Highlights
-
•
The Type I/Type III IFN regulated endoribonuclease.
-
•
2-5A activates RNase L.
-
•
Ribonuclease activity of RNase L.
-
•
RNase L cleavage products activate cellular signaling.
Keywords: Ribonuclease L, Oligoadenylate synthetase, Influenza A virus, Innate immunity, Inflammasome, Autophagy, 2′-5′ Phosphodiesterase, Cell migration
Abstract
Ever since the discovery of the existence of an interferon (IFN)-regulated ribonuclease, significant advances have been made in understanding the mechanism and associated regulatory effects of its action. What had been studied initially as a “unique” endoribonuclease is currently known as ribonuclease L (RNase L where “L” stands for latent). Some of the key developments include discovery of the RNase L signaling pathway, its structural characterization, and its molecular cloning. RNase L has been implicated in antiviral and antibacterial defense, as well as in hereditary prostate cancer. RNase L is activated by 2′-5′ linked oligoadenylates (2-5A), which are synthesized by the oligoadenylate synthetases (OASs), a family of IFN-regulated pathogen recognition receptors that sense double-stranded RNAs. Activated RNase L cleaves single stranded RNAs, including viral RNAs and cellular RNAs. The catalytic activity of RNase L has been found to lead into the activation of several cellular signaling pathways, including those involved in autophagy, apoptosis, IFN-β production, NLRP3 inflammasome activation leading to IL-1β secretion, inhibition of cell migration, and cell adhesion. In this review, we will highlight the newest advances in our understanding of the catalytic role of RNase L in the context of different cellular pathways and extend the scope of these findings to discussion of potential therapeutic targets for antimicrobial drug development.
1. Introduction
Endoribonucleases are essential for diverse physiological processes, including control of gene expression by regulating RNA processing and turnover, cellular stress responses, anti-microbial defenses, and inflammation. They often operate through closely controlled mechanisms to maintain normal cellular homeostasis. Ribonuclease L (RNase L) is a classic example of a tightly regulated endoribonuclease as it is controlled by IFN-inducible oligo-adenylate synthetases (OASs) and double-stranded RNAs (dsRNAs) [reviewed in [1], [2]]. RNase L (previously known as 2-5A dependent ribonuclease) is a metal ion-independent endoribonuclease (molecular weight of 83 KDa) that functions in the Type-I/-III IFN-regulated 2′5′-OAS-RNase L pathway [2].
