Highlights
-
•
The Umbrella Visualization (UV) is dedicated to the study of glycosylated proteins.
-
•
UV assesses the protein surface covered by a glycan during MD simulations.
-
•
UV is implemented in the UnityMol software, developed with Unity platform.
-
•
Bi and tri-antennary glycans have a different impact on the insulin receptor.
Keywords: UnityMol, Unity, Glycan flexibility, Covered surface, Molecular dynamics
Abstract
N-glycosylation is a post-translational modification heavily impacting protein functions. Some alterations of glycosylation, such as sialic acid hydrolysis, are related to protein dysfunction. Because of their high flexibility and the many reactive groups of the glycan chains, studying glycans with in vitro methods is a challenging task. Molecular dynamics is a useful tool and probably the only one in biology able to overcome this problem and gives access to conformational information through exhaustive sampling. To better decipher the impact of N-glycans, the analysis and visualization of their influence over time on protein structure is a prerequisite. We developed the Umbrella Visualization, a graphical method that assigns the glycan intrinsic flexibility during a molecular dynamics trajectory. The density plot generated by this method brought relevant informations regarding glycans dynamics and flexibility, but needs further development in order to integrate an accurate description of the protein topology and its interactions. We propose here to transform this analysis method into a visualization mode in UnityMol. UnityMol is a molecular editor, viewer and prototyping platform, coded in C#. The new representation of glycan chains presented in this study takes into account both the main positions adopted by each antenna of a glycan and their statistical relevance. By displaying the collected data on the protein surface, one is then able to investigate the protein/glycan interactions.
1. Introduction
In superior organisms, the extracellular matrix (ECM) is an environment embedding fibrous proteins, glycoproteins and polysaccharides synthesized and secreted by cells like fibroblasts [1]. In the animal reign, the ECM is divided into two main components: the basement membrane and the interstitial matrix. The basement membrane is a combination of proteins forming sheets used to anchor epithelial cells [2]. The interstitial matrix fills the space between cells [3]. Because of its mechanical properties, this environment provides structural support to cells and contributes to mechanisms such as cell migration or cancer metastasis.
The ECM is a very dynamical environment with ongoing remodelling or degradation, making it a source of many active molecules. Enzymes or matrix degradation products, i.e. matrikines [4], are able to interact with the nearby cells through receptors, mostly transmembrane proteins which are located at the cell surface [5], [6], [7], [8]. These proteins trigger molecular signals through the membrane, allowing cells to react to their environment. Understanding how these signaling processes take place and how cells are affected is essential in order to deepen our understanding of the biochemical role of the ECM. As an example, the insulin receptor (IR) is a protein belonging to the class II tyrosine kinase receptors family. IR is synthesized as a precursor of 210 kDa. The pro-protein is cleaved into an and a subunits linked by a disulfide bridge. The subunit is extracellular while the subunit contains an extracellular part about 190 amino acids, a transmembrane helix and a cytoplasmic domain. The mature receptor is present at the membrane as a dimer with 2 and subunits. Transmembrane proteins, as well as proteins composing the ECM, are modified after their synthesis by post-translational processes. Glycosylation is an enzymatic maturation process essential to the protein function, including ligand-receptor affinity and dimerisation [9]. This process involves the attachment of branched chains of saccharide residues on the protein surface. More precisely, N-glycosylations are defined by the covalent linking of a glycan chain to the amide nitrogen of asparagine predominantly at the sequons Asn-X-Thr and Asn-X-Ser, where X is any amino acid but not proline [10]. In the IR, 18 potential N-glycosylation sites are identified (14 in subunit and 4 in subunit). These chains are constituted of several types of saccharide residues connected by a linear glycosydic linkage and arranged around a common, rigid core with highly variable and flexible branches. N-glycans are classified into 3 types: high-mannose, complex and hybrid types. High-mannose types are glycans only elongated with mannose residues beyond the core structure. Complex types are extended with other monosaccharides. Hybrid glycans are bi/tri-antennary glycans with one high-mannose branch and one or two complex branches. Because these chains are bulky, they play an important role in protein folding, helping it to adopt its fully functional 3D structure. Glycosylations are sometimes bound by proteins, such as lectins [11], and act as recognition patterns in signaling pathways. Alterations of the glycosylation pattern are observed in pathological contexts and affect the protein functions in diseases such as cancers, diabetes or cardiovascular diseases [12].
Glycans are higly flexible because of the linear glycosidic linkage between each saccharide residues [13], granting them a high number of degrees of freedom. They are also very reactive structures because of the many hydroxyl groups composing these residues. Under experimental conditions, they are subject to a quick degradation because of this reactivity. Moreover, cells used in protein production and purification, such as E. coli, are not able to synthesize glycan structures found in animal organisms. It is therefore very difficult to study glycosylations with experimental methods.
Molecular dynamics (MD) is a useful tool to overcome this problem and give access to conformational information. By using empirical forcefields fitted on experimental observables, MD methods enable the description of molecules at the atomic level. Heavy computational resources and exhaustive sampling are used to calculate motions with a high precision. Considering the crucial impact of glycans on protein function, integrating glycosylations in in silico studies as well as developing tools dedicated to the study of MD simulations of glycosylated proteins is a key point towards the understanding and deciphering of protein function and regulation.
Our previous in silico studies investigating the impact of sialic acids on glycan’s structure and dynamics brought to light the lack of tools dedicated to glycan in the field [14]. This study was the beginning of the development of the Umbrella Visualization method. This method aims at evaluating the area covered by a glycan during a MD trajectory.
This representation helps appreciate the relative flexibility of the glycan during the trajectory. This method demonstrated, for the first time, the impact of sialic acids on the flexibility of glycan chains. Because this study was focused on isolated, solvated glycan chains, the results of this first application of the Umbrella Visualization were restricted to a 2D graphical representation.
We present here the latest developments of this method. The Umbrella Visualization now takes into account both the protein topology and the glycan motions above the protein surface.
The main goal of this original and unique approach is to fully understand how a glycan moiety will structurally impact the protein surface. The dynamic information collected during a MD simulation should help identify the most impacted areas and understand which parts of the protein would be accessible to other molecules such as solvent molecules or even other proteins.
First, we report the main tools already available in the field of glycosylated proteins. Many of these tools focus on the study of static, crystallographic structures. Visualization softwares such as PyMol [15] and VMD [16] integrate visualization tools dedicated to the representation of glycan with the aim of better understanding their conformations. We then present UnityMol, developed with the video game engine Unity. Using this software, we upgraded the Umbrella Visualization from a 2D method giving results as density plots to an integrated tool allowing the visualization of the impact of glycans onto a protein structure during a MD trajectory. The Umbrella Visualization is fully implemented in the UnityMol interface in order to provide a user-friendly, fully automated tool able to accommodate a wide variety of N-glycan structures. We tested this method on a glycosylated insulin receptor and highlighted the differential impact of bi-antennary and tri-antennary glycans on the protein.
This tool, in conjunction with insights of experimental studies, could help understand mechanisms involving N-glycans and design in vitro experiments aimed at understanding their impact on protein functions.
2. Related work
2.1. Glycosylation and modelling
The first step needed to study glycosylated proteins is to identify the glycosylation sites. Most of the time, only the first saccharides from a glycan are present on the .pdb structure files available on the Protein Data Bank at www.rcsb.org [17]. This can help identify some glycosylation sites but most of the time, glycan structures are not available: because of their great flexibility, glycans are not sufficiently ordered to be detected by classical methods used to determine protein structures such as X-ray crystallography [18]. It is thus necessary to use prediction tools in order to identify the glycosylation sites. Several tools are available as web-portals or standalone programs [13], [19], [20].
Before running MD simulation, it is useful to analyze and understand the structure of glycosylated proteins. Some tools can help identify conformational errors in structures [21], [22] while others are dedicated to the study of the torsion angles and saccharidic linkage [23], [24], [25]. Carbohydrates have many hydroxyl groups that are very reactive and can interact with their surroundings. This is why it is also important to understand how glycans will interact with the protein residues. GlyVicinity [26] is a tool that statistically analyzes the amino acids closest to a chosen type of carbohydrates.
