 1997;203(1):65–73. doi: 10.1016/s0378-1119(97)00493-9. [DOI] [PubMed] [Google Scholar]
- 32.Garmory H. S., Chanter N., French N. P., Bueschel D., Songer J. G., Titball R. W. Occurrence ofClostridium perfringensβ2-toxin amongst animals, determined using genotyping and subtyping PCR assays. Epidemiology and Infection. 2000;124(1):61–67. doi: 10.1017/S0950268899003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Czanderlova L., Hlozek P., Chmelar D., Lany P. Clostridium perfringens in suckling piglets with diarrhoea and its PCR typing and prevalence in the Czech Republic in 2001-2003. Veterinarni Medicina-Praha. 2006;51 [Google Scholar]
- 34.Van Damme-Jongsten M., Rodhouse J., Gilbert R. J., Notermans S. Synthetic DNA probes for detection of enterotoxigenic Clostridium perfringens strains isolated from outbreaks of food poisoning. Journal of Clinical Microbiology. 1990;28(1):131–133. doi: 10.1128/jcm.28.1.131-133.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McClane B. A. An overview of Clostridium perfringens enterotoxin. Toxicon. 1996;34(11-12):1335–1343. doi: 10.1016/s0041-0101(96)00101-8. [DOI] [PubMed] [Google Scholar]
- 36.Singh U., van Itallie C. M., Mitic L. L., Anderson J. M., McClane B. A. Caco-2 cells treated withClostridium perfringensEnterotoxin form multiple large complex species, one of which contains the tight junction protein occludin. The Journal of Biological Chemistry. 2000;275(24):18407–18417. doi: 10.1074/jbc.M001530200. [DOI] [PubMed] [Google Scholar]
- 37.Veshnyakova A., Protze J., Rossa J., Blasig I. E., Krause G., Piontek J. On the interaction of Clostridium perfringens enterotoxin with claudins. Toxins. 2010;2(6):1336–1356. doi: 10.3390/toxins2061336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fisher D. J., Miyamoto K., Harrison B., Akimoto S., Sarker M. R., McClane B. A. Association of beta2 toxin production with Clostridium perfringens type A human gastrointestinal disease isolates carrying a plasmid enterotoxin gene. Molecular Microbiology. 2005;56(3):747–762. doi: 10.1111/j.1365-2958.2005.04573.x. [DOI] [PubMed] [Google Scholar]
- 39.Sparks S. G., Carman R. J., Sarker M. R., McClane B. A. Genotyping of enterotoxigenic Clostridium perfringens fecal isolates associated with antibiotic-associated diarrhea and food Poisoning in North America. Journal of Clinical Microbiology. 2001;39(3):883–888. doi: 10.1128/JCM.39.3.883-888.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collie R. E., McClane B. A. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with non-food-borne human gastrointestinal diseases. Journal of Clinical Microbiology. 1998;36(1):30–36. doi: 10.1128/JCM.36.1.30-36.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cornillot E., Saint-Joanis B., Daube G., et al. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Molecular Microbiology. 1995;15(4):639–647. doi: 10.1111/j.1365-2958.1995.tb02373.x. [DOI] [PubMed] [Google Scholar]
- 42.Sheedy S. A., Ingham A. B., Rood J. I., Moore R. J. Highly conserved alpha-toxin sequences of avian isolates of Clostridium perfringens. Journal of Clinical Microbiology. 2004;42(3):1345–1347. doi: 10.1128/jcm.42.3.1345-1347.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.