Abstract
Wastewater treatment center (WTC) workers may be vulnerable to diseases caused by viruses, such as the common cold, influenza and gastro-intestinal infections. Although there is a substantial body of literature characterizing the microbial community found in wastewater, only a few studies have characterized the viral component of WTC aerosols, despite the fact that most diseases affecting WTC workers are of viral origin and that some of these viruses are transmitted through the air. In this study, we evaluated in four WTCs the presence of 11 viral pathogens of particular concern in this milieu and used a metagenomic approach to characterize the total viral community in the air of one of those WTCs. The presence of viruses in aerosols in different locations of individual WTCs was evaluated and the results obtained with four commonly used air samplers were compared. We detected four of the eleven viruses tested, including human adenovirus (hAdV), rotavirus, hepatitis A virus (HAV) and Herpes Simplex virus type 1 (HSV1). The results of the metagenomic assay uncovered very few viral RNA sequences in WTC aerosols, however sequences from human DNA viruses were in much greater relative abundance.
Keywords: Airborne viruses, Air sampling, Wastewater treatment plants, Viral metagenomics
Graphical abstract
Introduction
Wastewater treatment centers (WTCs) are, unsurprisingly, highly contaminated environments. Concentrations of viruses in effluent waters can be extremely high, sometimes reaching 1011 viruses/mL (La Rosa et al., 2010). Several studies have reported higher symptom and disease rates among this group of workers (Khuder et al., 1998). They experience respiratory symptoms, fevers, gastrointestinal symptoms, and headaches more often than the non-exposed population (Khuder et al., 1998, Smit et al., 2005, Van Hooste et al., 2010). The term “sewage worker's syndrome” was used for the first time in 1973 to describe the assemblage of these symptoms (Rylander et al., 1976).
Despite the fact that most WTC occupational symptoms can be associated with viral infections, only a few studies have investigated viruses as an occupational risk in these environments. Previous studies have demonstrated the presence of human pathogenic viruses in influent water, including noroviruses (Pouillot et al., 2015, Qiu et al., 2015) and rotaviruses (Baggi et al., 2001, Qiu et al., 2015) that cause gastroenteritis; adenoviruses (Osuolale and Okoh, 2015, Qiu et al., 2015), rhinoviruses and enteroviruses (Baggi et al., 2001, Qiu et al., 2015) that are responsible for the common cold, and even herpes simplex viruses, which can cause oral and genital sores and blisters (Bibby and Peccia, 2013). Among the viruses detected at WTCs, many of these pathogens are transmitted by aerosols (Tseng et al., 2010, Bonifait et al., 2015). Despite the importance of this route of transmission, surprisingly few studies have examined the potential exposure of WTC workers to pathogenic viruses through the air. Moreover, in these studies, only one or two viruses were analyzed (Romano et al., 1999, Uhrbrand et al., 2011, Masclaux et al., 2014). In Denmark WTCs, noroviruses (Noro) GI and GII were detected for the first time in air samples using personal samplers (Uhrbrand et al., 2011). Adenovirus (AdV), hepatitis E virus (HEV) and norovirus were also investigated in air samples from 31 Swiss WTCs. The researchers found AdV, noroviruses and HEV in 100%, 2% and 0%, respectively of the WTC aerosols tested (Masclaux et al., 2014). Enterovirus and reovirus were identified in 3.4% of the air samples from an aerosol study of WTCs in Italy (Romano et al., 1999).
To date, metagenomics has been used to characterize viral communities in aerosols in only three studies (Whon et al., 2012, Hall et al., 2013, Be et al., 2015). In all of these studies, RNA viruses were excluded, despite the fact that most human respiratory viruses have RNA genomes such as coronavirus, influenza virus, and metapneumovirus. To the best of our knowledge, this is the first time that a metagenomic approach has been used to characterize viral communities from WTC aerosols.
In Eastern Canada, WTC secondary treatment can differ from one plant to another. For example, some centers perform biological removal of residual matter (also called biofiltration) as the water exits the early stages of the treatment pipeline, while others conduct a secondary decantation as an alternative to biofiltration. Every day, WTC employees work in rooms or locations where different processing steps are taking place, some of which increase the risk of exposure to bioaerosols. In this study, we used various air samplers to evaluate the presence and abundance of some human pathogen viruses in aerosols in different locations of WTCs using quantitative polymerase chain reaction (qPCR). We also applied a viral metagenomics approach in one of the participating WTCs.
1. Material and methods
1.1. WTCs and site selection
Since very few reports exist on occupational airborne viral exposure, there is no consensus sampling strategy for the collection and purification of viral nucleic acids from aerosols. We therefore used both low and high flow rate sampling approaches.
Air samples were collected from four different WTCs from Eastern Canada during summertime. Indoor sites where wastewater treatment occurs and workers daily tasks are occurring were sampled. A total of 11 sites (or sampling locations) distributed among four WTCs are presented in this study. Pertinent details for each site are presented in Table 1 .
Table 1.
Description of wastewater treatment centers (WTCs) and sites of the study.
| WTCs | Sites | Tasks |
|---|---|---|
| WTCs 1 and 2 | Screening | Removal of big objects (e.g.,: plastics and paper) |
| Grit/fats, oils and greases (FOGs) removal) | Removal of granular matter and FOGs | |
| Biofiltration | Biological degradation of residual organic matter | |
| WTC3 | Primary screening | Removal of big objects |
| Secondary screening | Removal of residual big objects | |
| WTC4 | Screening | Removal of big objects |
| Grit/fats, oils and greases removal | Removal of granular matter and FOGs | |
| Secondary decantation | Removal of residual particles by decantation |
1.2. Air sampling methods
1.2.1. Sampling for qPCR analysis
In this study, two different samplers were used to collect samples for qPCR detection (Coriolis®μ and Marple). The samplers were positioned at the same location at each water treatment site. Although the sampling duration was dependent on the sampler model, sampling was always performed during the same day over the same 6-hour shift.
