Abstract
Multiprotein complexes catalyze vital biological functions in the cell. A paramount objective of the SPINE2 project was to address the structural molecular biology of these multiprotein complexes, by enlisting and developing enabling technologies for their study. An emerging key prerequisite for studying complex biological specimens is their recombinant overproduction. Novel reagents and streamlined protocols for rapidly assembling co-expression constructs for this purpose have been designed and validated. The high-throughput pipeline implemented at IGBMC Strasbourg and the ACEMBL platform at the EMBL Grenoble utilize recombinant overexpression systems for heterologous expression of proteins and their complexes. Extension of the ACEMBL platform technology to include eukaryotic hosts such as insect and mammalian cells has been achieved. Efficient production of large multicomponent protein complexes for structural studies using the baculovirus/insect cell system can be hampered by a stoichiometric imbalance of the subunits produced. A polyprotein strategy has been developed to overcome this bottleneck and has been successfully implemented in our MultiBac baculovirus expression system for producing multiprotein complexes.
Abbreviations: BEVS, baculovirus expression vector system; CFP, cyan florescent protein; CMV, cytomegalovirus; dpa, day of proliferation arrest; dsRed, red fluorescent protein; E. coli, Escherichia coli; EGFP, enhanced green fluorescent protein; EM, electron microscopy; FRET, fluorescence resonance energy transfer; HT, high throughput; kb, kilo base; kDa, kilo dalton; MOI, multiplicity of infection; NMR, nuclear magnetic resonance (spectroscopy); ORF, open reading frame; Ori, origin of replication; p10, p10 baculoviral late promoter; polh, polyhedrin baculoviral very late promoter; R6Kγ, bacteriophage R6Kγ; SDS–PAGE, sodium dodecylsulfate–polyacryamide gel electrophoresis; Sf9, Sf21, Spodoptera frugiperda cell lines 9 or 21, respectively; SLIC, sequence and ligation independent cloning; tcs, TEV protease cleavage site; TEV, tobacco etch virus, resp. a protease (N1A) from this virus; YFP, yellow fluorescent protein
Keywords: ACEMBL, Automation, Baculovirus, BEVS, High-throughput, Insect cells, Mammalian host, MultiBac, pET-MCN, Polyprotein, Recombineering, Robotics, Structural biology
1. Introduction
The structural proteomics initiative SPINE (Structural Proteomics IN Europe) was a highly successful European project aimed at production and structural characterization of mostly single proteins with important roles in human health. By building on these successes, SPINE2 (full name SPINE2-COMPLEXES) addressed more challenging biological systems which consists of many interlocking protein subunits assembled in complexes to exert their biological function. Multiprotein complexes are emerging as cornerstones of biological activity in the cell, and deciphering their structure and function is imperative for advancing research in health and disease (e.g. Alberts, 1998, Nie et al., 2009a). Many essential protein complexes, particularly in human cells are comparatively hard to come by, thus complicating their study. Low endogenous levels and sample heterogeneity often impede purification from native sources in the quality and quantity required for structural studies, especially by X-ray diffraction methods requiring highly purified material that can crystallize. Recombinant techniques can overcome several of the bottlenecks encountered, and can further provide the means to modify complexes at the gene level resulting in alteration of the protein complex subunits to meet the high quality requirements for structural analysis. Consequently, SPINE2-COMPLEXES placed considerable emphasis on developing and implementing technologies, protocols and reagents to facilitate protein complex production for structural studies, in prokaryotic and eukaryotic host organisms. In this contribution, we will review the high throughput platforms at Strasbourg and Grenoble as well as the practical considerations concerning setting up and running a eukaryotic expression facility. New approaches for eukaryotic protein production, including the benefits of polyprotein design, will also be discussed.
Owing to a large part to structural genomics efforts such as the SPINE project, affordable methods and equipment have been developed to automate molecular cloning of expression constructs, recombinant expression screening and purification. There are obvious advantages of automation in the molecular biology laboratory. Procedures can be carried out in parallel and scaled-up accordingly to process large amounts of samples at reasonable cost. Automation puts constraints on the robustness of protocols to be implemented which are certainly more stringent than regular manual laboratory intervention requires. In our experience, a good protocol yielding reproducible results, be it for cloning, transformation or expression screening, by no means guarantees that it can be scripted without further ado into a robotics routine. Rather, seemingly robust protocols for manual experimentation often have to be optimized further to work robustly in a parallelized manner on a robot. This is beneficial to the laboratory implementing automation in several ways. Not all experiments will be carried out on a robot even in the most well-equipped laboratory, and the general success rate of manual experimentation, at least in our laboratories, definitely increased when protocols working also robustly on robots become available to the researchers. In turn, automated processes on a robot can run in parallel for large numbers of reactions with unsurpassed precision, ensuring error-free operations once robust routines are implemented.
Automation in molecular biology laboratories can come in many forms ranging from simple modules for individual tasks that are integrated into a pipeline requiring frequent manual handling, to large systems which perform many tasks on an integrated robotic platform, with a minimum of manual intervention by the researcher. The high-throughput (HT) expression screening platform for co-expression in Escherichia coli at IGBMC Strasbourg and the ACEMBL platform at EMBL Grenoble represent two complementary solutions to some of the challenges of automated protein complex production (Fig. 1 ).
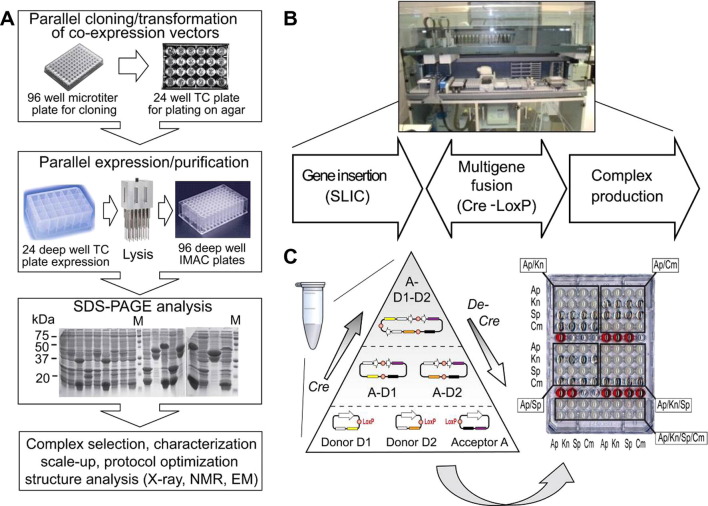
Fig.1.