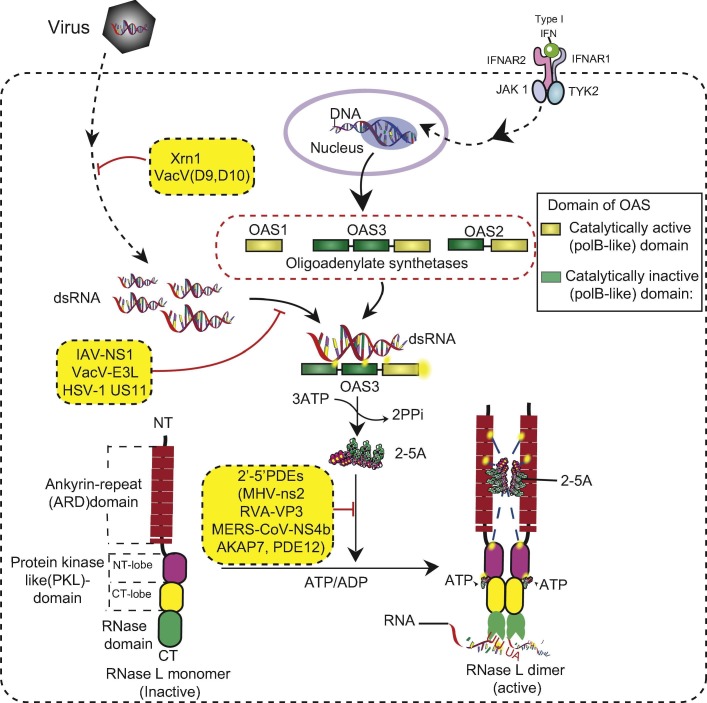
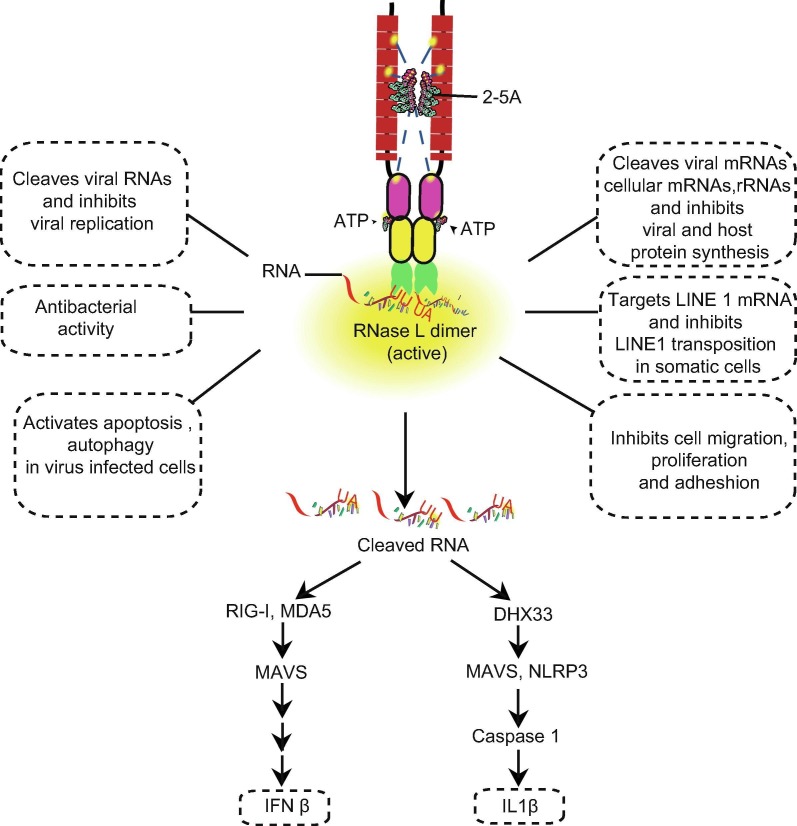
The OAS-RNase L (Fig. 1 ) pathway, discovered in the mid-1970s [3], [4], [5], [6], [7], [8], [9], [10], was one of the first IFN-dependent antiviral pathways to be characterized. OASs are IFN-I/-III-inducible genes that are expressed at very low basal levels in many cell types, except for immune cells such as macrophages [11], [12], [13]. Virus-infected cells induce the production of Type-I IFNs, which then activate JAK-STAT signaling [14], leading to the induction of hundreds of genes classified as members of the IFN-stimulated gene family, including the OASs [15]. OASs1-3 (but not OASL) act as pathogen recognition receptors that sense dsRNAs and activate the synthesis of 5′-phosphorylated 2′-5′ linked oligoadenylates from ATP. The 2′-5′ linked oligoadenylates [with a chemical structure of px5′A(2′p5′A)n, where x = 1–3; n > 2] are commonly referred to as 2-5A [5]. 2-5A acts as a second messenger and binds monomeric RNase L, and activates its dimer formation [16]. Naturally occurring 2-5A contains three phosphoryl groups in the 5′ end [5]; however, 2-5A monophosphorylated on the 5′ end has also been found to activate RNase L under in vitro conditions [16]. Active RNase L cleaves cellular and viral RNAs [17] within single-stranded regions, predominantly at the 3′-end of UN dinucleotides (preferentially after UU or UA), to produce RNA products that have a 5′-OH group and 2′,3′-cyclic phosphate at the 3′ end [18], [19], [20]. RNA degradation directly and indirectly activates subsequent events (Fig. 2 ), including the elimination of viral genomes, inhibition of cellular and viral protein synthesis [17]; and activation of several cellular signaling pathways, including those involved in autophagy [21], [22], apoptosis [23], [24], [25], senescence [26], IFN-β production [27], [28], [29], and NLRP3-inflammasome activation [30] as part of its antiviral mechanism.