The Coriolis®μ (Bertin Technologies, Montigny-le-Bretonneux, France) is a liquid cyclone that collects particles in 15 mL of Phosphate Buffer Saline (PBS). The flowrate was 200 L/min for 10 min, for a total of 2 m3 of air/sample. The volume of the recipient was readjusted to 15 mL after sampling to compensate for evaporation. Five mL of the total were concentrated in a final volume of 200 μL using tangential flow filtration devices (100 kDa, Millipore, Darmstadt, Germany) used to conduct viral qPCR analyses (0.67 m3 of air).
The second sampler, a size fractionating collector, the Marple Personal Cascade Impactor (Thermo Fisher Scientific, Waltham, USA) was used to collect information about aerosols size distribution of viruses containing bioaerosols. Due to material limitation, a maximum of two sampling locations per WTC were selected. In this sampler, air is accelerated by going through six radial slots of a first impactor stage in which each of the 8 stages impacts a subfraction of particles ranging from 0.5 to 21 μm of aerodynamic diameters that are ultimately collected on filters. After the 8th stage, another filter collects the remaining fine particles. It was used at a flowrate of 2 L/min for 5 hr, for a total of 0.6 m3 collected. Each of the 9 filters was eluted in 5 mL of PBS and 500 μL were concentrated in a final volume of 25 μL using tangential flow filtration devices (100 kDa, Millipore, Darmstadt, Germany). Nucleic acids were extracted for this subsample and then used for qPCR.
1.2.2. Viral metagenomics
For viral metagenomics approach, the sampling was accomplished with the SASS 2300 sampler (Research International, Washington, USA) a high flowrate (300 L/min) liquid cyclone that allow extended sampling periods and resulting in a larger air sample in a smaller collection liquid volume (5 mL). A total of 30 m3 of air was sampled (sampling time of 100 min).
1.3. Nucleic acid extraction
All nucleic acids were extracted and eluted in 100 μL with the QIAamp MinElute Spin Virus kit (Qiagen, Hilden, Germany) that co-purifies DNA and RNA as per instructions. Reverse transcription was performed on samples used for quantification of RNA viruses with the iScript™ cDNA Synthesis kit (Bio-Rad Laboratories, Mississauga, Canada) according to the manufacturers' instructions.
1.4. qPCR analyses
We selected viral qPCR primers based on two criteria: (1) whether they amplified human pathogens that can cause symptoms often encountered by WTC workers and (2) whether their presence in wastewaters had been demonstrated. Eleven viruses fit these criteria: influenza viruses (Inf) A and B, noroviruses (Noro) GI and GII, herpes simplex viruses (HSV) 1 and 2, human rhinovirus, enterovirus, human adenovirus, rotavirus (rota) and hepatitis A virus.
A volume of 2 μL of nucleic acids (DNA or cDNA) was used for each 20 μL qPCR reactions using the iQ™ Supermix kit (Bio-Rad Laboratories, Mississauga, Canada). The same thermocycler protocol was performed for all eleven viruses: an initial denaturation step of 3 min at 94°C, followed by 40 cycles of a denaturation step at 94°C for 15 sec followed by an annealing/extension step at 60°C for 1 min. All standard curves had efficiency between 90% and 110% and an R 2 above 0.98. Table 2 lists the sequences of the primers and probes used for the quantitative amplification assays.
Table 2.
Primers and probes used for qPCR detection of selected viruses.
| Viruses | RNA/ DNA |
Forward primers | Reverse primers | Probes | Reference |
|---|---|---|---|---|---|
| Inf A | RNA | GACCRATCCTGTCACCTCTGAC | AGGGCATTYTGGACAAAKCGTCTA | TGCAGTCCTCGCTCACTGGGCACG | (Selvaraju and Selvarangan, 2010) |
| Inf B | RNA | TCCTCAACTCACTCTTCGAGCG | CGGTGCTCTTGACCAAATTGG | CCAATTCGAGCAGCTGAAACTGCGGTG | (Selvaraju and Selvarangan, 2010) |
| Noro GI | RNA | CGYTGGATGCGNTTYCATGA | CTTAGACGCCATCATCATTYAC | AGATYGCGATCYCCTGTCCA | (Kageyama et al., 2003) |
| Noro GII | RNA | CARGARBCNATGTTYAGRTGGATGAG | TCGACGCCATCTTCATTCACA | TGGGAGGGCGATCGCAATCT | (Kageyama et al., 2003) |
| HSV-1 | DNA | CGCATCAAGACCACCTCCTC | GCTCGCACCACGCGA | TGGCAACGCGGCCCAAC | (Corey et al., 2005) |
| HSV-2 | DNA | CGCATCAAGACCACCTCCTC | GCTCGCACCACGCGA | CGGCGATGCGCCCCAG | (Corey et al., 2005) |
| HRV | RNA | GTGAAGAGCCSCRTGTGCT | GCTSCAGGGTTAAGGTTAGCC | TGAGTCCTCCGGCCCCTGAATG | (Hayden et al., 2003) |
| EV | RNA | GGCCCCTGAATGCGGCTAAT | CAATTGTCACCATAAGCAGCCA | CGGACACCCAAAGTAGTCGGTTCCG | (Donaldson et al., 2002, Meijer et al., 2012) |
| hAdV | DNA | GCCACGGTGGGGTTTCTAAACTT | GCCCCAGTGGTCTTACATGCACAT | TGCACCAGACCCGGGCTCAGGTACTCCGA | (Heim et al., 2003) |
| Rotavirus | RNA | ACCATCTWCACRTRACCCCTCTATGAG | GGTCACATAACGCCCCTATAGC | AGTTAAAAGCTAACACTGTCAAA | (Zeng et al., 2008) |
| HAV | RNA | GGTAGGCTACGGGTGAAAC | CCTCCGGCGTTGAATGGTTT | ACAGCGGCGGATATTGGTGAGTTGTTAAGA | (Qiu et al., 2014) |
W = A/T, S = C/G, K = G/T, R = A/G, Y = C/T and N = A, T, C or G.