SPINE2: automation for HT protein complex production. (A) The high throughput pipeline for screening co-expression constructs at IGBMC Strasbourg is shown in a schematic representation. Relying on semi-automated procedures, co-expression constructs are prepared in a parallel fashion by the cloning method of choice. This can be carried out in small volumes for example on a 96 well micro-titer plate. Constructs are transformed and plated on agar provided on 24 well tissue culture (TC) plates (top). Parallel expression of single colonies is performed in 2–4 ml miniculture volumes in deep well TC plates screening a variety of parameters (media, temperature, induction, etc.). Cells are pelleted and lysed by a multitip sonicator. Overproduced soluble protein complexes are purified from the cleared lysate by metal affinity purification on deep well plates using automated procedures (middle). The complexes retained on the affinity resin are analyzed by SDS–PAGE (below). Promising complex candidates are prioritized for scale-up, biochemical and biophysical characterization and structure determination (bottom). (B) The multigene cloning and expression pipeline ACEMBL at the EMBL Grenoble relies on fully automated protocols scripted into robotic routines that are implemented on a liquid handling workstation (a Tecan Evo II LHW is shown). One or several genes are inserted in specifically designed synthetic plasmids called Donors and Acceptors (Table 1) by sequence and ligation independent cloning (SLIC). These plasmids are next concatamerized by Cre–LoxP fusion into multigene constructs for protein complex production. The combination of SLIC and Cre–LoxP reactions for multigene vector construction is termed tandem recombineering (TR). The Cre-reaction is an equilibrium reaction and can also be exploited for disassembling constructs. (C) The Cre–LoxP fusion reaction is detailed in a schematic representation (left). In a single Eppendorf tube, Donors and one Acceptor (Table 1), each containing a LoxP sequence, are combined with Cre recombinase. Cre–LoxP fusion generates in this tube all possible combinations in an equilibrium reaction (arrows marked by Cre and De-Cre). After transformation and plating, desired Donor–Acceptor fusions are selected by the specific antibiotic combination conferred by the resistance markers on the plasmids. Identification of the desired recombinants can be carried out on a 96 well microtiter plate (right). D is Donor, A is Acceptor. A–D and A–D–D denote Donor–Acceptor fusions. LoxP sites are shown as colored balls. Ap, Cm, Kn, Sp stand for antibiotics ampicillin, chloramphenicol, kanamycin and spectinomycin. Antibiotics combinations are boxed. Red dye is added in wells on the microtiter plate for orientation. ACEMBL system Donors and Acceptors are listed in Table 1. C is adapted from Bieniossek et al. (2009) with permission of the publisher.
2. The SPINE2 high throughput platforms for expression of protein complexes in E. coli
2.1. The Strasbourg experience
The platform in Strasbourg is composed of individual modules, each for a specialized function in the recombinant protein expression process that integrates into the pipeline shown (Fig. 1A). Co-expression vectors are put together in a parallel fashion, for example in 48 or 96 well microtiter or PCR plates. Procedures involving restriction enzymes and ligation, or ligation independent cloning methods such as LIC (Novagen, Merck Biosciences), Gateway (Invitrogen) (Walhout et al., 2000) or In-Fusion (Clontech Takahara) (Berrow et al., 2009) can be applied, depending on the project and according to the preference of the user. Transformation is followed by plating on agar supplemented with antibiotic(s), optionally on six well or 24 well tissue culture (TC) plates at appropriate dilutions. Single colonies are used to inoculate liquid broth (LB) or terrific broth (TB) minicultures (2–4 ml volume) in 24 deep well TC plates. Autoinduction or addition of IPTG for induction can be chosen if expression plasmids relying on a lac operator are used (Studier, 2005). Lysis is carried out in the deep well plates by a multitip sonicator. The lysates are cleared by plate centrifugation and applied to affinity resin presented in a 96 deep well plate. At least one of the components of the complex studied is designed to incorporate a purification tag. SDS–PAGE analysis of the resin-bound material then reveals the outcome of the expression experiment. Promising complexes are selected and production is scaled up off-line for characterization by biochemical and biophysical methods and functional assays, followed by structure determination involving NMR, EM and X-ray crystallography.
This co-expression pipeline is at the core of the Strasbourg system and performs successfully with a number of co-expression systems developed by the Strasbourg laboratory during the SPINE and SPINE2 projects, as well as with co-expression systems from other laboratories (Busso et al., this issue). Specifically, this pipeline fully supports the pET-MCN/pET-MCP multi-expression system developed at IGBMC for protein complex expression in E. coli. This system relies on a cyclic multi-expression strategy based on several vectors that can be conjoined by restriction/ligation to yield multigene expression constructs (Romier et al., 2006, Perrakis and Romier, 2008, Diebold et al., this issue). This co-expression system has been instrumental for producing numerous complexes for high-resolution structure determination. Application of the pET-MCN/pET-MCP system has further revealed the paramount importance of tag placement on complex production efficiency, and the requirement to address this issue in a combinatorial manner. These results and the use of the pET-MCN/pET-MCP method are described in detail in an accompanying paper in this SPINE2 special issue by Diebold et al.
2.2. The Grenoble experience
High-resolution structure elucidation of single proteins often requires mutations, truncations or the elimination of low complexity regions that introduce heterogeneity in the sample and may preclude crystallization. For single proteins or small binary or trimeric complexes, this can be achieved still in a reasonable timeframe, for example by applying parallelized cloning, expression and purification approaches. As the size and number of protein subunits in a complex of interest increases, the work-load augments excessively and can become inhibitory, in particular if the recombinant multigene expression vectors required to produce this complex have to be reconstructed every time from scratch. This challenge was addressed at EMBL Grenoble by creating the ACEMBL platform for multigene expression vector construction and complex production.
3. The ACEMBL system facilitates high-throughput recombineering for protein complex production
ACEMBL is the result of deconstructing multigene assembly into two simple steps of gene integration and plasmid concatamerization, which can both be automated and performed by a liquid handling workstation (Fig. 1B). Sequence and ligation independent cloning (SLIC) was chosen for gene insertion (Li and Elledge, 2007). SLIC relies on the exonuclease activity of T4 DNA polymerase to produce sticky ends on DNA fragments that are annealed and joined together covalently upon transformation by the DNA repair machinery of the E. coli cell used. Gene insertion occurs in small (2 kb) synthetic circular DNA modules called Donor and Acceptor plasmids (Table 1 ). Donors contain a conditional origin of replication which makes their propagation dependent on expressing the pir gene in the host used for cloning. Acceptors contain a ColE1 derived, regular origin of replication. Donors are suicide vectors in all common cloning strains that are pir negative, and are only propagated in those strains if fused with an Acceptor providing a regular replicon (Bieniossek et al., 2009, Nie et al., 2009b).
Table 1.
ACEMBL systems for multiprotein complex expression.
 |
Fusions of Donors and an Acceptor, each containing one or several recombinant genes of interest, are catalyzed by Cre recombinase, which fuses DNA molecules containing a LoxP sequence. In an Eppendorf tube containing several Donors and one Acceptor, each with a different resistance marker, all possible multigene fusions are made if Cre enzyme is added, in an equilibrium reaction favoring disassembly (Fig. 1C) providing a convenient combinatorial option. Fusions containing the desired gene combinations are identified by their unique resistance marker combination via antibiotic challenge on a microtiter plate (Fig. 1C).
SLIC relies on one enzyme (T4 polymerase) and one protocol (annealing). The Cre–LoxP fusion relies also on one enzyme (Cre recombinase) and one protocol (incubation). This simplicity confers utter robustness and therefore lends itself to automation (Nie et al., 2009b). A welcome added value is the very low investment necessary in terms of material when using these enzymes, which can either be purchased at low cost (T4 DNA polymerase) or produced and purified efficiently in large amounts on site (Cre recombinase). Numerous multiprotein complexes were produced in E. coli using the ACEMBL system from plasmids constructed by SLIC/Cre–LoxP tandem recombineering (TR), notably including, among many others, important protein–RNA complexes and also large transmembrane assemblies such as the bacterial holotranslocon complex (Fig. 2 ). These examples show that the ACEMBL system greatly facilitates cloning and expression of protein complexes in E. coli.