Fig. 1.
The OAS–RNase L pathway. The OAS-RNase L pathway is activated by dsRNAs and IFN signaling. Among the OASs, OAS3 senses dsRNA more efficiently and induces the synthesis of 2-5A from ATP. 2-5A then binds to the RNase L monomer and activates its dimer formation, which is the active conformation of RNase L. Active RNase L degrades cellular and viral RNA, leading to the restriction of virus replication. Several cellular and viral proteins inhibit the OAS-RNase L pathway at different stages.
Fig. 2.
Role of RNase L in antiviral innate immunity and cellular pathways.
In this review we will first highlight recent discoveries on the ribonuclease function of RNase L, including its activation and targeting of certain RNAs; then comment on implications of several cellular events as potential targets for novel therapeutic intervention.
2. RNase L biochemistry and its activation
RNase L, first cloned in 1993 [31], is constitutively expressed in every human cell type in an inactive conformation. In mouse cells, its expression is slightly induced by IFN stimulation [32]. Human RNase L contains 741 amino acids and three domains: (i) the N-terminal ankyrin repeat domain (ARD), which contains one partial and eight complete ankyrin repeat domains; (ii) a protein kinase-like domain (PKL) or kinase homology domain that lacks the conserved DFG motif found in most protein kinases and contains a DFD sequence, similar to protein kinases Mnk1/2. So far, no kinase activity has been reported for PKL and hence RNase L is considered to be a pseudokinase. The PKL domain consists of a bi-lobal fold with a short N-terminal lobe (N-lobe) and a larger C-terminal lobe (C-lobe); and (iii) Extending as a rigid protrusion from the PKL C-lobe is the C-terminal of the nuclease domain. This kinase homology domain and the kinase extension of the nuclease (KEN) domain have structural similarity with IRE1, a metal ion-independent endoribonuclease activated during the endoplasmic stress response. The homology suggests that RNase L and IRE1 may share similar catalytic mechanisms [33].
The precise mechanism of RNase L activity was elucidated by the determination of the crystal structure of full-length dimeric RNase L in a complex with 2-5A and an ATP analog. Early structural studies identified two different 2–5A binding sites (2 and 4 ANKD) in the ankyrin domain (ANKD) [34]. In contrast, the crystal structure of 2-5A bound to full-length RNase L provided evidence that 2-5A binds both the ANK-repeat domain and the N-lobe of the PKL domain to facilitate its dimerization [35], [36]. ATP and ADP binding to the pseudokinase site leads to the formation of the single closed conformation, which is the activated form of RNase L. Detailed studies of ribonuclease activity and RNase L targets are highlighted in the following sections.
3. Cellular and viral inhibitors of OAS-RNase L
Several cellular and viral inhibitors of OAS-RNase L pathway have been identified (Table 1 ). These inhibitors act at different levels such as: sequestering dsRNA thus preventing or suppressing OAS activation, directly binding and inhibiting RNase L, and degrading 2-5A molecules hence inhibiting the dimerization and ribonuclease activity of RNase L. During viral infections, RNase L degrades both viral and cellular RNAs in single-stranded regions, including rRNA in intact ribosomes [37], [38]. Many, perhaps most viruses, have evolved or acquired strategies that antagonize the OAS-RNase L pathway to evade antiviral innate immunity (Fig. 1).
Table 1.