1.5. Metagenomics study
Samples from SASS 2300 samplers intended for metagenomics analyses were thawed from the − 80°C freezer and concentrated in Amicon 100 kDa tangential flow filtration devices (Millipore) to a final volume of 200 μL. Nucleic acids were subsequently extracted with the QIAamp MinElute Spin Virus kit (Qiagen, Hilden, Germany) as per instructions. Concentrated nucleic acids were eluted in a final volume of 87.5 μL to directly undergo the digestion of DNA with the RNase-Free DNase Set from Qiagen, followed by RNA purification with the RNeasy MinElute Cleanup kit from Qiagen. RNA samples were processed for library construction with the Kapa Stranded mRNA-Seq Kit (Kapa Biosystems, San Diego, USA) and then sequenced on the MiSeq sequencing platform (Illumina, Inc., paired-end 300 nucleotides reads) at the sequencing facility of the Centre hospitalier de l'Université Laval (CHUL). The reads were assembled into contigs by Ray Meta (Boisvert et al., 2012). The Ray Meta software uses a bloom filter to remove k-mers that are potentially background noise and sequencing errors. Data were subsequently analyzed via the web-based MG-RAST (Meyer et al., 2008) and MetaVir platforms (Roux et al., 2011).
2. Results
2.1. qPCR
Among all targeted viruses, two were detected; HAV and rotavirus. Table 3 shows where they were detected and at what concentration.
Table 3.
Viral amplifications from air samples using Coriolis high flow sampler in WTCs.
| WTCs | Sites | Virus (gene copies/m3 air) |
|---|---|---|
| WTC1 | Site 1 | Rota (3.2 × 104) |
| Site 2 | Rota (2.2 × 105) | |
| Site 3 | Rota (3.5 × 104), HAV (4.7 × 103) | |
| WTC2 | Site 1 | Rota (1.7 × 104) |
| Site 2 | Rota (2.2 × 105) | |
| Site 3 | Rota (1.8 × 105) | |
| WTC3 | Site 1 | – |
| Site 2 | – | |
| WTC4 | Site 1 | – |
| Site 2 | – |
Based on limit of detection of qPCR reactions, the limit of detection (LOD) for Coriolis sampler is estimated to 1.0 × 103 for both hepatitis A virus (HAV) and Rotavirus.
From the Marple samples, only rotavirus cDNA could be detected almost everywhere except in samples from site 1 of WTC 3. Samples from site 2 of WTC 1 seem to have viral material on larger airborne particles, and site 3 of WTC 4 presents the opposite situation, viral material on smaller airborne particles. The other samples show a spread of the viral particles among the stages of the sampler (Table 4 ).
Table 4.
Detection and quantification of rotaviruses with the Marple sampler.
| WTC 1 |
WTC 2 |
WTC 3 |
WTC 4 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Cut-Off (μm) | Stages | Site 2 | Site 3 | Site 2 | Site 3 | Site 1 | Site 2 | Site 2 | Site 3 |
| 21.3 | 1 | – | 6.1 × 105 | 6.6 × 105 | 2.7 × 106 | – | – | – | – |
| 14.8 | 2 | 2.4 × 105 | – | 1.3 × 105 | 8.0 × 105 | – | 6.6 × 104 | – | – |
| 9.8 | 3 | 1.2 × 104 | 4.5 × 104 | 7.3 × × 105 | 1.5 × 107 | – | 1.5 × 105 | – | 8.3 × 105 |
| 6 | 4 | 3.7 × 105 | – | 7.4 × 105 | 4.6 × 105 | – | – | – | – |
| 3.5 | 5 | 1.1 × 106 | 1.5 × 105 | 5.0 × 105 | 1.5 × 106 | – | 3.0 × 105 | – | 5.3 × 104 |
| 1.55 | 6 | – | – | 3.1 × 105 | 3.7 × 105 | – | 8.0 × 106 | – | – |
| 0.93 | 7 | – | – | 5.5 × 105 | 8.1 × 106 | – | 3.4 × 105 | 5.5 × 104 | – |
| 0.52 | 8 | 3.1 × 104 | – | 2.6 × 105 | 3.3 × 104 | – | 2.0 × 105 | – | 7.4 × 105 |
| 0 | F | – | – | 4.2 × 104 | 1.0 × 106 | – | 5.6 × 105 | 1.0 × 105 | – |
2.2. Metagenomics
The bigger samples obtained with the Sass 2300 sampler were used to produce viral metagenomes from three sites from WTC 4. The dataset for the three locations were as shown in Table 5 .
Table 5.