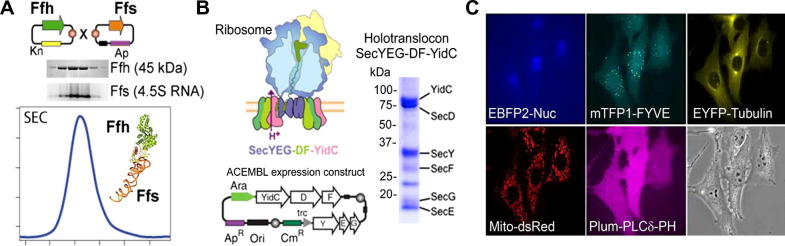
Fig.2.
Donor–Acceptor recombineering exploits. (A) Expression of the signal receptor particle SRP in E. coli was achieved by ACEMBL. A dual expression plasmid encoding for the protein subunit Ffh and the RNA subunit Ffs was created by fusing Donor and Acceptor plasmids containing the encoding genes. Co-expression resulted in SRP which eluted in a symmetric peak from a size-exclusion column. SDS–PAGE (Ffh) and agarose gel (Ffs) analysis of SEC fractions revealed the complex. SRP is depicted as a model (based on PDB entry 2IY3) in the inset. (B) E. coli was used as expression host to produce the SecYEG-DF-YidC holotranslocon. A schematic drawing of this transmembrane multiprotein complex bound to the ribosome is shown (top). A multigene ACEMBL construct was used to express and purify the holotranslocon (below). A gel section from SDS–PAGE of the complex purified from detergent solubilized membrane fractions shows all expected components (right). (C) Efficient multigene expression in mammalian cells. Five fluorescent marker proteins were inserted into Donors and an Acceptor containing mammalian cell active promoter elements. All cells produced all proteins at identical levels in all cells of the mammalian culture. EBFP2-Nuc stains the nuclei, mTFP-FYVE binds a phosphoinositol-phosphate (PI-3-P), tubulin is labeled with EYFP, Mito-dsRed localizes to mitochondria, and Plum-PLCδ-PH binds a phospho-inositol-biphosphate (PI-4,5-P2) The porcine aortic endothelial cells analyzed are shown in light field (bottom right). A and B are adapted from Bieniossek et al. (2009), C is adapted from a time-lapse movie in Kriz et al., 2010 (image credit to P. Berger, PSI Villigen, Switzerland).
3.1. Extending ACEMBL to eukaryotic expression systems
E. coli has been and remains the major host for heterologous production of proteins in many laboratories, notably when large quantities are required for biochemical and biophysical studies and structure analysis. Eukaryotic proteins and protein complexes, however, may impose requirements on the expression host which E. coli cannot support properly. These can include a specialized eukaryotic folding machinery, or the necessity for authentic processing and post-translational targeting and modifications to gain activity. Eukaryotic expression systems therefore have attracted considerable interest since some time as an alternative to expression in E. coli. Currently, two eukaryotic expression systems are increasingly being used for eukaryotic protein expression: the baculovirus expression vector system (BEVS) for protein production in insect cells, and mammalian expression systems for protein production in mammalian cells (Nettleship et al., 2010).
Introducing foreign genes into mammalian cells remains a challenge to date, especially if many genes should be introduced simultaneously. Genetic engineering of mammalian cells with transgenes, however, is essential in contemporary biology. Not only structural biology applications benefit from introducing foreign genes efficiently into mammalian cells. Reprogramming of somatic cells into stem cells by co-expressing an array of specific transcription factors is a prominent example. Efficient simultaneous monitoring of many parameters in living cells with fluorescent-protein sensors is an essential prerequisite for cell biology experiments, for example if multi-component pathways are to be followed precisely. Achieving robust results in all these experiments using mammalian cells has been a formidable task, requiring specialist knowledge and highly trained personnel. Existing approaches were hampered by many impediments, rendering it virtually impossible to generate mammalian cell populations in which every cell simultaneously expresses all desired genes, for labeling specific cell compartments, or for assembling into multiprotein complexes.
To tackle this bottleneck, the ACEMBL technology concept was extended to introducing multiple foreign genes simultaneously into mammalian cells by using a single multigene plasmid which is rapidly built from Donors and an Acceptor by tandem recombineering as outlined above (Kriz et al., 2010). This work was carried out in a collaboration between EMBL Grenoble and the Paul Scherrer Institute in Villigen, Switzerland. Highly efficient co-expression of currently five genes in a cell transfected with a Donor–Acceptor multigene construction was demonstrated (Fig. 2C). The generation of stable multigene expresser cell lines by using specific integration elements that can be readily introduced by the TR method was also achieved. The viability of the transfected cells was not adversely affected, as they show the expected behavior for example when stimulated with growth factors (ibid). Provision of mammalian cell compatible promoter and terminator DNA sequences on the ACEMBL plasmid modules originally conceived for E. coli expression, lead to the MultiMam system (Table 1). MultiMam consists of small synthetic Donors and Acceptors that are fully compatible with the robotic routines of the ACEMBL platform for multigene construct assembly by TR, thus setting the stage for efficient multigene expression, optionally transient or stable, in mammalian cells for structural molecular biology and numerous other applications.
Finally, the ACEMBL approach was implemented for the baculovirus/insect cell system by adapting our time-tested MultiBac technology (Berger et al., 2004, Fitzgerald et al., 2006, Bieniossek et al., 2008) for the requirements of the automated TR method (Table 1). The MultiBac methodology, the protocols and reagents, and the at times spectacular exploits, have been reviewed in some detail recently (Trowitzsch et al., 2010). We strove to maximally streamline procedures for reliable and robust use of the baculovirus expression vector system (BEVS) as a standard technique for protein expression in laboratories, not significantly more complicated than recombinant expression in E. coli, which has become routine practice virtually everywhere. Following our strategy of deconstructing multigene vector generation, we reengineered and streamlined our MultiBac plasmids by synthesizing small Donors and Acceptors containing baculovirus specific promoters and terminators. In addition to other characteristic features of the ACEMBL system such as homing endonuclease sites for inserting multiple expression cassettes, the new plasmids also contain DNA sequences required for integrating the expression cassettes of interest into the genome of our engineered MultiBac baculovirus by Tn7 transposition (Table 1). The classical head-to-head dual-expression-cassette design of the original MultiBac plasmids, or comparable plasmids such as pFastBacDual (Bac-to-Bac system, Invitrogen) can be easily recapitulated from the new Donors and Acceptors if desired, by iteratively inserting a second (or, by the same token, third, fourth, etc.) expression cassette utilizing homing endonuclease sites provided on all ACEMBL plasmids (Bieniossek et al., 2009, Nie et al., 2009b). Thus, a theoretically unlimited number of foreign genes can be incorporated into the recombinant baculovirus for heterologous multiprotein expression in infected insect cell cultures.