Inhibitors of the OAS-RNase L pathway.
| Target | Cellular | Viral |
|---|---|---|
| dsRNA | – | IAV NS1; VacV E3L, D9, D10; TGEV protein 7 |
| RNase L | ABCE1 | Poliovirus ciRNA; TMEV L∗ |
| 2-5A | AKAP7, ENPP1, PDE12 | MHV ns2; RVA VP3-CTD; MERS-CoV NS4b |
Inhibitors of RNase L are encoded by widely different types of viruses, indicating that RNase L targets a variety of viruses. The influenza A virus (IAV) non-structural 1 protein (NS1) contains an N-terminal RNA-binding domain which binds to and sequesters dsRNA, the activator of OAS [39], [40], [41]. RNase L inhibits IAV (NS1-mutant) replication by cleaving viral genomic RNA in sites encoding PB2, PB1, and PA proteins critical for viral replication [42]. In a similar mechanism, the herpes simplex virus type 1 (HSV1) US11 gene product [43] and vaccinia virus (VacV) E3L protein binds dsRNA to inhibit the innate immune response [44], [45], [46]. Recently two separate studies [47], [48] found that VacV encodes decapping enzymes D9 and D10 which are capable of degrading methylated mRNA cap structures, making them susceptible to the cellular 5′ exonuclease Xrn1. Infection with D9, D10 catalytic mutant viruses significantly increased dsRNA-induced activation of OAS and PKR, similar to Xrn1 mutation. In addition, protein 7 of transmissible gastroenteritis coronavirus (TGEV) binds to protein phosphatase 1 catalytical subunit, and it inhibits eIF2α dephosphorylation by protein kinase R (PKR) and rRNA degradation by RNase L [49]. Other viral proteins bind to monomeric RNase L to prevent dimerization. The L∗ protein of Theiler’s murine encephalomyelitis virus (TMEV, a picornavirus) binds to the ankyrin domain of murine RNase L and inhibits 2-5A binding and RNase L activation. Interestingly the L∗ protein is species-specific and binds to murine but not to human RNase L [50]. A phylogenetically conserved highly structured RNA within the open reading frame of poliovirus and other group C enteroviruses also serves as a competitive inhibitor of RNase L (ciRNA). CiRNA does not impair the 2-5A binding but instead acts as a competitive inhibitor of the RNASE domain, and the inhibitory function requires a putative E-motif and H-H kissing loop [51], [52].
An increasing number of viruses have been identified to encode for 2′,5′-phosphodiesterases (PDEs) that directly degrade the 2-5A activator molecules [reviewed in [53]]. These 2′,5′-PDEs belong to the 2H-phosphoesterase superfamily of proteins, characterized by two conserved His-X-Thr/Ser motifs in their active site [54]. The non-structural 2 (ns2) protein of group 2a coronavirus mouse hepatitis virus (MHV), a member of the viral Lig-T family of 2H-phosphoesterases, was necessary for replication of A59 strain of MHV in livers of infected mice as well as liver pathogenesis [55]. ns2 degraded 2-5A in vitro. A catalytically mutant histidine virus A59-ns2H126R had significantly lower titers compared to wild type virus in vivo. In contrast, in RNase L knockout B6 mice, both viruses replicated similarly, suggesting that ns2 directly acts through OAS-RNase L [56]. Similarly, rotavirus A (RVA) encodes for the viral protein 3 (VP3), which is structurally homologous in its C-terminal domain to MHV-ns2 [54], [57], [58]. Although from evolutionarily non-related viruses, VP3 degraded 2-5A in similar manner as ns2 in vitro and in intact cells, whereas mutation of the two conserved histidines in the PDE catalytic site of VP3 prevented 2-5A degradation. Furthermore, the VP3 C-terminal domain restored the function of catalytically mutant ns2 when inserted into a non-essential gene ns4 in a chimeric MHV [57]. Another viral 2′,5′-PDE is the NS4b protein of Middle East respiratory syndrome coronavirus (MERS-CoV). Structurally homologous to MHV-ns2, nuclear protein NS4b also degrades 2-5A and was able to restore the function of a mutant ns2 in a chimeric MHV. Mutation of the NS4b PDE region activated RNase L, resulting in rRNA cleavage [59].