Sequencing and assembly output details.
| Sites | Number of reads | Number of contigs | Total size of assembly | Average contig length | N50 of assembly | Median contig length | Largest contig |
|---|---|---|---|---|---|---|---|
| Site 1 | 18741055 | 45130 | 101595404 | 2251 | 8800 | 772 | 1635750 |
| Site 2 | 764755 | 602 | 536721 | 891 | 811 | 588 | 15090 |
| Site 3 | 604133 | 268 | 240403 | 897 | 849 | 622 | 8222 |
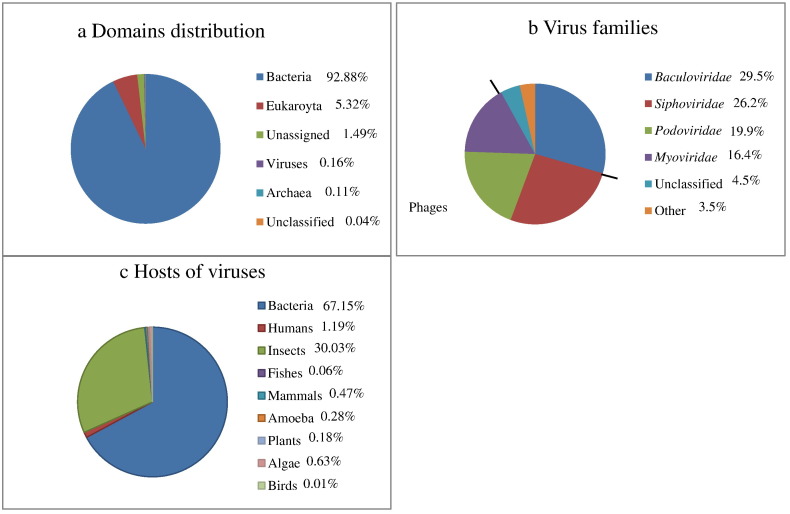
In Fig. 1a, the classification of assigned sequences is at a domain taxonomic level. In Fig. 1b, the classification of virus-assigned sequences (n = 9996) was divided by Family: Families with less than 1% representation were classified in the group “other”. Fig. 1c represents the hosts of viruses based on the Fig. 1b assignments. Bacteria represented the most abundant organisms in all three samples with an average of almost 93% of all sequences. Among viral sequences, more than 62% were from phages in the order Caudovirales. 1.2% of the viruses' hosts were human (see Fig. 1ca). The classification of these sequences to the lowest taxonomic level possible is shown in Fig. 2 . Table S1 (Appendix A) shows the taxonomy of the viruses in the group “other” to the family level. (See Fig. 3).
Fig. 1.
Air samples from WTC 4 (a) classification of assigned sequences at a domain taxonomic level, (b) classification of virus-assigned sequences by Family, (c) hosts of viruses based on the panel b assignments.
Fig. 2.
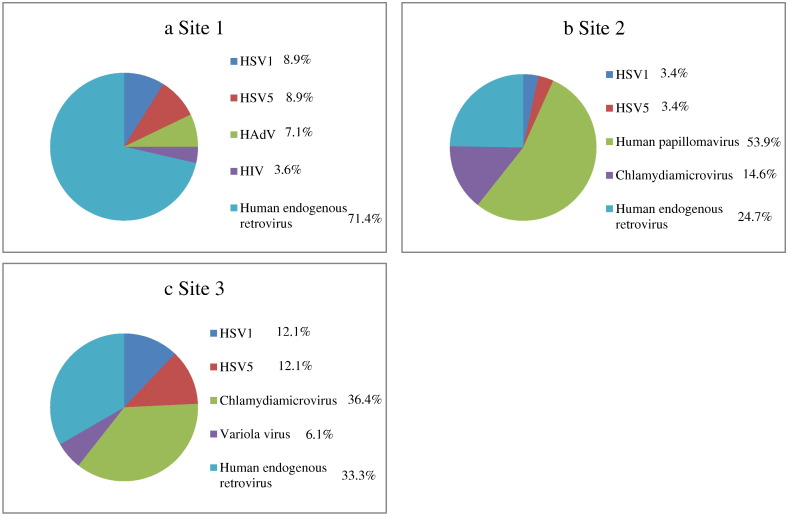
Distribution of human viruses in samples from sampling locations 1 (a), 2 (b), and 3 (c) from WTC 4.
Fig. 3.
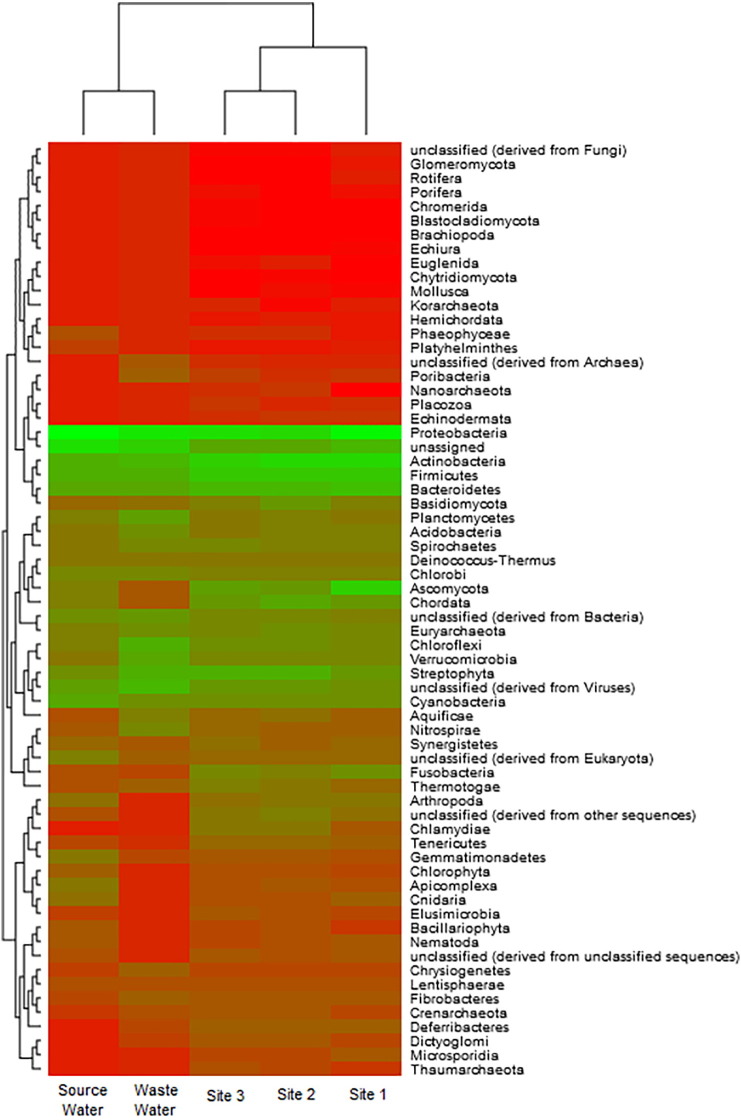
Heat map comparing metagenomic results from sites 1, 2 and 3 of WTC 4 with samples from wastewater and source water in North America (public databases). The red scale represents low abundance cases, as the green one represents high abundance cases.