The ACEMBL systems shown in Table 1 are available from ATG-biosynthetics (www.atg-biosynthetics.com), and are called MultiColi (E. coli system), MultiMam (mammalian system) and MultiBacTurbo (BEVS).
4. Tips and tricks for eukaryotic protein production by BEVS
When setting up the eukaryotic expression facility (EEF) at EMBL Grenoble for baculovirus expression using our MultiBac BEVS, we strove to control the expenditures of equipment and consumables while maintaining the superior performance of the system (Bieniossek et al., 2008, Trowitzsch et al., 2010). We use regular screw-capped Erlenmeyer flasks (250 ml to 2.5 L volume) and common table-top shakers in a room constantly kept at 27 °C, instead of the significantly more expensive equipment commonly encountered in baculovirus expression facilities (spinner flasks, magnetic stirrers, incubators, fermentors, wave bioreactors, etc.). The last major cost factor when operating the facility is the media used for cell culture. We undertook to test a number of commercially supplied media (liquid and dry powder), as well as own formulations based on literature from the early days of baculovirus expression and insect cell culture. We systematically compared cell viability and growth curves, variation from batch-to-batch, and protein production efficacy of the media tested, in relation to the cost and effort incurred in their purchase and/or preparation. To our surprise, when informed about our ongoing study, commercial suppliers commenced offering significant discount rates on their products, effectively disqualifying our own media formulations, which tended to suffer from variations between batches prepared. The outcome of our study is compiled in Fig. 3 . Formerly, we used SF900 II SFM serum free media (Gibco Life Technologies, Invitrogen) with excellent results for all operations (transfection, virus preparation, amplification and storage, protein expression) in the EEF. Among those received, the most competitive offer was for Hyclone (Thermo Fisher). The Gibco media proved to be clearly superior to Hyclone in supporting cell growth to higher densities, however, in the region relevant to our protocols (0.5–2.5 million cells per ml), both media performed equally well (Fig. 3A). We assayed protein expression using a MultiBac virus expressing YFP and a test protein to infect Sf21 cells adapted to the media analyzed. Both YFP expression levels (Fig. 3B) and expression and solubility of the test protein (Fig. 3C) were equivalent. We conclude that Hyclone media supports baculovirus expression in the EEF to our full satisfaction, at a fraction of the costs hitherto incurred.
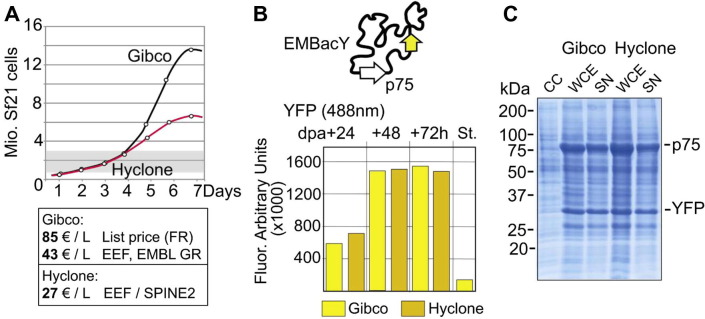
Fig.3.
Sf21 growth and protein production: controlling facility costs. Media performance of several suppliers was assessed in SPINE2. (A) Sf21 growth curves for SF900 II SFM from Gibco (Life Technologies, Invitrogen) and Hyclone (Thermo Fischer Inc.) are shown. Both are serum free media. The Gibco media supports growth to high cell densities. Both media perform equally in the region (shaded in gray) relevant for protein complex expression in the laboratory (0.5–2.5 Mio cells/ml). Hyclone per liter cost is at a third of the Gibco list price and 40% below the best offer to the SPINE2 consortium and the eukaryotic expression facility (EEF) at EMBL Grenoble. (B) A recombinant baculovirus (EMBacY-p75) simultaneously expressing YFP and a test protein (p75) was used to infect Sf21 cells adapted to the Gibco and Hyclone media, respectively. YFP expression per 1 Mio cells in both media was identical. St stands for a defined fluorescent protein standard (used to calibrate for 100,000 arbitratry units), dpa stands for day of proliferation arrest in the infected culture. (C) Coomassie blue stained SDS–PAGE of the test expression shows comparable levels of recombinant protein for both media. CC stands for uninfected cell control, WCE for whole cell extract, SN for cleared lysate after sonication and centrifugation. Bands corresponding to YFP and p75 are indicated.
A recurring subject of discussion when considering co-expression of proteins by BEVS is the utility of the co-infection approach, where recombinant baculoviruses, each containing one foreign gene of interest, are used to co-infect an insect cell culture for producing the ensemble of proteins encoded by the baculoviruses. As widely documented, co-expression of subunits of a multiprotein assembly is often mandatory for production of high-quality complexes. Proper folding and/or solubility of the subunits may critically depend on each other, thus ruling out reconstitution of individually purified components to achieve the complex. The realization of this necessity was originally the impetus for creating MultiBac, where all DNAs encoding a protein complex are assembled into and expressed from a single composite multigene baculovirus (Berger et al., 2004).
Nonetheless, co-expression of two or more proteins in the baculovirus system can also be achieved by co-infection of the insect cells with several baculoviral stocks encoding the individual subunits. This approach can offer advantages, in particular for exploratory screening of putative interaction partners prior to large scale expression. Co-infection also offers a possibility to adjust individual virus titers by simply using more or less volume of a particular virus for infection, thus choosing the ratio between viruses and thereby possibly also the amounts of proteins produced in the culture. Simply speaking, if single-gene baculoviruses are at hand, why not use them for testing in a straight-forward experiment whether or not the encoded proteins interact, at least analytically, prior to up-scaling? Undoubtedly, even such seemingly straight-forward experiments need to be carried out with professional care to avoid false interpretations for example due to a virus titer “going off”, i.e. absence of protein caused by a weak virus must not be confused with absence of interaction or incorporation into a complex.
In a pilot experiment, the Strasbourg group addressed the requirements for co-infection experiments using baculoviruses. Results of this experiment are shown in Fig. 4 , illustrating the limits and the difficulty to optimize co-infection experiments for achieving tangible results. Insect cells were co-infected with two viruses expressing fluorescent proteins dsRed and EGFP, respectively. Cells infected either with dsRed or EGFP or both were counted using fluorescence microscopy (Fig. 4A). These experiments already demonstrate that, in order to maximize co-expression, optimization is absolutely required. The best ratio between viruses needs to be determined experimentally, by varying the multiplicity of infection (MOI) i.e. the number of infectious virus particles of each kind counted against the insect cells present in the infected culture. The percentage of cells that are productively co-infected varies significantly with the amount and ratio of individual viruses used for co-infection. Importantly, in these experiments, even after optimization, only a fraction (50%) of all insect cells effectively expressed both proteins simultaneously (Fig. 4B). Taken together, these results certainly suggest that co-infection experiments, seemingly easy, may need significant investment of time and effort and should be approached with caution to arrive at interpretable and reliable outcomes.