Once the glycosylation sites are identified, it is necessary to build the glycan structure on the site of interest. Even if the first saccharide residues are present in the .pdb file, the remaining part of the glycan has to be reconstructed, avoiding any aberrant structures or steric clashes with the protein. SHAPE is a tool enabling a user to build any glycan structure [27]. SHAPE also predicts several energetically-favorable conformers using a genetic algorithm.
SWEET II on the glycosciences.de portal is another tool that can be used to build a glycan structure [28]. This glycan can then be used on the GlyProt page. GlyProt allows a user to upload the .pdb file and add the glycan created with SWEET II on one or more glycosylation sites [29]. Glycam-Web (http://glycam.org/) and Charmm-gui with the Glycan-Reader tool [30] are also web-servers including a builder dedicated to glycosylated-protein structure.
Very recently, the DoGlycan set of tools [31] was developed in order to perform the same function with the Glycam forcefield, which is dedicated to the study of carbohydrate structures.
2.2. Visualization
Many of the main visualization software integrate the visualization of saccharides in all-atom representations but are often limited to licorice, balls and sticks or van der Waals (VdW) sphere representations. But, few of them propose ways of visualization that highlight the positions and structures of glycosylations on the proteins’ surface.
PyMOL includes a plugin called Azahar [32] which is able to build a glycan from a template list of saccharide structures. The plugin adds three specific display modes aimed at simplifying the representation of glycan structures: the cartoon and wire representations show the cycles as non-flat polygons linked by rods and the bead representation shows each cycle as a sphere. This plugin also gives the possibility to compute relevant structural values in order to analyze both static structures or MD simulations.
With the 3D-SNFG plugin [33], the VMD software implements its own graphical representation based on the nomenclature developed before the second edition of the Essentials of Glycobiology textbook [34], [35]. Each monosaccharide is represented by a 3D object based on the corresponding symbol.
UnityMol, an open source platform developed with the Unity engine (http://unity3d.com/) in C# language [36] is dedicated to the visualization of biological molecules and uses HyperBalls [37] representation. In the latest versions called SweetUnitymol [38], this software integrates display modes and representations dedicated to saccharides such as ribbons representation or the possibility to color saccharide depending on its nature: for each saccharide, the associated color is determined by the symbol found in the SNFG nomenclature.
The tools described above offer different ways of building, analyzing and visualizing a glycosylated-protein structure. Mazola et al. [39] introduced a workflow leading to the building of a glycosylated system ready for MD simulations. However, the forcefields and nomenclatures can differ from one to another, making it hard to switch between them. Moreover, these tools are mostly dedicated to the building of glycosylated protein systems and the analysis of static, crystallographic structures. Despite the possibility to obtain MD simulations data on glycans and glycosylated proteins, there is a lack of tools dedicated to the analysis of such trajectories.
3. Background
3.1. 2D Umbrella Visualization
It is well known that sialic acids, charged saccharides often found at the extremities of glycan chains, are involved in protein function’s regulation [40] or dysfunctions such as in type 2 diabetes [41]. To better understand the influence of sialic acid on protein’s function and structure, a molecular dynamics study on sialylated and non-sialylated glycan chains has been initiated. Trajectories for four glycan chains were computed, with glycans bearing two and three branches, with and without sialic acids [14]. For each system, the starting carbohydrates structures were built using the Avogadro software. 500 ns of trajectories were generated with the OPLS-AA forcefield and the Gromacs package 4.6.3. NPT ensemble was used with a 310 K temperature and 1 bar pressure.
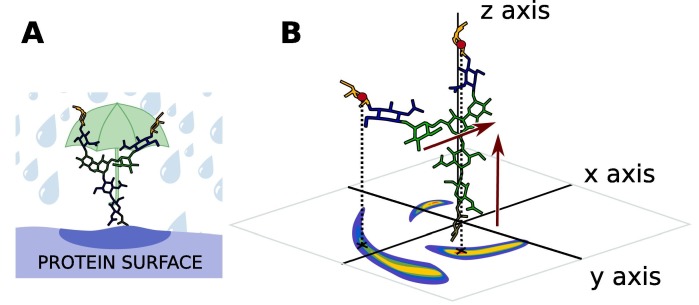
In order to plot and evaluate the relative flexibility of glycan chains with and without sialic acids, the Umbrella Visualization was developed. The Umbrella Representation considers the glycan acting as an umbrella protecting the protein surface so that an hypothetical molecule would not be able to interact directly with this part of the protein (Fig. 1 A). The glycan is divided into vectors representing its different components: two vectors describe the glycan core. The other vectors describe the branches of the glycan. The first step is to measure the distance between the centers of mass of the saccharide residues defining these vectors for every trajectory step. This is done thanks to the Gromacs [42], [43] software and the distance module. Files listing the coordinates of these vectors during the simulation are generated and analyzed.
Fig. 1.
A) Schematic principle of the Umbrella Visualization: The glycan here is shown in sticks representation and colored by residue type. The Umbrella Representation considers the glycan acting as an umbrella over the protein surface. B) Implementation of the Umbrella Visualization process: The glycan is represented in sticks. The core of the Glycan, shown in green, is aligned along the z axis thanks to vectors describing the core (red arrows). The center of mass (red dots) of the last saccharide residue on the branches is projected on the xy plane. The final result is a density plot reflecting the relative flexibility of the glycan during a MD trajectory. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
An in-house program reads those coordinates in order to place the chain on an orthogonal coordinate system, with the asparagine amino acid at the glycan’s base set on the origin. The two core vectors as described on Fig. 1B, are used in order to orient the glycan within this coordinate system. First, the angle between the inner-core vector and the z-axis is calculated so this core can be oriented along this axis. Then the angle between the second vector and the x axis is calculated and the glycan chain is rotated to make this vector coplanar with the xz plan. Finally, the xy coordinates of the last common saccharide residue for both types of glycans (sialylated and non-sialylated) are written and displayed on a density graph.
The non-sialylated bi-antennary glycan showed increased flexibility and explored a much larger region of the plot compared to the sialylated bi-antennary glycan. Desialylation impacted the tri-antennary glycan differently: only one of the three branches showed a significant change in flexibility. Though the influence of sialic acids varies depending on the branching of the glycan, it is clear that sialic acids have an impact on the flexibility and stability of glycosylation chains. The Umbrella Visualization, combined with clustering methods and measurement of angles values during the trajectory, was thus able to demonstrate this influence [14].
3.2. UnityMol
In order to better understand how glycans and sialylation impact proteins, it was then decided to transform this 2D-analysis method into a visualization method aimed at identifying which regions of the protein surface are impacted by a glycan during a MD trajectory.
For this purpose, we have decided to work with an existing open source software, UnityMol, to implement this new functionality. By integrating the Umbrella Visualization to a software already used by the scientific community, we hope to make it user-friendly and broadly available.
As stated previously, UnityMol is a molecular editor, viewer and prototyping platform, coded in C# with the Unity game engine. It already has all the classic visualization modes used in molecular modeling and is even able to automatically detect saccharide residues in order to provide user-friendly visualization modes dedicated to the representation and study of saccharide chains. It also provides a plug-in reading MD trajectories. The Unity engine makes it easy to work either with built-in tools or to implement new features. The wide use of this platform also warrants support from an active community and provides online resources and tutorials. This engine supports different platforms such as Android, Windows, Linux or Mac platforms. Exporting the executable file for these platforms is done at compilation, and saves a great amount of time by avoiding further developments. The Unity graphical interface also displays a Game Mode that allows a user to run its program without compiling it. This feature is especially useful in order to rapidly and easily test adjustments brought to the code. Unity is also known for being relatively user-friendly, partially thanks to its interface, but it is also important to note that every parameters and components are accessible via scripting in C# or Javascript languages, making it a very versatile tool. Thus, the UnityMol prototype developed with this engine is easily modifiable and extensible and provides the means necessary for the implementation of the Umbrella Visualization.
4. System design
The aim of this implementation is to evaluate the covered protein surface during a MD simulation by taking into account the glycan’s motions and the protein topology. This enables the display of the 3D structural information. First, the general principle of the shadow projection is detailed. The second part describes the method used to align the plane under the glycan. Then the merging of the information obtained from all the trajectory frames is explained. Finally, the modifications brought to the Unitymol interface in order to implement the Umbrella Visualization are detailed.