Several host PDEs can also degrade 2-5A (Fig. 1, Table 1). The A kinase anchoring protein 7 (AKAP7) is a cellular member of the 2H-phosphoesterase superfamily [54], [60]. AKAP7 degraded 2-5A in vitro with rates similar to its viral homologues, ns2 of MHV and VP3 of RVA. The 2H central domain of the murine AKAP7 restored replication of ns2 mutant MHV chimeric virus whereas MHV chimeras with a histidine-to-arginine mutation in the His-X-Thr motif of AKAP7 central domain failed to replicate in bone marrow macrophages of B6 mice [61]. Due to structural homology, it was proposed that AKAP7 could be the evolutionary precursor of viral 2′,5′ PDE members [58], [61]. In addition, phosphodiesterase 12 (PDE12) is a member of the exonuclease-endonuclease phosphatase family that localizes in mitochondria and regulates mRNA turnover by removing the polyA tails [62], [63]. Despite its mitochondrial localization, several studies have shown PDE12 to be an antagonist of OAS-RNase L. It directly cleaves the phosphodiester bonds of 2-5A in vitro. Decreased expression of PDE12 in cells reduced viral titers of VacV [64], but for EMCV, in the absence of PDE12 expression, greater multiplicity of infection was required for a cytopathic effect comparable to wild type cells [65]. Furthermore, the plasma membrane ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1) was also shown to have a 2′-5′ nuclease activity by degrading 2-5 in vitro [66]. ATP binding cassette E1 (ABCE1), also known as RNase L inhibitor (RLI), is another mammalian protein that inhibits the OAS-RNase L pathway. ABCE1 was reported to directly bind to RNase L, preventing 2-5A binding [67] and it is induced during viral infection by EMCV and HIV [68], [69], [70].
In summary, viruses and host cells encode a variety of proteins that inhibit the OAS-RNase L pathway. In contrast, as discussed below, 2-5A is the only known physiological activator of the pathway.
4. Activation of the OAS-RNase L pathway
Since the discovery of the OAS-RNase L pathway, a significant amount of research has been directed toward identifying its activators and inhibitors [reviewed in [53]]. As discussed above, several inhibitors, including viral proteins, viral RNA antagonists, and host proteins, have been identified within the last few decades, but little is known about potential activators of this pathway. So far, 2-5A is the only well-characterized and known physiological activator of RNase L. In addition, studies on some small molecule activators have been published, but their potency is much less than that of 2-5A, as sub-nanomolar concentrations of 2-5A lead to the activation of RNase L [71].
2-5A synthesis occurs when dsRNAs bind to and activate IFN-inducible oligoadenylate synthetases (OAS1-3). The human OASs include OAS1, 2, 3, and L, and additional isoforms due to alternative splicing. OASs 1-3 are encoded on chromosome 12q24.1 arm and possess 2′-5′ nucleotidyl transferase activity. Human OAS L is catalytically inactive and is encoded on chromosome 12q24.2 [reviewed in [72]]. Each OAS protein contains a core polymerase β (pol-β)-like nucleotidyl transferase domain [73] that has structural similarity with polyA polymerase, CCA-adding enzyme, and cGAS [74]. However, only the OASs and cGAS are known to polymerize nucleotides in the 2′-5′ orientation. Although OAS and cGAS have structural similarity, they differ in their substrate recognition. cGAS recognizes cytosolic dsDNA (B-form) and catalyzes cyclic guanosine monophosphate–adenosine monophosphate (cGAMP) [74], whereas OASs 1-3 are activated by dsRNA (A-form) [75] and catalyze the synthesis of 2-5A. On the other hand, human OAS L has a catalytically inactive OAS domain at the N-terminal and contains two ubiquitin-like domains in the C-terminus [76], which are not present in any of the other OAS proteins. Human OAS L was shown to promote antiviral activity by enhancing RIG-I activation [77].