Human endogenous retroviruses were recovered from all three samples. Sequences from HSV1 and HSV5, two types of herpes simplex viruses causing latent infections, were also found in all three sites. Chlamydiamicrovirus sequences were identified in both site 2 and site 3 samples, and were the most highly represented human virus in site 3. Human papillomavirus, a virus that causes latent sexually transmissible infections, was the most common human virus detected in site 2. Variola virus sequences were found to be in the air sample from site 3 only.
We used a heatmap generated with MetaVir to compare our metagenomic data from WTC aerosols with published metagenomic data from WTC-related locations in order to gain insight into the primary source of airborne microbes. In this analysis, we included the complete dataset to construct the map, and thus bacteria, archaea, fungi, and viruses were included.
3. Discussion
Using qPCR, few viruses were detected with the tested sampling devices. Rotavirus was detectable more frequently with the Marple compared to the Coriolis. Rotavirus is a double-stranded RNA virus and as RNA is extremely fragile and rapidly degraded by RNases found ubiquitously in nature, there is a chance that samplers with a flow rate higher than 200 L/min tend to lead to the degradation of less robust RNA virus species during collection (Cao et al., 2011, Toulouse et al., 2014). The Marple and the Coriolis are based on two completely different capture mechanisms; filtration and liquid impaction. Both of these mechanisms may induce particle loss (bouncing and re-aerosolization). The different sizes and characteristics of the eleven selected viruses likely also plays a role in the capture efficiency of each sampler, as some enveloped or non-enveloped viruses are more or less resistant to desiccation caused by the sampling times and rates. Marple samples also suggest a wide range of particles sizes carrying viruses, suggesting lower and upper respiratory tract exposure. In both cases, viruses can either remain in the lungs or upper tract or be subsequently swallowed.
Overall, rotaviruses seem to be abundant in the air of WTCs based on qPCR detection. The sites that showed the highest concentrations of rotaviruses were also the sights that had the highest concentration of total viruses (e.g., biofiltration sites of WTCs 1 and 2). Inhalation of pathogenic viruses causing respiratory infections has been studied over the years since it represents a preferential route of transmission for some viruses. Rotavirus is known to cause gastroenteritis. The infectious dose of this virus through the oral route is below 100 viral particles (Payne et al., 2011), which was 104 times less than the concentrations of genome copies detected by cubic meter in this study, although we did not test for viral infectivity. It is clear from our data that additional studies should be performed to determine the concentration of infectious viruses in WTC aerosols. The impact on human health of aerosol exposure to viruses such as HAV has not been documented, as they are not known to be transmitted through the airborne route. However, the possibility of these viruses infecting a host via the air pathway cannot be ruled out, and raises another avenue of future research. As mentioned before, WTC workers experience symptoms such as fever, nausea and vomiting that could be linked to infections by HAV. Like HAV, the possibility of rotavirus transmission via the air has not been examined to date, nevertheless, the fact that these viruses have been detected in air samples from a hospital (Dennehy et al., 1998) raises the likelihood of transmission via aerosol. In addition, a recent study performed by our team showed that norovirus, a virus responsible for gastroenteritis outbreaks worldwide, can be present in the air (Bonifait et al., 2015), suggesting that perhaps other viruses that cause the same disease, like rotaviruses, could follow the same pattern. Establishing whether aerosol transmission of these viruses is a viable route is critical if we are to better understand the occupational risks of WTC workers.
In this study, Noroviruses GI and GII were not detected although WTC workers personal air samplers were positive in previous studies (Uhrbrand et al., 2011, Masclaux et al., 2014). This may be due to differences in flowrate between our samplers and the personal samplers. The flowrate of personal samplers, which use gelatin filters, is much less than the devices we used (except for the Marple). It is possible that norovirus, an RNA virus that is easily damaged by a greater airflow, was removed during sampling in our study. hAdV was not detected Coriolis and Marple samples, this virus was observed in a previous study (Masclaux et al., 2014). Enterovirus was not detected at all, which is consistent with the literature data where only 2 out of 118 samples were positive (Romano et al., 1999).
In addition to the direct inhalation of viruses in aerosols, workers are exposed to viruses that can be trapped in the upper respiratory tract or the lungs and swallowed afterwards. Although this study did not aim to document the surface contamination by viruses, workers can also be exposed to viral particles deposited on surfaces via aerosol deposition, i.e., the fomite transmission route. In this pathway, aerosolized viruses adsorb to charged fomites on a surface. The fomite with the pathogen can then come into contact with a worker, his or her hand for example. The worker is subsequently infected when he/she touches his/her mouth and/or eyes. Many viruses remain infectious for extended periods of time after deposition. For example, HAV and rotavirus remain infectious for more than two months on fomites (Boone and Gerba, 2007). This route of infection has been proven for many viral pathogens, including rotavirus (Butz et al., 1993, Hota, 2004), HAV (Sundkvist et al., 2000, Aitken and Jeffries, 2001), and AdV (Abad et al., 1994, Aitken and Jeffries, 2001, Hamada et al., 2008) among others, and yet remains an underestimated and understudied risk for workers.