4.1. General principle
One of the many advantages of the Unity engine is the possibility to build prefabs that can be used for development purposes. Prefabs are assets storing GameObjects components and setting their properties to desired values. This way, when the prefab is instantiated into a scene, the GameObjects components are already positioned and oriented as desired. Among these components, C# or Javascript scripts can be assigned to a GameObject component and used in order to dictate a behavior to this component. Prefabs are very versatile: even if all the instances of a prefab will share its properties and ensure a certain homogeneity between the instances, individual instances are independently editable and it is thus possible to change some individual properties as needed. Building a prefab and assigning scripts to its component is a good way to build a template for an asset that would be instantiated several times into a Unity scene.
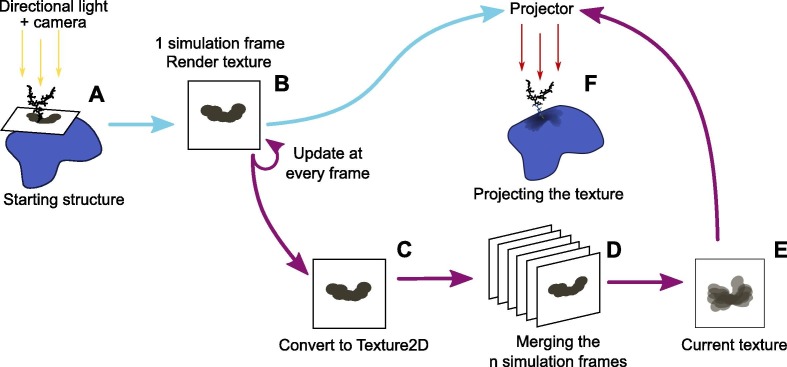
By using these built-in tools and components from the Unity engine, we designed a Unity prefab able to capture the glycan shadow and project it onto the protein surface. This prefab is composed of a camera, a projector and a directional light oriented perpendicular to a flat mesh defining a plane (Fig. 2 A). These components are organized together to project the shadow of a glycan on the protein.
Fig. 2.
Unity workflow: The blue arrows describe the workflow to project only one frame. The purple arrows describe the workflow to project the shadow processed from the whole simulation. A directional light projects the glycan’s shadow over an oriented plane (A). This shadow is rendered into a render texture by a camera (B) and either displayed on the surface for a static structure or, with a MD trajectory, converted into a Texture2D object (C). This object is read and combined with the images from previous frames (D). All the results acquired along the MD simulation are merged together into a final Texture2D (E) object which is then used in the projector, thus displaying it onto the protein surface (F). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Cameras are also defined as GameObjects components and can be used into prefabs. Cameras usually capture and display a view to the user but can also render this view into a texture. To this end, a Render Texture is assigned to a camera. Unity also provides layers that allow for lights and cameras to not illuminate or not render elements from the layers defined in their respective “culling masks”.
The Projector prefab from the Unity Standard Assets package projects a desired material or texture onto all objects intersecting its frustum. Projectors have dedicated shaders in which the projected texture is assigned.
One of the many features of the Unity engine is the possibility to quickly instantiate GameObjects of basic shapes such as cubes, spheres or flat planes. As we need a flat surface to project the glycan shadow, the prefab integrates a plane positioned under the glycan. Different types of lights, traditionally used to illuminate a scene, are also available on the Unity engine. As glycan’s shadow size must be independent from the light to plane distance, we thus use a directional light. This type of light illuminates scenes with parallel rays, similar to sun rays. The light’s orientation is perpendicular to the plane’s surface, reproducing an orthogonal space used to orient the glycan.
Once the shadow is displayed, the VdW representation mode has been used in order to reflect the atomic volume of the glycan. The VdW representation mode is based on the VdW radius, a theoretical value representing the atom’s volume. In order to only display the shadow of the glycan on the plane, the glycan atom’s mesh is set to Shadow Only mode. Doing so makes this mesh also invisible to UnityMol’s main camera but it is possible to copy the glycan atoms mesh and set it on another layer in order to see the shadow and the atoms at the same time with the prefab’s camera ignoring the glycan mesh. Likewise, to prevent the camera to capture parts of the surface going through the plane, the protein surface is set on a dedicated layer and a ”culling mask” on this layer is applied to the prefab’s camera. This is often due to the irregularity of the protein surface and the plane being close to the surface. This set-up is able to save the glycan’s shadow into a render texture (Fig. 2B).
Finally, the shadow-projector uses the texture generated by the prefab displaying the glycan shadow. This texture is then projected onto the protein surface, highlighting parts of the protein surface impacted by the glycan (Fig. 2F). In order to avoid displaying unnecessary elements to the UnityMol’s user, the lit-plane prefab is set on another layer and a culling mask is applied on this layer on UnityMol’s main camera and light. The plane is thus not visible to the user and only the molecules are displayed on the screen. Using a layer dedicated to the prefab’s elements also prevents the main light to interact with the lit plane prefab, which would alter the glycan shadow.
With a similar goal, the camera’s and projector’s views are set on orthographic mode. With this mode, 3D objects are represented in two dimensions by the means of a parallel projection of the render texture’s pixels. The picture projected on the protein surface will not vary with the relative distance to the protein surface. This workflow projects the glycan’s shadow onto the protein surface for a static structure. However, this tool is dedicated to the study of data extracted from MD. We have thus to take into account the whole length of a MD trajectory, as described in Section 4.3.
4.2. Orienting the prefab
Whereas the 2D Umbrella Visualization aligned the glycan core along the z axis in order to evaluate the branches’ motions, in UnityMol we use the glycosylated asparagine residue as a reference point to place and orient the lit plane prefab. By doing so, we take into account every motion of the glycan on the protein surface, including motions and tilts coming from the core of the glycan.
With the Unity engine, every GameObject has a Transform component. This components has three properties: the Position property determines the position of the Transform in x, y, and z coordinates. The Rotation components determines the rotation, in degrees, on the x, y and z axis. Finally, the Scale component determines the relative size of the object along the x, y and z axis. For the Umbrella Visualization, we use the Position and Rotation properties in order to move and orient the prefab around the glycosylated asparagine.
A normal to a surface is determined by the vector perpendicular to a surface, pointing outward. They are used on meshes to determine how an object will be illuminated by comparing the orientation of the normal to the light’s vector. Here, normals define the lit plane’s rotation in order to orient it under the glycan. We also use normals pointing outward from the protein surface in order to refine the intensity of the picture projected depending on the orientation of said surface.
Quaternions are based on complex numbers and are used to define and store rotations. Most of the time, their components are not directly modified via scripting, but are re-defined thanks to already existing vectors. Quaternions are easily build from vectors, allowing to compute rotations in a three dimensional space with ease.
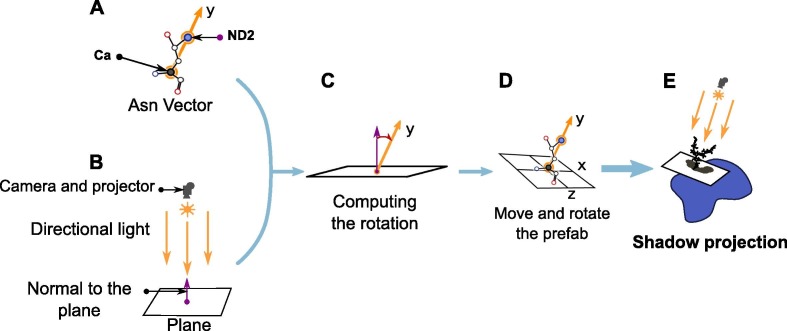
As described in Fig. 3A , we define a vector going from the alpha carbon of the asparagine residue to the nitrogen atom involved in the glycosidic linkage (called the Asn vector). This vector is used as the y axis of our coordinate system. When the plane is instantiated in the Unity scene, its Transform component doesn’t contain any rotation. The plane normal is thus defined by a vertical vector (0, 1, 0) (Fig. 3B). The rotation going from this normal to the Asn vector is computed for the starting structure and stored into a quaternion (Fig. 3C). The coordinates of the alpha carbon are used in the Transform component of the prefab, hence positioning the center of the plane at this atom’s position. The previously built quaternion is then applied on the prefab’s Transform component, thus orienting the plane under the glycan with respect to the asparagine’s residue (Fig. 3D). As the Asn vector is computed only on the starting structure, it is necessary to align the MD trajectory on the glycan in order to ensure that the glycan and Asparagine residue both stay in the same area during the trajectory read.