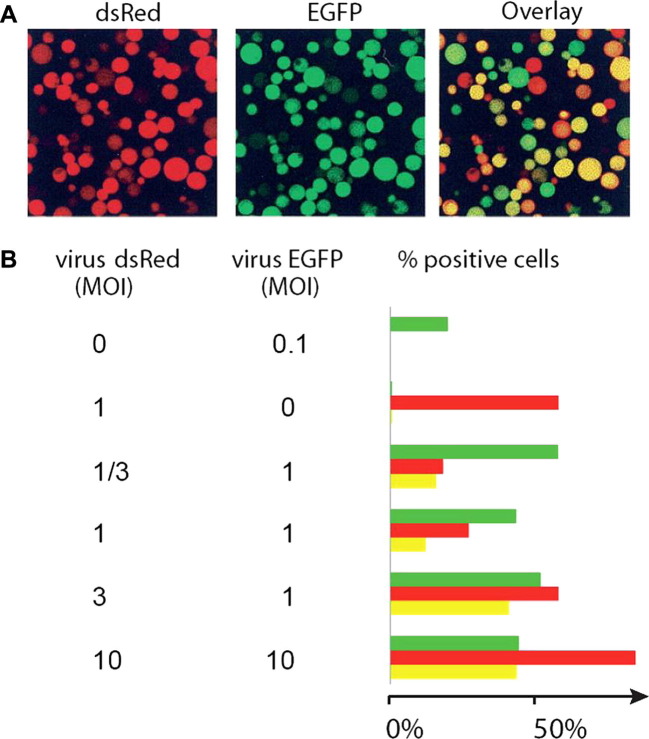
Fig.4.
Limits of multigene expression by baculovirus co-infection. Infection of Sf9 cells with one or two baculoviruses (expressing dsRed protein or EGFP, respectively) 48 h post-infection are shown. (A) Micrographs of the same field of Sf9 cells showing specific fluorescence signals for dsRed (left), and for EGFP (middle) and an overlay of the two (right). Multiplicity of infection (MOI) was one for both viruses. Cells that express dsRed are colored red, cells expressing EGFP are colored green. Cells that effectively co-express both dsRed and EGFP appear yellow in the overlay. (B) The influence of virus titers on co-expression efficacy was analyzed, and the results are shown in a graphical representation. The green, red and yellow bars represent the proportion of cells that expressed dsRed, EGFP or both dsRed and EGFP, respectively. At least 1500 cells in each condition were analyzed. Saturating co-infection with both viruses was not achieved in this experiment.
5. Polyproteins: a new concept for producing protein complexes
Since its introduction in 2004, the MultiBac system has been prolifically used by us and many other laboratories, including users and visitors in the EEF at EMBL Grenoble to generate what in the past would have been considered difficult targets. Many proteins and their complexes were produced successfully, often for the first time (Trowitzsch et al., 2010). MultiBac relies on infecting insect cells with a single virus containing all foreign genes of choice. When logging and interpreting the outcomes of many complex expressions, we arrived at two important conclusions. Firstly, even though all genes were present on one MultiBac baculovirus used for complex expression, it still occurred occasionally that one or the other subunit was expressed at much lower levels than the remaining subunits, thus reducing the yield of intact complex purified. This feature of inhibitory imbalance in individual expression levels appeared to be exacerbated in the case of large complexes with many subunits, and proved to be difficult to rationalize (although it appeared to affect mostly proteins larger than 100 kDa). Secondly, we observed that we could express very large single proteins (400–500 kDa and larger) efficiently by using MultiBac.
We then considered a viral strategy. For example, SARS coronavirus, which is the virus that causes severe acute respiratory syndrome (SARS), is a positive and single stranded RNA virus belonging to a family of enveloped coronaviruses (Peiris et al., 2003). Genome expression is realized by translation of two large open reading frames (ORFs), which are two polyproteins. The polyproteins are then processed into 16 smaller subunits by proteases contained within the polyproteins. One polyprotein of SARS coronavirus is 700 kDa in size (Gorbalenya et al., 2006).
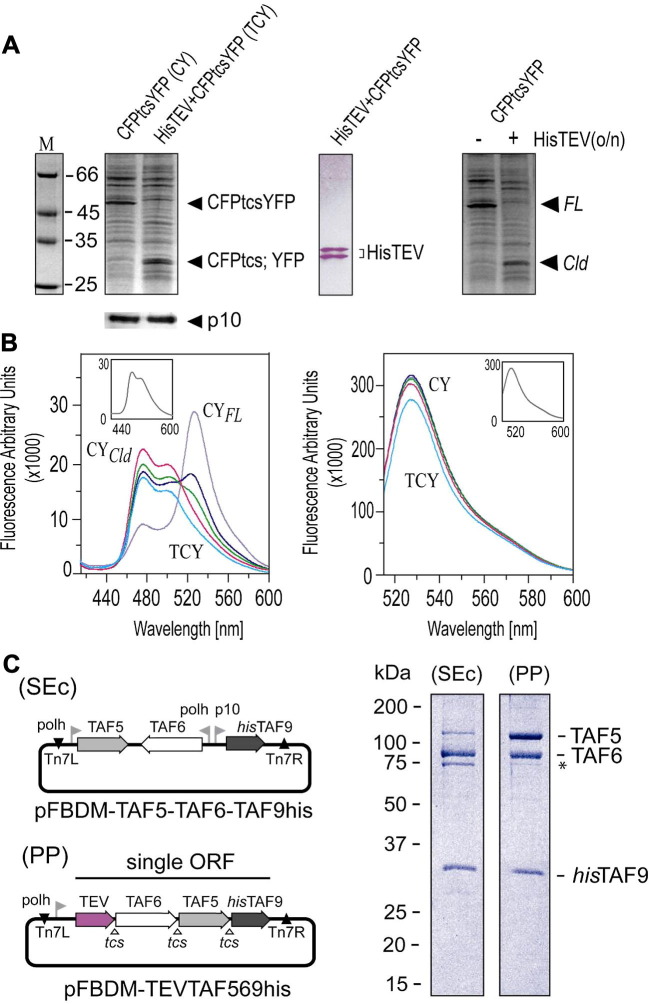
We asked whether a similar approach could be exploited to produce protein complexes from large polyproteins by the baculovirus system, thus potentially overcoming the limitations imposed by stoichiometric imbalance of the subunits produced (Fig. 5 ). In a first approach, we expressed by MultiBac a fusion protein (CFPtcsYFP) of cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP), connected by a short linker incorporating the amino acids that are recognized and cleaved by protease N1A from tobacco etch virus (TEV). CFP and YFP represent a Förster pair which can be used for fluorescent resonance energy transfer (FRET) measurements. If CFP and YFP are closely spaced as in our fusion protein, excitation of CFP at a wavelength of 400 nm will result in excitation of YFP by Förster transfer, leading to an emission spectrum characteristic of YFP. Cleared lysates of insect cells infected by the virus encoding for CFPtcsYFP were analyzed by SDS–PAGE showing an over-expressed band at 50 kDa. Co-expression of TEV protease resulted in complete cleavage of the fusion protein as evidenced by SDS–PAGE. The expression of TEV protease interfered neither with cell proliferation nor with the baculoviral life cycle upon infection. In a separate experiment, the fusion protein could also be quantitatively cleaved by adding purified TEV protease to the lysate and incubation overnight, resulting in appearance of the same bands as in the TEV/CFPtcsYFP co-expression experiment (Fig. 5A). Analysis of the lysates revealed efficient FRET for the fusion protein. Interestingly, exclusively the spectrum of CFP was observed in the TEV/CFPtcsYFP co-expression experiment upon excitation at 400 nm. FRET is an extremely sensitive tool for measuring close proximity of suitable fluorophores. The absence of any residual YFP signal in the lysate of cells co-expressing TEV and fusion protein confirm complete cleavage of the fusion at the TEV protease site. We can confidently rule out the possibility of cleavage occurring during or immediately after cell lysis, as addition of TEV protease to a lysate containing fusion protein only gradually reverted the YFP emission spectrum to the CFP spectrum overnight (Fig. 5B). Excitation at 488 nm revealed the presence of comparable amounts of YFP protein in all samples analyzed.