Our metagenomic analysis focused on RNA viruses because they are responsible for the majority of human respiratory and enteric illnesses suffered by WTCs' workers and the DNA virus community is typically dominated by viruses that infect bacteria (i.e., bacteriophages) (Lundholm and Rylander, 1983, Uhrbrand et al., 2011, Paez-Espino et al., 2016. However, the fact that the majority of sequences that we were able to identify were not viral in origin was not surprising. While some viral metagenomic libraries are constructed with enriched or purified viral nucleic acids, in this study, we chose to sequence unpurified and unenriched samples directly in order to avoid removing viruses adsorbed to large agglomerates of particles in the air (data not shown). The most likely reason for the low number of RNA viral sequences detected in our study is that we did not achieve an adequate depth of sequencing to fully characterize this relatively minor component of the total sequences in our samples and why the viruses targeted by qPCR were not present in our metagenomic results.
Rotaviruses and HAV were detected by qPCR from samples collected only with the Coriolis and the Marple devices. This suggests that low flowrate samplers may result in a better recovery of RNA viruses in these conditions. High flowrate samplers are preferable when sampling rare events as they can generate more concentrated samples than moderate to low flowrate devices. Nonetheless, the greater rate of airflow and the extended sampling period of these devices can damage the particles and organisms collected by the sampler (Toulouse et al., 2014). Some RNA viruses are particularly prone to degradation because of the fragility of their envelope and the instability of RNA due to the ubiquity of RNAses in the environment (Li, 2013).
The most abundant family of RNA viruses based on our metagenomic data was Retroviridae, a family that includes the Human Immunodeficiency Virus. However, a particularity of these viruses is that they possess a proviral stage in their replication cycle where they insert their genetic material in the form of DNA into the chromosome of their host. Given the likely presence of contaminating DNA in our metagenomes, we do not know if the sequences identified in our samples are from the DNA provirus, retroviral messenger RNA or the encapsidated retrovirus RNA genome. Five contigs were homologous to Caulimoviridae, a family of single-stranded RNA viruses that infects plants and insects. This is a plausible result considering that the WTC inflow is a combination of domestic, industrial and agricultural surface runoff and storm water that includes many potential insect and plant viral hosts. We must also underline the fact that the classification of these data is based on the homology of our sequences to a database and thus the presence of a particular virus must be confirmed independently, something that was beyond the scope of this study.
Herpes virus was the most abundant human DNA virus pathogen identified by metagenomics in this study. Its detection is not surprising considering how widespread it is in the human population. In fact, 65% of the population of the United States possesses antibodies to HSV1 (Xu et al., 2002). Interestingly, HSV1 was identified in our metagenomic data for site 4, and was not detected by qPCR. The presence of sequence homology to Variola virus, a pathogen responsible for smallpox, was unexpected, but is presumably due to the presence of the Vaccinia virus, the active agent in the smallpox vaccine that has a genome with high sequence similarity to the genome of Variola virus. The Vaccinia virus, the active agent in the smallpox vaccine, is in circulation in bovine populations. We must again state that the presence of these viruses must be identified independently (Assis et al., 2013).
Sequences with homology to adenoviruses were also abundant in the WTC aerosol metagenome. In corroboration with our results, human adenoviruses have also been found in recent studies based on the viral metagenomic characterization of wastewaters (Bibby and Peccia, 2013). Among the dsDNA phages identified herein, Chlamydiamicrovirus and its host, Chlamydia, a taxon of bacteria that are obligate parasites of eukaryotic cells and that are known to cause pneumoniae, eye trachoma, sexually transmissible infections and psittacosis, were detected as well.
In the future, a more complete and broader risk assessment study could be performed in WTCs using symptoms, epidemiological data that could be linked and metagenomic analyses of viruses in aerosols and surfaces to better understand the impacts of the presence of viral particles on health problems.
4. Heatmap
As this was the first time that viral metagenomics had been applied to air samples from WTCs, we supposed that viruses, and also all organisms found in the air, would originate from treated water. Although we hypothesized that the wastewater and aerosol datasets would be most similar, this comparison was ultimately inconclusive. Nevertheless, our results suggest that WTC-related microbial communities are demonstrably distinct from their most likely sources (i.e., wastewater and source water).
Conflict of interests
The authors have no conflict of interests to declare.
Acknowledgments
Acknowledgements
This study was funded by the Fondation IUCPQ-J.-D. Bégin-P.-H. Lavoie (2014) and Institut de recherche Robert-Sauvé en santé et sécurité du travail (IRSST) (grant number 2010-050). EB received a graduate program scholarship from IRSST. CD was a Fonds de recherche du Québec-Santé (FRQ-S) senior scholar. We would like to thank WTCs workers and managers for their participation to the study.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jes.2017.07.015.
Appendix A. Supplementary data
Families represented in the group “other” of graph B with their respective percentage.