Fig. 3.
Positioning and orienting the lit plane prefab with the glycosylated asparagine. A) The coordinates of the alpha carbon and terminal nitrogen atoms define a vector (Asn vector, orange arrow). This vector defines a (x, y, z) coordinate system. B) Instantiated prefab, the normal to the plane is the purple arrow and is defined by a (0,1,0) vector. C) We compute the rotation needed in order to go to the Asn vector from the normal to the plane. D) The center of the plane is moved to the C coordinates and the rotation defined at the previous step is applied to the prefab. E) Once the prefab is correctly positioned, we project the shadow on the plane and save it thanks to the camera. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Once the plane is instantiated and correctly positioned, the next step is to collect the information for every frame of a molecular dynamics trajectory and compute the statistical, final result (Fig. 3D).
4.3. Frame merging
MD trajectories can be constituted of several thousands of frames to visualize and analyze. Our goal is to provide a tool allowing the user to understand the behavior of the glycan and to see which parts of the proteins are more affected by the glycan’s positions and motions. We have to combine or merge the information from all the simulation frames and display the result.
To this end, a script is included on the lit-plane prefab. When the Umbrella Visualization is activated, an integer array is created. The array size is defined by the number of pixels of the final texture. When the simulation frames are loaded, for each frame, the prefab collects the glycan shadow and saves it in a render texture (Fig. 2C). The scripts converts this render texture into a temporary Texture2D object in order to read the pixels from this texture. Then, for each pixel, the script will detect if the pixel is black (a shadow pixel) or not. The corresponding element in the array will be incremented by 1, thus counting the number of times a pixel is shadowed by the glycan during the trajectory (Fig. 2D). After having read all the frames, the array’s values for each pixel are normalized. The higher the count, the darker the pixel will be, highlighting the parts of the protein that are most often obscured by the glycan (Fig. 2E). Pixels color are defined by three components: r, g and b. These components determine the final color of the pixel by adjusting the red, green and blue hues (r, g and b). Using the values from the array, we are able to define the r, g and b elements of the accumulation texture’s pixels. Thus, throughout the trajectory, the accumulation of the different positions of the glycan is visualized and at the end of the trajectory, the distribution of the conformations of the glycan is obtained.
During the MD reading, the current shadow is projected onto the surface of the protein so that the user can visualize the motions of the glycan above the protein surface during the trajectory. At the end of the read, the projected texture is switched in order to display the final result compiling the information from all the individual frames.
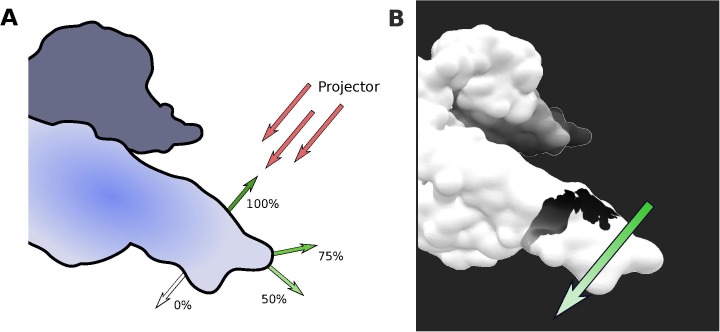
In order to prevent the overestimation of the impact of the glycan on parts of the protein that are too far from the glycosylation site, the distance from the glycan is evaluated and the rendering of the projected texture is modified accordingly. The projector’s shader modulates the projected shadow’s intensity by comparing the direction of the projector and the direction of the normals to the protein surface. The intensity of the shadow is altered depending on the angle between those two vectors, as described in Fig. 4 A.
Fig. 4.
Fading of the projected shadow on the protein surface. A) Variation of the intensity of the projecting shadow depending on the direction of the normal to the surface. When this normal is facing the projector, the shadow is not faded out. When the normals are facing the same direction as the projector, the shadow is not visible. For other normals, the intensity of the shadow is adjusted proportionally. B) Insulin receptor protein surface with a single-structure shadow projected on the surface. The shadow intensity is also modulated by the distance to the projector. The green and white arrow highlight this directional fading. This snapshot is extracted from the 250 ns MD simulations (for details, refer to Section 5.1). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The projector also allows the use of a “falloff” texture, meaning that it can modulate the projected shadow’s intensity depending on the distance to the projector (Fig. 4B). As described on the next section, the UnityMol interface integrates a box in order to deactivate the falloff texture.
4.4. User interface
One of the aim of the Umbrella Visualization is to make this method accessible to every UnityMol users and enable the production of pictures for analysis and illustration purposes. This implies to link the described process to the UnityMol interface and works on display modes highlighting clearly the covered parts of the protein surface.
We expanded the UnityMol sugar menu, adding four new boxes. The first one (Enable box) starts the Umbrella Visualization once the data are loaded. Detection of the data type (single structure file or molecular dynamics trajectory) is automated so that no other input from the user is needed.
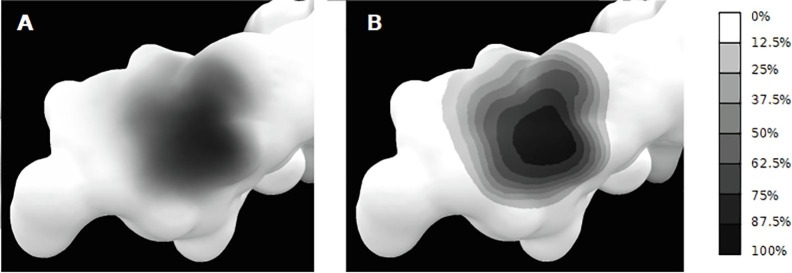
Depending on the input data, the limits of the covered surface may be blurry. In order to have a clear view of this surface, we added a display mode available through the Contour button. This display mode modifies the result texture in order to define clear intensity zones, as seen on the Fig. 5 . Eight zones are delimited, ranging from black zones (most covered parts) to white/transparent zones (least covered parts). These zones are defined thanks to the number of times a pixel is obscured during the trajectory, as described previously. This gives a clear view of the protein surface impacted by the glycan during the MD trajectory.
Fig. 5.
Display modes available for the Umbrella Visualization. A) classical display, the glycan’s shadow is very detailed but blurry. B) The Contour display mode gives a better idea of the covered surface by highlighting different zones. The darker parts correspond to the most covered area during the trajectory. The scale to the right gives the thresholds for each zones. These pictures are obtained averaging the data extracted from 12,500 frames of MD simulations of a glycosylated insulin receptor (for details, refer to Section 5.1).
Once the calculation is finished and the shadow displayed, the Save Result button saves the projected texture in a separate file, giving access to the 2D information besides the 3D information. The raw results can be saved as well as the Contour result.
Finally the Falloff box enables the fading of the projected shadow with the distance to the projector. Disabling this option gives access to the full, unaltered projected result while activating it could help highlight the influence of the glycan on parts of the protein near the glycosylation site.
5. Preliminary results and application on a biologically relevant system
In order to further our studies on the insulin receptor, we used the Umbrella Visualization on the insulin receptor ectodomain (extracellular part of the protein) dimer. After detailing our systems and the forcefields used in the MD trajectories, we tested the Umbrella Visualization on a small samples of frames of a mono-glycosylated IR in order to evaluate the relevance of this method. Then, thanks to the identification of two glycosylation sites on previous studies, we ran a comparative preliminary study on the impact of bi-antennary and tri-antennary glycans on the insulin receptor. Finally, we focus on the performances of this method by comparing the quality and time needed to compute the results depending on the number of frames to analyze.