Among the OASs, OAS1 contains one OAS domain (catalytically active), and OAS2 and OAS3 contain two and three OAS domains respectively, but only the C-terminal OAS domain is catalytically active [reviewed in [12], [72], [73]]. The OAS active site contains three aspartic acids, which coordinate with two Mg2+ ions required for enzyme activity. OAS3 has higher affinity for long dsRNA segments. Both the catalytic and non-catalytic domains of OAS3 are involved in dsRNA binding, but only the C-terminal domain is involved in 2-5A synthesis [73]. OAS1, 2 and 3 all synthesize 2-5A in vitro, but in intact cells, OAS3 synthesis of 2-5A predominates [11], [73], [78]. In a recent study we used individual OAS knockout human cell lines and identified OAS3 as the principle enzyme required for optimal RNase L activation and its antiviral mechanisms [11]. OAS1 and OAS2 knockout cells produced 2-5A in amounts comparable to the parental cells in response to dsRNA or virus infection, whereas OAS3 knockout cells were completely deficient in 2-5A synthesis and lacked significant RNase L activation [11]. These findings support prior studies in which OAS3 was shown to exert antiviral activity against Dengue virus in an RNase L-dependent manner [79], indicating that OAS3 synthesizes active 2-5A in sufficient amounts for RNase L activation. OAS1 and OAS2 probably have antiviral functions that support OAS3 activity, but they might possibly have additional or alternative functions as well. Recombinant OAS1 protein was reported to inhibit EMCV replication in an RNase L-independent manner [80], [81], indicating that this protein may have an alternative antiviral mechanism.
Besides dsRNA sensing and IFN signaling, very little information is available on possible relationships between other cellular pathways and OAS-RNase L activation. The nucleotide-binding oligomerization domain-containing protein 2 (NOD2), a member of the NOD-like receptor family (NLR), interacts with OAS2 (P69) and leads to the enhanced activation of RNase L in the poly(rI):poly(rC)-induced human monocytic leukemia cell line THP1 [82]. NOD2 was also shown to bind ssRNA and activate IRF3 [83].
5. RNase L cleavage
Ribosomal RNA cleavage is one of the hallmarks of RNase L activation [37], [38]. RNase L cleaves cellular and viral RNAs within single-stranded regions after a UN dinucleotide (with a preference in the order of UU > UA ≫ UG > UC). Studies focused on identifying the RNA targets of RNase L have provided some insight on cellular and viral targets [18], [42], [84], [85]. A recent microarray-based study identified certain target mRNAs regulated by RNase L activation. mRNAs that encode ribosomal proteins were found to be more prominent targets [84]. However, the study identified only finite transcripts that were either upregulated or downregulated during RNase L activation. Another investigation developed a new approach for transcriptome-wide profiling of RNase L-dependent decay (RLDD) and mapped hundreds of direct target and nontarget mRNAs in human cells [85]. This study excluded rRNAs and focused mainly on the mRNA targets of RNase L. AUUUA/UAUAU motif enrichment was greater in target RNA compared to the non-target RNAs. RLDD also targeted several miRNA-regulated transcripts, indicating that RNase L may mimic the effect of some miRNAs involved in the suppression of cell proliferation and adhesion.
None of these studies focused on RNase L cleavage sites on viral RNAs, however. In a recent modified RNA sequencing study by Cooper and colleagues [18], [42], RNase L target sites in the cellular RNAs (rRNA, snRNA such as U6 and U3) and also specific mapping of the viral RNAs (HCV, IAV, polio virus) were precisely mapped. The method used exploits the 2′-3′ cyclic phosphate at the 3′ end of the cleaved RNA (which is a characteristic of metal ion-independent RNases) and employs 2′-3′ cyclic phosphate cDNA synthesis with Illumina sequencing to map RNase L targets from either in vitro cleaved RNAs or RNAs from virus-infected cells. Unlike many studies that actively exclude rRNAs from RNA sequencing experiments to enrich for mRNA and other RNA populations, these two studies intentionally retained rRNAs to precisely locate RNase L-target and RNase L non-target cleavage sites in rRNAs. This method specifically identified two prominent RNase L-dependent cleavage sites in 18S RNA UU541 and UU743 [18], [42]. This method also uncovered other RNase L-dependent and -independent cleavage sites within the ribosomal RNAs, most of which were mapped in association with