References
- Abad F.X., Pinto R.M., Bosch A. Survival of enteric viruses on environmental fomites. Appl. Environ. Microbiol. 1994;60(10):3704–3710. doi: 10.1128/aem.60.10.3704-3710.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken C.D., Jeffries J. Nosocomial spread of viral disease. Clin. Microbiol. Rev. 2001;14(3):528–546. doi: 10.1128/CMR.14.3.528-546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis F.L., Borges I.A., Mesquita V.S., Ferreira P.C., Trindade G.S. Vaccinia virus in household environment during bovine vaccinia outbreak. Brazil. Emerg. Infect. Dis. 2013;19(12):2045. doi: 10.3201/eid1912.120937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggi F., Demarta A., Peduzzi R. Persistence of viral pathogens and bacteriophages during sewage treatment: lack of correlation with indicator bacteria. Res. Microbiol. 2001;152(8):743–751. doi: 10.1016/s0923-2508(01)01255-4. [DOI] [PubMed] [Google Scholar]
- Be N.A., Thissen J.B., Fofanov V.Y., Allen J.E., Rojas M., Golovko G. Metagenomic analysis of the airborne environment in urban spaces. Microb. Ecol. 2015;69(2):346–355. doi: 10.1007/s00248-014-0517-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K., Peccia J. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ. Sci. Technol. 2013;47(4):1945–1951. doi: 10.1021/es305181x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert S., Raymond F., Godzaridis E., Laviolette F., Corbeil J. Ray meta: scalable de novo metagenome assembly and profiling. Genome Biol. 2012;13(12) doi: 10.1186/gb-2012-13-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifait L., Charlebois R., Vimont A., Turgeon N., Veillette M., Longtin Y. Detection and quantification of airborne norovirus during outbreaks in healthcare facilities. Clin. Infect. Dis. 2015;61(3):299–304. doi: 10.1093/cid/civ321. [DOI] [PubMed] [Google Scholar]
- Boone S.A., Gerba C.P. Significance of fomites in the spread of respiratory and enteric viral disease. Appl. Environ. Microbiol. 2007;73(6):1687–1696. doi: 10.1128/AEM.02051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butz A.M., Fosarelli, Dick P.J., Cusack T., Yolken R. Prevalence of rotavirus on high-risk fomites in day-care facilities. Pediatrics. 1993;92(2):202–205. [PubMed] [Google Scholar]
- Cao G., Noti J.D., Blachere F.M., Lindsley W.G., Beezhold D.H. Development of an improved methodology to detect infectious airborne influenza virus using the NIOSH bioaerosol sampler. J. Environ. Monit. 2011;13(12):3321–3328. doi: 10.1039/c1em10607d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey L., Huang M.L., Selke S., Wald A. Differentiation of herpes simplex virus types 1 and 2 in clinical samples by a real-time taqman PCR assay. J. Med. Virol. 2005;76(3):350–355. doi: 10.1002/jmv.20365. [DOI] [PubMed] [Google Scholar]
- Dennehy P.H.N., Crowley S.M., Saracen B.A., Cheryl L. The American Pediatric Society and The Society for Pediatric Research; New Orleans: 1998. Detection of Rotavirus Rna in Hospital air Samples by Polymerase Chain Reaction (PCR) [Google Scholar]
- Donaldson K.A., Griffin D.W., Paul J.H. Detection, quantitation and identification of enteroviruses from surface waters and sponge tissue from the Florida Keys using real-time RT-PCR. Water Res. 2002;36(10):2505–2514. doi: 10.1016/s0043-1354(01)00479-1. [DOI] [PubMed] [Google Scholar]
- Hall R.J., Leblanc-Maridor, Wang M.J., Ren X., Moore N.E., Brooks C.R. Metagenomic detection of viruses in aerosol samples from workers in animal slaughterhouses. PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0072226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada N., Gotoh K., Hara K., Iwahashi J., Imamura Y., Nakamura S. Nosocomial outbreak of epidemic keratoconjunctivitis accompanying environmental contamination with adenoviruses. J. Hosp. Infect. 2008;68(3):262–268. doi: 10.1016/j.jhin.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Hayden F.G., Herrington D.T., Coats T.L., Kim K., Cooper E.C. Efficacy and safety of oral pleconaril for treatment of colds due to picornaviruses in adults: results of 2 double-blind, randomized, placebo-controlled trials. Clin. Infect. Dis. 2003;36(12):1523–1532. doi: 10.1086/375069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim A., Ebnet C., Harste G., Pring-Akerblom P. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 2003;70(2):228–239. doi: 10.1002/jmv.10382. [DOI] [PubMed] [Google Scholar]
- Hota B. Contamination, disinfection, and cross-colonization: are hospital surfaces reservoirs for nosocomial infection? Clin. Infect. Dis. 2004;39(8):1182–1189. doi: 10.1086/424667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama T., Kojima S., Shinohara M., Uchida K., Fukushi S., Hoshino F.B. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 2003;41(4):1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuder S.A., Arthur T., Bisesi M.S., Schaub E.A. Prevalence of infectious diseases and associated symptoms in wastewater treatment workers. Am. J. Ind. Med. 1998;33(6):571–577. doi: 10.1002/(sici)1097-0274(199806)33:6<571::aid-ajim8>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- La Rosa G., Pourshaban M., Iaconelli M., Muscillo M. Quantitative real-time PCR of enteric viruses in influent and effluent samples from wastewater treatment plants in Italy. Ann. Ist. Super. Sanita. 2010;46(3):266–273. doi: 10.4415/ANN_10_03_07. [DOI] [PubMed] [Google Scholar]
- Li R. CRC Press; Boca Raton, FL: 2013. Forensic Biology. [Google Scholar]