5.1. Molecular dynamics set-up
To start our studies on the insulin receptor and the impact of glycosylation, we first ran docking experiments with the insulin receptor and the neuraminidase I subunit of the elastin receptor complex [8]. Correlating a map of the consensus sequence for N-glycosylation with the docking results led to the identification of two potential glycosylation sites on the insulin receptor dimer accessible to the neuraminidase I protein. Trajectories generated with the GROMACS software (5.0.2 package) and the OPLS-AA forcefields, using the NPT ensemble (P = 1 bar and T = 310 K) were used for this study. Each dimer was glycosylated on two sites, thus creating two systems: the IR with two non-sialylated bi-antennary glycans and the IR with two non-sialylated tri-antennary glycans. 250 ns of trajectories were generated for each system.
The MD trajectory used in the 5.2 is a sample of 250 frames extracted from the IR-dimer with two non-sialylated bi-antennary glycans.
The results presented in the Section 5.3 were computed on 12500 frames from a 250 ns trajectory (1 frame every 20 ps). These data were also used in order to evaluate the performances of the Umbrella Visualization (Section 5.4).
5.2. Relevance of the Umbrella Visualization
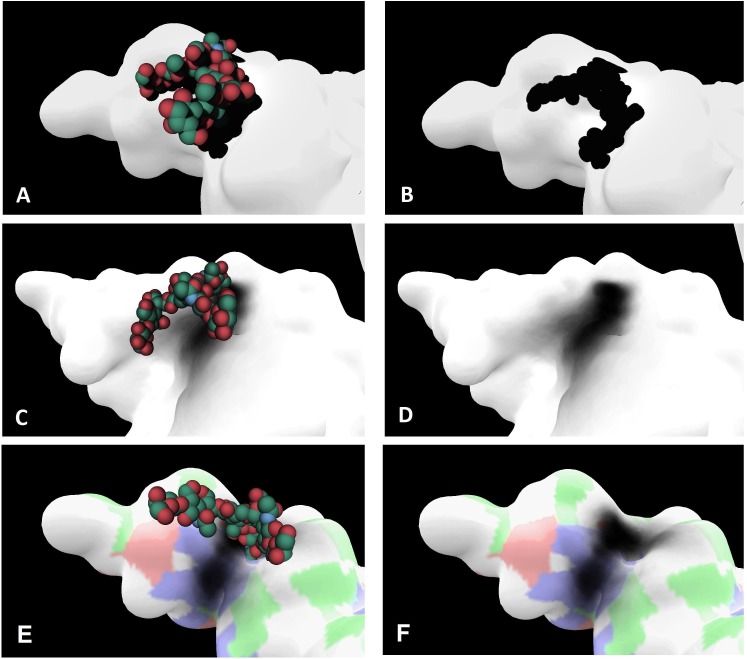
In order to evaluate the relevance of Umbrella Visualization, we used this method on the insulin receptor ectodomain dimer with one bi-antennary glycan per subunit. The Fig. 6 shows the result of the Umbrella Visualization with one frame displayed (A and B panels). The protein surface is displayed in white while the glycan’s atoms are represented in VdW spheres. Each sphere corresponds to an atom from the glycan (For clarity, the hydrogen atoms are not displayed). The green spheres are carbon atoms, the red spheres are oxygen atoms. Thanks to the VdW representation, the shadow size matches the volume of the glycan and helps approximate the surface covered for this specific conformation. On the A and B panels, the glycan is oriented above a shallow groove of the protein. Such a conformation on the .pdb file could indicate a preferential conformation due to favorable interactions with the protein surface. To confirm or reject this hypothesis, we ran a MD simulation of the glycosylated insulin receptor ectodomain dimer.
Fig. 6.
Relevance of the Umbrella Visualization: On the A, B, C and D panels, the protein surface is shown in white. On the E and F panel, this surface is white with colored patches. The shadow projected on the C, D, E and F panels is the result obtained over the 250 frames (dark grey/black areas). The glycan is displayed on VdW mode. A) Starting structure used for the MD trajectory. The glycan is located near a shallow groove on the protein surface. B) Shadow of the glycan projected on the protein surface on the starting structure. C) This conformation is extracted from the MD simulation and shows how the glycan is positioned on the groove, in a conformation similar to the starting structure. D) On this particular example, it is possible to see that, during this MD trajectory, the glycan’s motions are mostly restricted to an area along a small groove on the protein surface. E) and F) show the glycan and the projected shadow over the groove with the surface colored depending on the residues underneath. Blue patches are for basic residues, red patches are for acid residues, green patches are for polar residues, white patches are for non-polar residues. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The C and D panels show the projected shadow on a 250 frames trajectory. Because we normalize the r, g, b values with the number of frames, the result is statistically relevant. On this example, we can conclude that during most of the trajectory, as suggested by the .pdb structure, the glycan is extended above the groove but doesn’t explore the surrounding part of the protein surface. This could indicate that particular protein residues in this region have favorable interactions with the saccharide residues of the glycan, hence favoring this conformation. These results thus corroborate the hypothesis made with the static structure. The Fig. 6E and F show that the groove covered by the glycan during most of the 250 frames is composed of positively charged. In order to investigate the nature of residues interacting with the glycan, a color scale corresponding to the physico-chemical nature of residues located under the surface is applied. Thus, red, blue and green patches show the presence of acid, basic and polar residues, respectively. The superimposition of this information and the shadow of the glycan emphasize what are the properties of the residues below that interact with the glycan.
In the context of protein/protein interactions, this is a valuable information as basic, positively charged residues can be involved in these interactions. On this case, these residues would be able to interact with the many hydroxyl groups of the sugar residues, hence stabilizing the position of the glycan in the groove, hindering the interaction of other molecules with this patch. This could highlight how a glycan could influence and modulate protein/protein interactions.
5.3. Comparative study on the impact of bi-antennary and tri-antennary glycans
In order to evaluate the impact of different glycans on the insulin receptors, we ran molecular dynamics simulation on two systems: a glycosylated insulin receptor with bi-antennary glycans and a glycosylated receptor with tri-antennary glycans. We used the Umbrella Visualization on the computed trajectories and compared the results to highlight the differences in the covered protein surface.
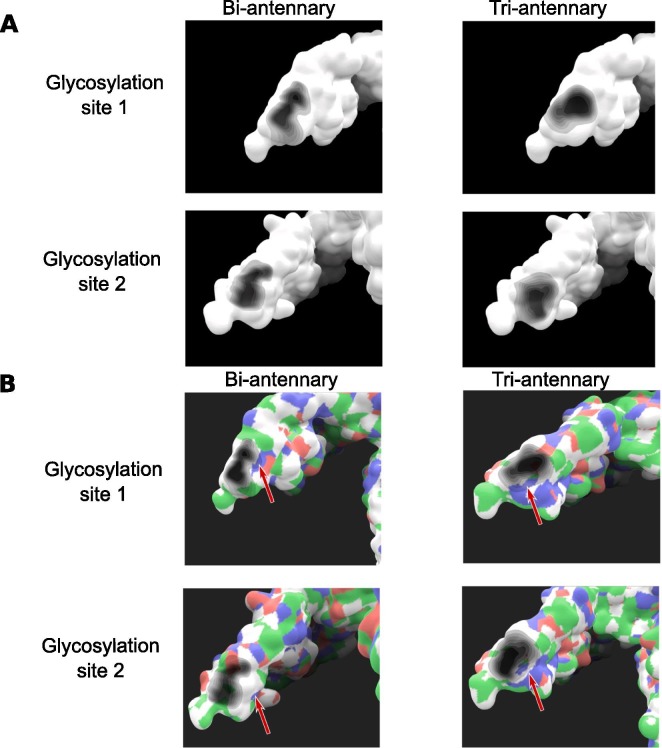
On both insulin receptor structures, the global shape of the projected shadow is very similar between the two glycosylation sites, showing a similar behavior for both glycans. Howewer, the number of branches seems to have a higher impact on the covered surface. For the bi-antennary glycans, the shadow is elongated lengthwise on the protein (Fig. 7 A). The tri-antennary glycans covered surface takes a more rounded or oval shape, suggesting a higher coverage over the length of the trajectory. This is consistent with the higher number of branches on the tri-antennary glycan.
Fig. 7.