Fig.5.
Protein complex expression from a polyprotein. (A) A fusion protein of CFP and YFP, linked by amino acid residues corresponding to a cleavage site for TEV protease, was constructed and expressed using the MultiBac system, alternatively with and without TEV protease co-expressed. A gel section from SDS–PAGE is shown, with the band corresponding to the fusion protein indicated. Co-expression of TEV protease results in loss of this band and the appearance of bands at half of its molecular weight (left). Expression of the baculoviral protein p10 is shown as an internal control (below). Presence of TEV protease in the co-expression was confirmed by Western blot using antibody specific for the 6 histidine N-terminal tag of the protease, revealing a doublet (middle). Addition of TEV protease to the lysate of the fusion protein resulted likewise in cleavage at the specific proteolytic site (right). FL denotes uncleaved, full-length fusion protein, Cld denotes protein cleaved at the TEV protease site, tcs stands for TEV protease cleavage site. (B) Excitation of CFP in the CFPtcsYFP pair results in FRET and an emission maximum characteristic of YFP (curve marked CYFL). Addition of TEV protease to the lysate containing the fusion gradually converts the curves (red, purple, blue) to a spectrum resembling CFP emission by eliminating FRET. An emission spectrum of CFP expressed alone is inset for comparison. The spectrum of lysate from the co-expression of TEV and the fusion (TCY) is identical to CFP only (left). Spectra of all lysates excited at the YFP excitation wavelength (488 nm) confirm that all samples tested contain comparable amounts of flourescent protein (line color as before). For comparison, the spectrum of YFP expressed alone is inserted (right). (C) A 400 kDa human transcription factor complex composed of proteins TAF5, TAF6 and TAF9 (with 6 histidine tag) was expressed by using the MultiBac system from single expression cassettes (SEc) or, alternatively, from a polyprotein (PP) joining TEV protease and the three subunits by TEV protease cleavage sites followed by purification by IMAC. Sections from SDS–PAGE are shown on the right. Bands corresponding to TAFs are indicated. A band corresponding to a degradation product of TAF6 is marked with an asterisk. Expression from single cassettes results in complex significantly depleted of TAF5. Polyprotein expression, in contrast, results in stoichiometrically balanced sample and reduction of degradation. Protein was purified from equivalent amounts of cells infected by SEc or PP expressing virus, respectively.
We have reported a 400 kDa transcription factor subcomplex of human general transcription factor TFIID composed of two copies each of the TBP associated factors (TAFs) 5, 6 and 9 (Fitzgerald et al., 2007). This complex was expressed by MultiBac from a single virus containing three separate expression cassettes (Fig. 5C). The smallest subunit, TAF9, contained a 6 histidine tag. Purification of the complex by IMAC revealed substoichiometric production of the largest subunit, TAF5 (100 kDa). The eluate contained an excess of TAF6–TAF9 dimer. Purification of the intact complex was nonetheless possible by a further affinity step taking advantage of a calmodulin binding peptide present on TAF5 (Fitzgerald et al., 2007). However, a large portion of the protein produced was lost due to the observed stoichiometric imbalance. We now created a single ORF encoding for TEV protease, TAF6, TAF5 and TAF9. Again, TAF9 contained a 6 histidine tag in this polyprotein. Surprisingly and to our delight, expression of the polyprotein by MultiBac not only revealed stoichiometrically balanced sample production, but also indicated that TAF5 production from the polyprotein had in fact “caught up” to the levels previously obtained for TAF6 and TAF9 (Fig. 4C).
5.1. The pPBac vector for polyprotein expression
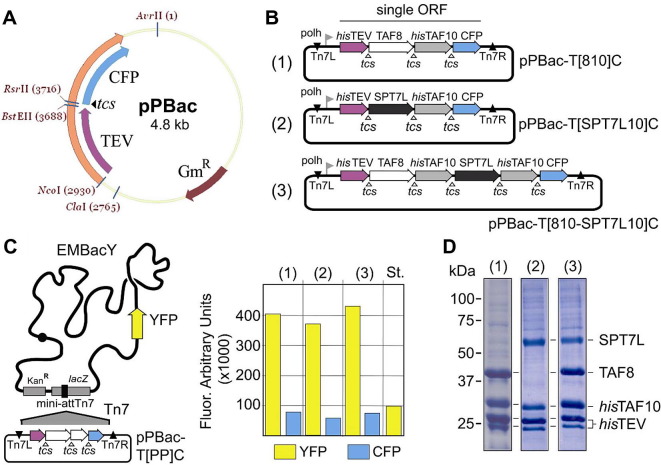
Encouraged by our success with the TFIID subcomplex, we went on to generate a plasmid, pPBac, specifically designed for producing polyproteins with the Multibac system (Fig. 6 ). In addition to elements required for Tn7 transposition into the MultiBac baculoviral genome, pPBac contains an expression cassette flanked by a polyhedrin promoter and a terminator derived from the SV40 polyA signal sequence. The cassette contains an ORF encoding for TEV protease, followed by a short linker DNA sequence with a BstEII restriction site and a RsrII restriction site. The ORF is completed by the gene encoding for CFP, cloned in frame with TEV, and containing a TEV protease cleavage site at its N-terminus. ORFs encoding for polyproteins are inserted into pPBac by using the BstEII and RsrII cleavage sites. Both enzymes are asymmetric cutters with recognition sequences encompassing seven nucleotides each. LIC or SLIC procedures can likewise be used to insert ORFs into pPBac linearized at the BstEII and/or RsrII sites, maintaining the reading frame. Polyproteins produced from pPBac will start with TEV protease and end with CFP. Monitoring CFP emission by using a fluorescence spectrophotometer will report in “real-time” on the expression level of the polyprotein in probes taken from the expressing cell culture. TEV protease expressed from pPBac contains a 6 histidine tag to allow for detection of TEV with an antibody specific for the polyhistidine tag. By analyzing the position of TEV in a Western blot, expression of the polyprotein as well as (in)complete cleavage can be monitored. Completely processed polyprotein will only produce a TEV specific signal in the blot at the molecular weight corresponding to the protease. Incomplete processing would result in a staining pattern resembling a ladder. For more than 40 expressions from pPBac, we thus confirmed by Western blotting quantitative cleavage of all polyproteins produced to date (IB, unpublished).
Fig.6.