- Lundholm M., Rylander R. Work related symptoms among sewage workers. Br. J. Ind. Med. 1983;40(3):325–329. doi: 10.1136/oem.40.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux F.G., Hotz P., Gashi D., Savova-Bianchi D., Oppliger A. Assessment of airborne virus contamination in wastewater treatment plants. Environ. Res. 2014;133:260–265. doi: 10.1016/j.envres.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Meijer A., van der Sanden S., Snijder s B.E., Jaramillo-Gutierrez G., Bont L., van der Ent C.K. Emergence and epidemic occurrence of enterovirus 68 respiratory infections in The Netherlands in 2010. Virology. 2012;423(1):49–57. doi: 10.1016/j.virol.2011.11.021. [DOI] [PubMed] [Google Scholar]
- Meyer F., Paarmann D., D'Souza M., Olson R., Glass E.M., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A., Wilkening J., Edwards R.A. The metagenomics RAST server — a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinforma. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuolale O., Okoh A. Incidence of human adenoviruses and Hepatitis A virus in the final effluent of selected wastewater treatment plants in Eastern Cape Province, South Africa. Virol. J. 2015;12:98. doi: 10.1186/s12985-015-0327-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D., Eloe-Fadrosh E.A., Pavlopoulos G.A., Thomas A.D., Huntemann M., Mikhailova N., Rubin E., Ivanova N.N., Kyrpides N.C. Uncovering Earth's virome. Nature. 2016;536(7617):425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- Payne D.C., Wikswo M., Parashar U.D. VPD Surveillance Manual. 5th ed. C. f. D. C. a. Prevention; Atlanta: 2011. Chapter 13: Rotavirus; p. 13-11. [Google Scholar]
- Pouillot R., Van Doren J.M., Woods J., Plante D., Smith M., Goblick G. Meta-analysis of the reduction of norovirus and male-specific Coliphage concentrations in wastewater treatment plants. Appl. Environ. Microbiol. 2015;81(14):4669–4681. doi: 10.1128/AEM.00509-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu F., Cao J., Su Q., Yi Y., Bi S. Multiplex hydrolysis probe real-time PCR for simultaneous detection of hepatitis A virus and hepatitis E virus. Int. J. Mol. Sci. 2014;15(6):9780–9788. doi: 10.3390/ijms15069780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Lee B.E., Neumann N., Ashbolt N., Craik S., Maal-Bared R., Pang X.L. Assessment of human virus removal during municipal wastewater treatment in Edmonton, Canada. J. Appl. Microbiol. 2015;119(6):1729–1739. doi: 10.1111/jam.12971. [DOI] [PubMed] [Google Scholar]
- Romano M., Montesi F., Rausa G. Treatment plants: viruses in aerosols. G. Ital. Med. Lav. Ergon. 1999;21(1):9–12. [PubMed] [Google Scholar]
- Roux S., Faubladier M., Mahul A., Paulhe N., Bernard A., Debroas D., Enault F. Metavir: a web server dedicated to virome analysis. Bioinformatics. 2011;27(21):3074–3075. doi: 10.1093/bioinformatics/btr519. [DOI] [PubMed] [Google Scholar]
- Rylander R., Andersson K., Belin L., Berglund G., Bergstrom R., Hanson L.A., Lundholm, Mattsby M.I. Sewage worker's syndrome. Lancet. 1976;2(7983):478–479. doi: 10.1016/s0140-6736(76)92583-6. [DOI] [PubMed] [Google Scholar]
- Selvaraju S.B., Selvarangan R. Evaluation of three influenza A and B real-time reverse transcription-PCR assays and a new 2009 H1N1 assay for detection of influenza viruses. J. Clin. Microbiol. 2010;48(11):3870–3875. doi: 10.1128/JCM.02464-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit L.A., Spaan M.S., Heederik D. Endotoxin exposure and symptoms in wastewater treatment workers. Am. J. Ind. Med. 2005;48(1):30–39. doi: 10.1002/ajim.20176. [DOI] [PubMed] [Google Scholar]
- Sundkvist T., Hamilton G.R., Hourihan B.M., Hart I.J. Outbreak of hepatitis A spread by contaminated drinking glasses in a public house. Commun. Dis. Public Health. 2000;3(1):60–62. [PubMed] [Google Scholar]
- Toulouse M.J., Turgeon N., Veillette M., Duchaine C. AIRMON 2014—The 8th International Symposium on Modern Principles for Air Monitoring and Biomonitoring, Marseille, France. 2014. Fluorimetric neuraminidase assay to monitor airborne respiratory viruses. [Google Scholar]
- Tseng C.C., Chang L.Y., Li C.S. Detection of airborne viruses in a pediatrics department measured using real-time qPCR coupled to an air-sampling filter method. J. Environ. Health. 2010;73(4):22–28. [PubMed] [Google Scholar]
- Uhrbrand K., Schultz A.C., Madsen A.M. Exposure to airborne noroviruses and other bioaerosol components at a wastewater treatment plant in Denmark. Food Environ. Virol. 2011;3(3–4):130–137. [Google Scholar]
- Van Hooste W., Charlier A.M., Rotsaert P., Bulterys S., Moens G., van Sprundel M. Work-related helicobacter pylori infection among sewage workers in municipal wastewater treatment plants in Belgium. Occup. Environ. Med. 2010;67(2):91–97. doi: 10.1136/oem.2008.040436. [DOI] [PubMed] [Google Scholar]
- Whon T.W., Kim M.S., Roh S.W., Shin N.R., Lee H.W., Bae J.W. Metagenomic characterization of airborne viral DNA diversity in the near-surface atmosphere. J. Virol. 2012;86(15):8221–8231. doi: 10.1128/JVI.00293-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Schillinger J.A., Sternberg M.R., Johnson R.E., Lee F.K., Nahmias A.J. Seroprevalence and coinfection with herpes simplex virus type 1 and type 2 in the United States, 1988–1994. J Infect Dis. 2002;185(8):1019–1024. doi: 10.1086/340041. [DOI] [PubMed] [Google Scholar]
- Zeng S.Q., Halkosalo A., Salminen M., Szakal E.D., Puustinen L., Vesikari T. One-step quantitative RT-PCR for the detection of rotavirus in acute gastroenteritis. J. Virol. Methods. 2008;153(2):238–240. doi: 10.1016/j.jviromet.2008.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Families represented in the group “other” of graph B with their respective percentage.