Results obtained with the Umbrella Visualization on the glycosylated Insulin Receptor. A) Comparison of the different shadows computed. The global shape for the bi-antennary glycans is more stretched along the protein. Tri-antennary glycans display a wider shadow. This is consistent with the fact that this glycan has one extra branch able to explore a wider space. B) Patch of basic residues located near the glycan (red arrows). The glycan’s shadow tends to hover around this part of the surface. This could indicate the role of these amino acids in stabilizing the glycan. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Coloring the protein surface with the amino acid’s properties shows that the glycan is located on a part of the protein surface composed of apolar residues (colored in white). Though most of the covered surface is located on this apolar zone, a small patch made by basic residues is partially covered by the glycan (red arrows in the Fig. 7B). Both bi-antennary and tri-antennary glycans seem to cover part of this patch during the trajectory. However, the tri-antennary glycans cover up a larger portion of this patch. This might indicate that the glycan interacts with these amino acid in order to stabilize its conformation over the protein and limits the explored space during the trajectory. Indeed, along the trajectories, the glycans don’t explore much of the space on the opposite side of this patch. For the bi-antennay glycan, this could also explain why the elongated shape of the covered surface is oriented towards it.
5.4. Performances
The investigated system on this study is a dimer of the insulin receptor ectodomain with one glycan per monomer. This corresponds to a number of 26030 atoms, not counting the water molecules used to calculate the motions along the trajectory. The Umbrella Visualization analysis was performed with a 3.7 GHz Intel Xeon W-2145 CPU machine with 64 Gb of DDR4 RAM and a Nvidia GeForce GTX 1080Ti graphic card.
The performances of our methodology were calculated through the analysis of three different sets of frames: 250, 2500 and 10,000 frames. We used 512 pixels wide texture. Using a smaller texture saved time (18 min for a 256 pixels wide texture). The main limitation is the number of atoms contained in the structure. The dimer simulated here is a relatively large system, thus processing the data for every frame of the simulation requires more computational resources than a smaller system. In spite of that size, this method still process a significant number of frames in a reasonable time (roughly 30 min), especially when considering the amount of time necessary to collect MD data (almost 8 days accumulating 32 ns/day using 480 CPUs).
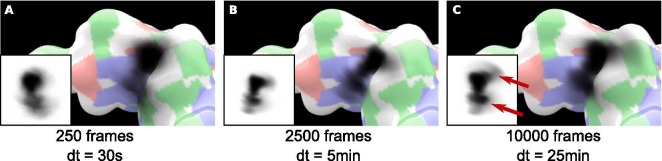
As indicated on Fig. 8 , the rendering of the Umbrella Visualization through 250, 2500 and 10,000 frames took approximately 30 s, 5 min and 25 min respectively. As expected, more information is available with a higher number of frames. Though the more prominent conformations are already displayed on the 250 frames sample (Fig. 8A), the two other samples bring to light the impact of less represented conformations (Fig. 8B and Fig. 8C). In every case, the combined frames show that the glycan have a tendency to contact the patch of basic residues on the groove.
Fig. 8.
Results of the Umbrella Visualization on three MD samples. The flat textures projected onto the surfaces are shown as insets. A) Results extracted from 250 frames. The overall tendency is already visible, highlighting the importance of the basic residues. B) Results extracted from 2500 frames. C) Results extracted from 10000 frames. By increasing the number of frames used for the computation, the lower area (bottom red arrow) becomes more compact and darker while a light spread zone (top red arrow) appears in the upper area. This clearly indicates that with a low amount of frames, the role of zones may be overestimated while others are missed. The dt label is the elapsed time for the calculation of these results. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
6. Discussion and concluding remarks
The Umbrella Visualization is a new method of analysis using a camera and a projector to project the shadow of a glycan on the protein surface. Shadows rendering of the glycan is performed and processed by counting the number of times a pixel is obscured by the glycan and determine the color of the final texture depending on this number. During the trajectory read, the individual frame’s shadows are projected on the protein surface, highlighting all the single events of the simulation. As the Umbrella Visualization is also available for single-structure .pdb files, this helps to put into evidence rare events occurring during a trajectory or a representative conformation, for demonstration and illustration purposes.
The integration of the Umbrella Visualization on the UnityMol interface overcomes the lack of flexibility and the cumbersome process of the former 2D version [14] since it offers a fully automated and user-friendly method able to accommodate all N-glycan structures. Saving the computed results is possible thanks to the Save result button. Moreover, the UnityMol interface provides tools to save pictures of the projected results on the protein surface.
The design of clear pictures for illustration purposes is offered through two display modes related to the projected shadow: the native computed results and a simplified representation delimiting clear zones depending on the glycan’s impact on the protein surface. Because comparing results between trajectories can be harder on a 3D protein surface, the projected texture can be saved in a picture file. As illustrated in this study, this helps emphasize interesting results and comparisons between the results since both the 2D information and the 3D information are available.
The relevance and performance of the method was evaluated considering different number of frames within a MD trajectory: namely 250, 2500 and 10,000 frames. We were able to see significant results even with a small numbers of frames that allowed us to highlight the main conformational tendencies of the glycan of interest. The largest samples showed more details and hinted at less represented conformations.
This emphasizes the high importance of the number of frames. Because of the statistical nature of the Umbrella Visualization, working with a low number of frames could lead to the overestimation of some conformations and the underestimation of others, meaning that we could miss relevant information. The choice of trajectory length is thus a key point regarding the significance of the Umbrella Visualization results. Nonetheless, this first experiment showed that this method has the potential to give informative results that could also either help design in vitro experiments and/or corroborate data from other calculations.
The Umbrella Visualization was applied in order to investigate the impact of glycans on the insulin receptor. We ran molecular dynamics simulation on the glycosylated protein and analyzed the trajectories. The Umbrella Visualization showed the different behaviour between the bi-antennary and tri-antennary glycans on the insulin receptor surface. We were able to see the impact of the third branch, extending the surface covered during the trajectory. Common behaviours between these MD trajectories also help to highlight a patch of basic residues that could be involved in protein/glycans interactions.
Although the glycan structures we used here are very common, we also know that, in some pathological contexts such as cancers, alteration of the glycosylation process leads to the synthesis of abnormal structures. Adding more branches could heavily influence the way proteins (With the insulin receptor, a known interaction partner is the neuraminidase I, for instance) interact with each other. For example, the profiling of the N-glycome of breast cancer cells showed that tri-antennary and tetra-antennary glycans were detected only in cancerous cells [44]. Investigating how multi-antennary glycans impact protein surfaces could bring new light on the influence of disruption of the glycosylation process.
We also showed that the glycans had a tendency to shift toward a patch of basic residues above the protein surface. These residues would be able to interact with the many hydroxyl groups of the sugar residues, hence stabilizing the position of the glycan above this region. Knowing how basic residues can be key amino acids in protein/protein interactions [45], it would be interesting to investigate the role of the residues composing this patch. We can imagine that these basic residues play a role in stabilizing the glycan in order to allow other proteins such as the neuraminisade I to interact with it. By correlating these results with the docking experiment previously ran, we bring new informations regarding the interaction with the neuraminidase I and the glycosylated insulin receptor.
We have described in previous studies how the insulin receptor’s function was altered by the hydrolysis of sialic acids [8] and that these saccharide had an impact on the flexibility of glycans[14], the next step will be to investigate how this change in flexibility impacts the insulin receptor function. By applying the Umbrella Visualization on an insulin receptor bearing fully sialylated glycans, we hope to point out important interactions involved in this mechanism. Because sialic acids are negatively charged, they have the potential to interact strongly with positively charged residues. It would be interesting to see if the basic patch we highlighted in our results has any influence on sialylated glycans.
The results presented in this paper showed the relevance and potential of the Umbrella Visualization in molecular modeling studies. In the near future, we plan to improve this method with further developments.
Currently, this methodology is usable for the study of one glycan at a time. Because some proteins are heavily glycosylated, we will optimize this to enable the visualization and analysis of multi-glycosylated proteins, meaning that their molecular weight is increased. This process heavily relies on the use of a prefab, so it is possible to instantiate one object several times in the same UnityMol session while applying transformations to components of single instances of the prefab. Each instance of the plane assigned to a glycan can thus be oriented accordingly. The Umbrella Visualization method could help investigate how glycans modulate protein-protein interfaces in biological contexts. For example, it has been shown that the human coronavirus NL63, a virus causing severe lower-respiratory-tract infections, uses a glycan shield to protect surface epitopes from the host immune system [46]. The role and impact of this shield could be unraveled with the Umbrella Visualization.