BEVS pPBac vector for polyprotein expression of complexes. A new plasmid, pPBac, for expressing polyproteins with the baculovirus system was constructed. The plasmid map is shown (A). pPBac contains a gene for TEV protease linked to a CFP gene, spaced apart by a BstEII recognition site, followed by a RsrII site and a TEV protease cleavage site. pPBac contains all elements required for Tn7 transposition into a recipient baculovirus. (B) Polyprotein constructs for producing TAF8 and TAF10 (1), TAF10 and SPT7L (2) , or TAF8, SPT7L and two copies of TAF10 (3) were generated as shown. (C) Constructs were inserted via Tn7 transposition into EMBacY baculovirus expressing YFP from the backbone (left). Tracking YFP and CFP expression simultaneously evidenced production of YFP and of the polyprotein in the expressions (right). St stands for a standard sample (100,000 counts) used to calibrate the spectrophotometer. (D) SDS–PAGE sections of IMAC batch purifications are shown. All polyprotein specimens are processed completely resulting in stoichiometrically balanced product. Bands corresponding to SPT7L, TAF8, TAF10 and TEV protease are observed. Samples are numbered 1–3 as in B and C.
Recently, a small TAF containing complex (SMAT) was discovered in nuclear extracts from HeLa cells, containing TAF8, TAF10 and SPT7L, a subunit specific for the SAGA coactivator complex (Demény et al., 2007). We used pPBac to produce the subunits encoding for SMAT from polyproteins. Two copies of TAF10 were proposed to exist in SMAT. We therefore created a corresponding polyprotein that contained two copies of the TAF10 gene in the ORF (Fig. 6B). The polyproteins were inserted into EMBacY, a baculovirus where we had inserted YFP as a single expression cassette into the backbone (Bieniossek et al., 2008, Trowitzsch et al., 2010). YFP fluorescence can be used to monitor virus performance in infection experiments using EMBacY. We monitored in parallel the emission of YFP (from the virus backbone) at 488 nm excitation, and of CFP (from the polyprotein) at 400 nm excitation (Fig. 6C). YFP and CFP intensities were similar for the three polyproteins produced, indicating comparable virus performance and polyprotein expression levels. CFP intensities were consistently lower as compared to the YFP levels owing to the lower (10%) quantum yield of the former fluorophore. The polyproteins were again completely processed, resulting in stoichiometrically virtually balanced expression of the subunits (Fig. 6D), thus compellingly validating our approach and the utility of the pPBac vector for polyprotein expression by BEVS.
6. Conclusions
Technology drives discovery. In this contribution, we have described the complementary HT pipelines for protein complex production implemented during SPINE2-COMPLEXES at IGBMC in Strasbourg and at the EMBL in Grenoble. The SPINE2C consortium provided a rich environment for developing expression technologies, and several co-expression systems have been designed, validated and successfully implemented during the SPINE2 project, each with its own merit (Busso et al., this issue, Diebold et al., this issue). The ACEMBL system at EMBL Grenoble was originally designed for rapidly generating multigene vectors for protein complex expression in E. coli. The ACEMBL tandem recombineering procedures, scripted in robotics routines and implemented on a robot, are equally applicable to generating multigene expression constructs for producing heterologous protein complexes in mammalian cells, and the original suite of prokaryotic Donor and Acceptor plasmids has now been complemented by equivalents for mammalian expression. A broad range of applications will benefit from this methodology besides structural biology. For instance, fluorescent markers can be used to genetically tag specific proteins to study cellular processes and entire pathways, with interesting possibilities to enhance screening in mammalian cells for pharmacological applications. Further, we have redesigned and streamlined our MultiBac system for baculovirus/insect cell expression of complexes such that it can be accommodated in the ACEMBL pipeline. In summary, we now have ACEMBL compatible multi-expression systems for protein production in E. coli, mammalian and baculovirus/insect cells, which represent the three major host systems used in contemporary structural molecular biology for recombinant protein production.
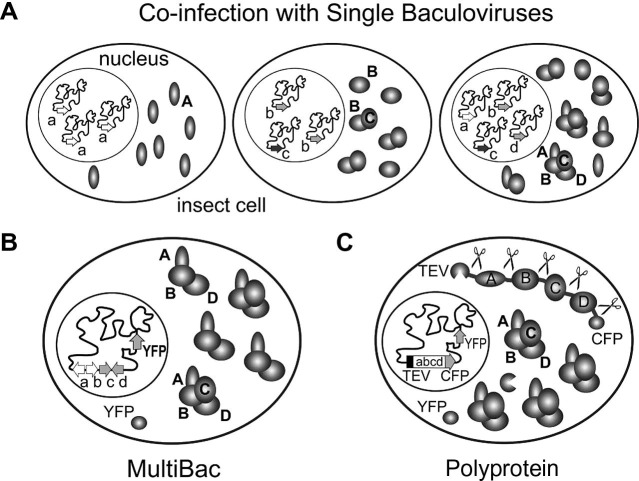
Notably, we have strikingly extended the BEVS complex expression tool-box by implementing a novel logic for multisubunit complex expression in insect cells by using polyproteins (Fig. 7 ). The potential of this technology, also for commercial applications has been recognized (Berger and Richmond, 2006). More recently, a comparable polyprotein approach was also implemented for delivering foreign genes at defined ratios into mammalian cells (Chen et al., 2010). It will be interesting to see whether or not E. coli and mammalian overproduction systems can likewise benefit from the polyprotein approach for structural biology applications. We anticipate strategies relying on tandem recombineering, with Donor and Acceptor plasmids each modified similar to pPBac, for producing recombinant protein complexes with a very large number of subunits efficiently by co-expressing polyproteins. Each of the polyproteins can contain a different fluorescent marker at the C-terminal end, which would allow for directly following expression levels during complex production by multichannel fluorescence emission measurements.
Fig.7.
Multiprotein complex expression strategies by baculovirus. (A) Co-infection of baculoviruses each encoding for a different protein subunit of a complex results in heterogenous infection patterns in the ensemble of cells in the expression culture, ranging from cells infected with only one virus (left) to cells infected with all viruses (right). Balanced expression is difficult to achieve. (B) Expressing all subunits from a single baculovirus (for example by the MultiBac system) containing separate expression cassettes reliably achieves expression of all components in each individual cell in the culture. Inherent differences in individual protein expression levels can impede complex purification and drastically reduce yields if one subunit is in severely short supply. (C) Expression of all components of a multiprotein complex from a single polyprotein. Transcription of a long ORF containing all genes results in a single polypeptide encoding for all subunits and TEV protease. A fluorescent protein at the C-terminal end of the polyprotein allows real-time evaluation of the expression level. TEV protease releases itself from the polyprotein and proceeds to tailor the remaining cleavage sites in the polyprotein. This can lead to stoichiometrically balanced production of the complex components and superior complex assembly. Letters A–D denote protein subunits encoded by genes labeled a to d. TEV protease, CFP and YFP are indicated. Sites of cleavage activity by TEV protease are marked (scissors).