It is also important to note that the surface receiving the shadow is based on the initial .pdb file loaded in UnityMol and doesn’t change during the course of the trajectory (reduced computational work). Because proteins are also dynamical objects, it would be interesting to implement a quicker surface calculation and alter the surface shape for each frame. This would indicate how the protein evolves and adapts to the glycan motions. This could also impact the regions on which the shadow is projected and thus better highlight the influence of the glycan on the protein.
In a similar fashion, the lit plane prefab’s position and orientation is defined by the atom’s position on the starting structure and is not moving during the MD trajectory. This means that, in order to have significant results, the MD trajectory must be centered on the glycan so that it stays on the same area during the trajectory. While this is easily done with most MD software, this could still introduce bias on the results. To avoid those bias and generate results as precise as possible, we wish to recompute the plane’s position and orientation for every trajectory frame.
Local topology such as groove, bulges or the presence of other protein domains can also influence how this interaction would take place. On this first version of the Umbrella Visualization, the directional light is oriented perpendicularly to the plane, reproducing an orthogonal space. This introduces a bias into the results as it doesn’t reflect how another molecule would approach a protein in order to interact or form a complex. To correct this, we wish to allow the prefab to move around the glycan, following a given position defined by the user, and then visualize the results with this new orientation.
The Umbrella Visualization brings valuable structural information about the impact of glycans, but doesn’t offer any quantifiable value that would help evaluate more concretely the space occupied by glycans. However, the 2D graphical method previously developed took the form of a density plot [14], giving statistical informations but no structural information. Correlating the results of the 2D graphical method and the results of the 3D visual method is a good way to provide quantifiable data to support the visualization results.
To support this, we also plan to add methods to bring more informations to the Umbrella Visualization results, such as the occupied volume during the trajectory: as glycans are bulky, the space they explore and occupy during a MD trajectory could also inform us on their influence on molecular complex formation.
More generally, correlating the Umbrella Visualization results with other calculations could bring new insight into the impact of glycans on proteins. Among the methods used in molecular modeling, free-energy calculations are used to characterized interactions between partners[47]: it is possible to evaluate the contribution from each residue from both partners. It would be interesting to combine the Umbrella Visualization with free-energy calculation to emphasize how the protein’s amino acids influence the glycan’s behaviour and favor interactions with the glycan’s hydroxyl groups. By displaying both the results of such calculation and the results of the Umbrella Visualization on the protein surface, it would be possible to correlate the binding free energy with the glycan’s motions.
Molecular modeling is also able to help designing in vitro experiments. Molecular docking, for example, is used to design new drugs thanks to its ability to test several molecules without having to synthesize them [48]. Identification of the key interactions can further the understanding on how a drug will interact with a protein and influence its function. In a similar way, with the Umbrella Visualization, we wish to provide a tool able to help the design of studies focused on glycosylations. By highlighting which parts of the protein are affected by glycans, it is possible to identify important protein residues that are good candidates for point mutations in order to understand their impact on glycans and protein structure and dynamics.
In conclusion, we propose a new method available on the UnityMol viewer called the Umbrella Visualization. This method is dedicated to the study of glycosylated protein and highlights regions of the protein surface hidden by the glycan during a molecular dynamics trajectory. These regions are thus inaccessible to other molecules, emphasizing the impact of glycans on protein surfaces. By implementing this method into the UnityMol interface, the Umbrella Visualization becomes user-friendly and efficient, bringing us first results about the impact of glycans on the insulin receptor.
Acknowledgements
This work was supported by grants from the Region Grand Est and the ERDF (European Regional Development Fund). The authors thank the HPC-Regional Center ROMEO, the Multiscale Molecular Modeling Platform (P3M) and the national HPC-CINES Center for providing CPU-time and support. The chair MAgICS is aknowledged for financial and technical support.
References
- 1.Halper J., Kjaer M. Basic components of connective tissues and extracellular matrix: elastin, fibrillin, fibulins, fibrinogen, fibronectin, laminin, tenascins and thrombospondins. Adv. Exp. Med. Biol. 2014;802:31–47. doi: 10.1007/978-94-007-7893-1_3. [DOI] [PubMed] [Google Scholar]
- 2.Pozzi A., Yurchenco P.D., Iozzo R.V. The nature and biology of basement membranes. Matrix Biol. 2017;57–58:1–11. doi: 10.1016/j.matbio.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Theocharis A.D., Skandalis S.S., Gialeli C., Karamanos N.K. Extracellular matrix structure. Adv. Drug. Deliv. Rev. 2016;97:4–27. doi: 10.1016/j.addr.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Maquart F.X., Siméon A., Pasco S., Monboisse J.C. Regulation of cell activity by the extracellular matrix: the concept of matrikines. J. Soc. Biol. 1999;193:423–428. [PubMed] [Google Scholar]
- 5.Bonnans C., Chou J., Werb Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell. Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawecki C., Bocquet O., Schmelzer C.E.H., Heinz A., Ihling C., Wahart A., Romier B., Bennasroune A., Blaise S., Terryn C., Linton K.J., Martiny L., Duca L., Maurice P. Identification of CD36 as a new interaction partner of membrane NEU1: potential implication in the pro-atherogenic effects of the elastin receptor complex. Cell. Mol. Life Sci. 2019;76:791–807. doi: 10.1007/s00018-018-2978-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romier B., Ivaldi C., Sartelet H., Heinz A., Schmelzer C.E.H., Garnotel R., Guillot A., Jonquet J., Bertin E., Guéant J.-L., Alberto J.-M., Bronowicki J.-P., Amoyel J., Hocine T., Duca L., Maurice P., Bennasroune A., Martiny L., Debelle L., Durlach V., Blaise S. Production of elastin-derived peptides contributes to the development of nonalcoholic steatohepatitis. Diabetes. 2018;67:1604–1615. doi: 10.2337/db17-0490. [DOI] [PubMed] [Google Scholar]
- 8.Blaise S., Romier B., Kawecki C., Ghirardi M., Rabenoelina F., Baud S., Duca L., Maurice P., Heinz A., Schmelzer C.E., Tarpin M., Martiny L., Garbar C., Dauchez M., Debelle L., Durlach V. Elastin-derived peptides are new regulators of insulin resistance development in mice. Diabetes. 2013;62:3807–3816. doi: 10.2337/db13-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang J.B., Hernandez J., Leduc R., Frost S.C. Alternative glycosylation of the insulin receptor prevents oligomerization and acquisition of insulin-dependent tyrosine kinase activity. Biochim. Biophys. Acta. 2000;1499:74–84. doi: 10.1016/s0167-4889(00)00109-9. [DOI] [PubMed] [Google Scholar]
- 10.Gavel Y., von Heijne G. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 1990;3:433–442. doi: 10.1093/protein/3.5.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagae M., Yamaguchi Y. Sugar recognition and protein-protein interaction of mammalian lectins conferring diverse functions. Curr. Opin. Struct. Biol. 2015;34:108–115. doi: 10.1016/j.sbi.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Stanley P., Taniguchi N., Aebi M. N-glycans. In: Varki A., Cummings R.D., Esko J.D., Stanley P., Hart G.W., Aebi M., Darvill A.G., Kinoshita T., Packer N.H., Prestegard J.H., Schnaar R.L., Seeberger P.H., editors. Essentials of Glycobiology. third ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY): 2015. pp. 99–111. [Google Scholar]
- 13.Chuang G.-Y., Boyington J.C., Joyce M.G., Zhu J., Nabel G.J., Kwong P.D., Georgiev I. Computational prediction of N-linked glycosylation incorporating structural properties and patterns. Bioinformatics. 2012;28:2249–2255. doi: 10.1093/bioinformatics/bts426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guillot A., Dauchez M., Belloy N., Jonquet J., Duca L., Romier B., Maurice P., Debelle L., Martiny L., Durlach V., Baud S., Blaise S. Impact of sialic acids on the molecular dynamic of bi-antennary and tri-antennary glycans. Sci. Rep. 2016;6:35666. doi: 10.1038/srep35666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.L.L.C. Schrodinger, The PyMOL Molecular Graphics System, Version 1.8, 2015.