During the SPINE2 project, we have also optimized the performance of the eukaryotic expression facility (EEF) which uses our Multibac system for protein complex production. The controlled reduction of expenditures by purchasing appropriate equipment and, importantly, identifying media that performs well but is available at a fraction of the previous costs, has resulted in a prototype facility which can be duplicated in many laboratories in Europe, including labs with comparatively small operating budgets. The robust protocols and procedures we developed for baculovirus expression have, at least in our view, reduced the previously perceived “complexity” of this eukaryotic expression technology to the level of E. coli expression. These developments have set the stage to welcome many scientists, from academia and industry, for intensive training visits at the EEF in Grenoble, for example through the EC funded MultiBac trans-national access program in the FP7 I3 project P-CUBE (www.p-cube.eu).
Conflict of interest
The authors declare competing financial interest. I.B. and T.J.R. are authors on patents and patent applications describing parts of the technologies discussed in this contribution.
Acknowledgments
We thank Yuichiro Takagi, Wassim Abdul Rahman, Philipp Berger, Daniel Fitzgerald, Michel O. Steinmetz, Daniel Frey, Darren Hart, Yvonne Hunziker and all members of the Berger and Schaffitzel laboratories for helpful discussions, and Hubert Bernauer and Alexander Craig (ATG-biosynthetics), Marcel Boeglin (IGBMC Imaging Center) and Isabelle Kolb-Cheynel (IGBMC Baculovirus Expression Facility) for advice and expert support. S.T. is a European Commission (EC) Marie Curie post-doctoral fellow. C.B. is supported by a Swiss National Science Foundation Advanced Researcher fellowship (SNSF, Switzerland). I.B. acknowledges support from the Agence Nationale de la Recherche (ANR, France), the Centre National de la Recherche Scientifique (CNRS), the SNSF, the EMBL through the EIPOD program, and the EC through Framework Program 7 (FP7) projects INSTRUCT and P-CUBE. The present work was supported by EC projects SPINE2-COMPLEXES and 3D Repertoire (FP6).
References
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- Berger I., Fitzgerald D.J., Richmond T.J. Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 2004;22:1583–1587. doi: 10.1038/nbt1036. [DOI] [PubMed] [Google Scholar]
- Berger, I., Richmond, T.J., 2006. New logic for recombinant expression of multiprotein complexes using polygenes. PCT/EP2006/010608, WO2007/054250 (granted 2009).
- Berrow N.S., Alderton D., Owens R.J. The precise engineering of expression vectors using high-throughput In-Fusion PCR cloning. Methods Mol. Biol. 2009;498:75–90. doi: 10.1007/978-1-59745-196-3_5. [DOI] [PubMed] [Google Scholar]
- Bieniossek, C., Richmond, T.J., Berger, I., 2008. MultiBac: multigene baculovirus-based eukaryotic protein complex production. Curr. Protoc. Protein Sci. Unit 5, 20 (Chapter 5). [DOI] [PubMed]
- Bieniossek C., Nie Y., Frey D., Olieric N., Schaffitzel C., Collinson I., Romier C., Berger P., Richmond T.J., Steinmetz M.O., Berger I. Automated unrestricted multigene recombineering for multiprotein complex production. Nat. Methods. 2009;6:447–450. doi: 10.1038/nmeth.1326. [DOI] [PubMed] [Google Scholar]
- Busso, D., Peleg, Y., Romier, C., Salim, L., Troesch, E., Perrakis, A., Celie, P.H.N. Expression of protein complexes using multiple E. coli protein co-expression Systems: A benchmarking study. J. Struct. Biol., this issue (SPINE2 Special). [DOI] [PubMed]
- Chen X., Pham E., Truong K. TEV protease-facilitated stoichiometric delivery of multiple genes using a single expression vector. Protein Sci. 2010;19:2379–2388. doi: 10.1002/pro.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demény M.A., Soutoglou E., Nagy Z., Scheer E., Jànoshàzi A., Richardot M., Argentini M., Kessler P., Tora L. Identification of a small TAF complex and its role in the assembly of TAF-containing complexes. PLoS One. 2007;21:e316. doi: 10.1371/journal.pone.0000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold, M.L., Fribourg, S., Koch, M. Metzger, T., Romier, C. Deciphering correct strategies for multiprotein complex assembly by co-expression: application to complexes as large as the histone octamer. J. Struct. Biol., this issue (SPINE2 Special). [DOI] [PubMed]
- Fitzgerald D.J., Berger P., Schaffitzel C., Yamada K., Richmond T.J., Berger I. Protein complex expression by using multigene baculoviral vectors. Nat. Methods. 2006;3:1021–1032. doi: 10.1038/nmeth983. [DOI] [PubMed] [Google Scholar]
- Fitzgerald D.J., Schaffitzel C., Berger P., Wellinger R., Bieniossek C., Richmond T.J., Berger I. Multiprotein expression strategy for structural biology of eukaryotic complexes. Structure. 2007;15:275–279. doi: 10.1016/j.str.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriz A., Schmid K., Baumgartner N., Ziegler U., Berger I., Ballmer-Hofer K., Berger P. A plasmid-based multigene expression system for mammalian cells. Nat. Commun. 2010;1 doi: 10.1038/ncomms1120. [DOI] [PubMed] [Google Scholar]
- Li M.Z., Elledge S.J. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- Nettleship J.E., Assenberg R., Diprose J.M., Rahman-Huq N., Owens R.J. Recent advances in the production of proteins in insect and mammalian cells for structural biology. J. Struct. Biol. 2010;172:55–65. doi: 10.1016/j.jsb.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Nie Y., Viola C., Bieniossek C., Trowitzsch S., Vijayachandran L.S., Chaillet M., Garzoni F., Berger I. Getting a grip on complexes. Curr. Genomics. 2009;10:558–572. doi: 10.2174/138920209789503923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y., Bieniossek C., Frey D., Olieric N., Schaffitzel C., Steinmetz M.O., Berger I. ACEMBLing multigene expression vectors by recombineering. Nat. Protoc. 2009;4 doi: 10.1038/nprot.2009.104. [DOI] [PubMed] [Google Scholar]
- Peiris J.S. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis A., Romier C. Assembly of protein complexes by co-expression in prokaryotic and eukaryotic hosts: an overview. Methods Mol. Biol. 2008;426:247–256. doi: 10.1007/978-1-60327-058-8_15. [DOI] [PubMed] [Google Scholar]
- Romier C., Ben Jelloul M., Albeck S., Buchwald G., Busso D., Celie P.H., Christodoulou E., De Marco V., van Gerwen S., Knipscheer P., Lebbink J.H., Notenboom V., Poterszman A., Rochel N., Cohen S.X., Unger T., Sussman J.L., Moras D., Sixma T.K., Perrakis A. Co-expression of protein complexes in prokaryotic and eukaryotic hosts: experimental procedures, database tracking and case studies. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1232–1242. doi: 10.1107/S0907444906031003. [DOI] [PubMed] [Google Scholar]
- Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Trowitzsch S., Bieniossek C., Nie Y., Garzoni F., Berger I. New baculovirus expression tools for recombinant protein complex production. J. Struct. Biol. 2010;172:45–54. doi: 10.1016/j.jsb.2010.02.010. [DOI] [PubMed] [Google Scholar]
- Walhout A.J., Temple G.F., Brasch M.A., Hartley J.L., Lorson M.A., van den Heuvel S., Vidal M. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]