- 16.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(33–38):27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 17.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wormald M.R., Petrescu A.J., Pao Y.-L., Glithero A., Elliott T., Dwek R.A. Conformational studies of oligosaccharides and glycopeptides: complementarity of NMR, X-ray crystallography, and molecular modelling. Chem. Rev. 2002;102:371–386. doi: 10.1021/cr990368i. [DOI] [PubMed] [Google Scholar]
- 19.Hamby S.E., Hirst J.D. Prediction of glycosylation sites using random forests. BMC Bioinf. 2008;9:500. doi: 10.1186/1471-2105-9-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R. Gupta, E. Jung, S. Brunak, Prediction of N-glycosylation sites in human proteins, in preparation (2004).
- 21.Lutteke T., Frank M., von der Lieth C.-W. Data mining the protein data bank: automatic detection and assignment of carbohydrate structures. Carbohydr. Res. 2004;339:1015–1020. doi: 10.1016/j.carres.2003.09.038. [DOI] [PubMed] [Google Scholar]
- 22.Lutteke T., von der Lieth C.-W. pdb-care (PDB carbohydrate residue check): a program to support annotation of complex carbohydrate structures in PDB files. BMC Bioinf. 2004;5:69. doi: 10.1186/1471-2105-5-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bohne-Lang A., Lang E., Frster T., von der Lieth C.W. LINUCS: linear notation for unique description of carbohydrate sequences. Carbohydr. Res. 2001;336:1–11. doi: 10.1016/s0008-6215(01)00230-0. [DOI] [PubMed] [Google Scholar]
- 24.Frank M., Lutteke T., von der Lieth C.-W. GlycoMapsDB: a database of the accessible conformational space of glycosidic linkages. Nucleic Acids Res. 2007;35:287–290. doi: 10.1093/nar/gkl907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lutteke T., Frank M., von der Lieth C.-W. Carbohydrate Structure Suite (CSS): analysis of carbohydrate 3D structures derived from the PDB. Nucleic Acids Res. 2005;33:D242–D246. doi: 10.1093/nar/gki013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rojas-Macias M.A., Lutteke T. Statistical analysis of amino acids in the vicinity of carbohydrate residues performed by GlyVicinity. Methods Mol. Biol. 2015;1273:215–226. doi: 10.1007/978-1-4939-2343-4_16. [DOI] [PubMed] [Google Scholar]
- 27.Rosen J., Miguet L., Perez S. Shape: automatic conformation prediction of carbohydrates using a genetic algorithm. J. Cheminform. 2009;1:16. doi: 10.1186/1758-2946-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bohne A., Lang E., von der Lieth C.-W. W3-SWEET: carbohydrate modeling by internet. J. Mol. Model. 1998;4:33–43. [Google Scholar]
- 29.Bohne-Lang A., von der Lieth C.-W. GlyProt: in silico glycosylation of proteins. Nucleic Acids Res. 2005;33:W214–219. doi: 10.1093/nar/gki385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jo S., Song K.C., Desaire H., MacKerell A.D., Im W. Glycan reader: automated sugar identification and simulation preparation for carbohydrates and glycoproteins. J. Comput. Chem. 2011;32:3135–3141. doi: 10.1002/jcc.21886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danne R., Poojari C., Martinez-Seara H., Rissanen S., Lolicato F., Rág T., Vattulainen I. doGlycans-tools for preparing carbohydrate structures for atomistic simulations of glycoproteins, glycolipids, and carbohydrate polymers for GROMACS. J. Chem. Inf. Model. 2017;57:2401–2406. doi: 10.1021/acs.jcim.7b00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arroyuelo A., Vila J.A., Martin O.A. Azahar: a PyMOL plugin for construction, visualization and analysis of glycan molecules. J. Comput. Aid Mol. Des. 2016;30:619–624. doi: 10.1007/s10822-016-9944-x. [DOI] [PubMed] [Google Scholar]
- 33.Thieker D.F., Hadden J.A., Schulten K., Woods R.J. 3D implementation of the symbol nomenclature for graphical representation of glycans. Glycobiology. 2016;26:786–787. doi: 10.1093/glycob/cww076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varki A., Cummings R.D., Aebi M., Packer N.H., Seeberger P.H., Esko J.D., Stanley P., Hart G., Darvill A., Kinoshita T., Prestegard J.J., Schnaar R.L., Freeze H.H., Marth J.D., Bertozzi C.R., Etzler M.E., Frank M., Vliegenthart J.F., Látteke T., Perez S., Bolton E., Rudd P., Paulson J., Kanehisa M., Toukach P., Aoki-Kinoshita K.F., Dell A., Narimatsu H., York W., Taniguchi N., Kornfeld S. Symbol nomenclature for graphical representations of glycans. Glycobiology. 2015;25:1323–1324. doi: 10.1093/glycob/cwv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varki A., Cummings R.D., Esko J.D., Freeze H.H., Stanley P., Bertozzi C.R., Hart G.W., Etzler M.E., editors. Essentials of Glycobiology. second ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY): 2009. [PubMed] [Google Scholar]
- 36.Lv Z., Tek A., Da Silva F., Empereur-mot C., Chavent M., Baaden M. Game on, science – how video game technology may help biologists tackle visualization challenges. PLoS One. 2013;8 doi: 10.1371/journal.pone.0057990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chavent M., Vanel A., Tek A., Levy B., Robert S., Raffin B., Baaden M. GPU-accelerated atom and dynamic bond visualization using hyperballs: a unified algorithm for balls, sticks, and hyperboloids. J. Comput. Chem. 2011;32:2924–2935. doi: 10.1002/jcc.21861. [DOI] [PubMed] [Google Scholar]
- 38.Perez S., Tubiana T., Imberty A., Baaden M. Three-dimensional representations of complex carbohydrates and polysaccharides–SweetUnityMol: a video game-based computer graphic software. Glycobiology. 2015;25:483–491. doi: 10.1093/glycob/cwu133. [DOI] [PubMed] [Google Scholar]
- 39.Mazola Y., Chinea G., Musacchio A. Integrating bioinformatics tools to handle glycosylation. PLoS Comput. Biol. 2011;7 doi: 10.1371/journal.pcbi.1002285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schauer R. Sialic acids as regulators of molecular and cellular interactions. Curr. Opin. Struct. Biol. 2009;19:507–514. doi: 10.1016/j.sbi.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dotz V., Lemmers R.F.H., Reiding K.R., Hipgrave Ederveen A.L., Lieverse A.G., Mulder M.T., Sijbrands E.J.G., Wuhrer M., van Hoek M. Plasma protein N-glycan signatures of type 2 diabetes. Biochim. Biophys. Acta. 2018;1862:2613–2622. doi: 10.1016/j.bbagen.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 42.Berendsen H.J.C., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 43.Pronk S., Pall S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M.R., Smith J.C., Kasson P.M., van der Spoel D., Hess B., Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee L.Y., Thaysen-Andersen M., Baker M.S., Packer N.H., Hancock W.S., Fanayan S. Comprehensive N-glycome profiling of cultured human epithelial breast cells identifies unique secretome N-glycosylation signatures enabling tumorigenic subtype classification. J. Proteome Res. 2014;13:4783–4795. doi: 10.1021/pr500331m. [DOI] [PubMed] [Google Scholar]
- 45.Sinha N., Smith-Gill S.J. Electrostatics in protein binding and function. Curr. Protein Pept. Sci. 2002;3:601–614. doi: 10.2174/1389203023380431. [DOI] [PubMed] [Google Scholar]
- 46.Walls A.C., Tortorici M.A., Frenz B., Snijder J., Li W., Rey F.A., DiMaio F., Bosch B.-J., Veesler D. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016;23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Christ C.D., Mark A.E., van Gunsteren W.F. Basic ingredients of free energy calculations: a review. J. Comput. Chem. 2010;31:1569–1582. doi: 10.1002/jcc.21450. [DOI] [PubMed] [Google Scholar]
- 48.Sledz P., Caflisch A. Protein structure-based drug design: from docking to molecular dynamics. Curr. Opin. Struct. Biol. 2018;48:93–102. doi: 10.1016/j.sbi.2017.10.010. [DOI] [PubMed] [Google Scholar]