the single-stranded region at the ribosome surface. In addition, these methods allowed for the precise mapping of endoribonuclease cleavage sites in viral RNAs from IAV (ΔNS1) infected cells [18], [42]. The influenza A virus has eight negative sense RNA (genome) segments (PB1, PB2, PA, HA, NA, NP, M and NS segments) encoding one or multiple viral proteins required for viral replication and pathogenesis. The endoribonuclease cleavages were prominent at discrete locations in PB2, PB1, PA, NP and M RNA segments, and RNase L knockdown reduced the amount of cleavage [42]. Large numbers of cleavage sites were detected within or adjacent to some areas of increased synonymous-site conservation. It was noted that RNase L-mediated cleavage was more prominent in the M positive-strand than in the other positive strand IAV RNAs, which has a uridine-rich 15-base sequence (200AUUUUAGGAUUUGUG214). The cleavage sites in M RNA lay within the area of increased synonymous-site conservation, 50 bases downstream from the M4 splice donor sequence. Cleavage sites also occurred more frequentlyin the PA, PB1, and PB2 segments of the IAV genome (−ve strand) than in other IAV genomic RNAs, suggesting that the viral nucleocapsid does not protect IAV RNAs from RNase L-mediated cleavage within the ribonucleoprotein complexes. Prominent cleavages were mapped within specific regions of IAV RNAs, including some areas of increased synonymous-site conservation such as the PB1-positive and -negative strands, NP-positive strands, and PA-negative strands. PB2 negative-strand RNA was cleaved in discrete regions without increased synonymous-site conservation. Compared to the other segments, the HA and NA segments have less synonymous-site conservation and therefore are less susceptible to RNase L-mediated cleavage. Because the viral packaging signals tend to be uridine-rich sequences within the areas of increased synonymous-site conservation [86], [87] and on the basis of patterns of synonymous-site of conservation and RNase L target sites, it might be interesting to consider that the viral RNA packaging signals are potential targets of RNase L. Although the study by Cooper et al. precisely mapped RNase L cleavage sites in viral RNAs, for cellular RNAs only highly abundant cellular RNAs like 28SRNA, 18SRNA, and U6 RNA were precisely mapped.
6. Role of RNase L in autophagy
RNase L has been implicated in the induction of autophagy during viral infection [21], [22]. Activation of autophagy conferred antiviral resistance during the early stages of EMCV infection but promoted viral replication during later phases. Both protein kinase R and c-jun kinase were shown to be involved in autophagy activation; however, the exact mechanism by which RNase L mediates autophagy is yet to be discovered [21]. It has been found that RNase L activation is essential for autophagy induction as a result of rRNA cleavage, which induces stress due to the rRNA degradation. Ribosome turnover might be a potential cause of RNase L-mediated autophagy induction [22]. RNase L activation and RNase L cleavage products participate in crosstalk between autophagy and apoptosis, as RNase L cleavage products can activate the switch to apoptosis by caspase 3-dependent cleavage of Beclin 1 and hence may trigger cytochrome c release from mitochondria [88].
7. Role of RNase L in innate immunity
RNase L activation by dsRNA signaling or viral infection contributes to IFN-β production, indicating its important role in innate immunity. The ribonuclease function of RNase L is essential for its effect on IFN-β production [27], [28], [29]. RNase L-mediated cleavage products of both cellular and viral RNAs (less than 200 nt) are involved in the activation of pathogen recognition receptors such as RIG-I and MDA5 and hence activate downstream signaling cascades for IFN-β production [28], [29]. These RNA species are designated as suppressor of virus RNA (svRNA) with a 2′-3′-cyclic phosphate [18] and 5′-hydroxyl (5′-OH) at the termini. The presence of the terminal phosphate on the cleaved RNA is essential for its recognition by RLRs (RIG-like receptors, RIG-I or MDA5) as calf intestinal phosphatase treatment diminished svRNA ability to produce IFN-β [28]. Moreover, mice deficient in RNase L had several-fold reduced levels of IFN-β induction after infection with RNA viruses (EMCV and Sendai virus) [29]. In MEF cells infected by HSV-2, a DNA virus, RNase L also induced production of IFN-β [89]. However, in mouse primary macrophages, RNase L negatively regulated IFN-β production after poly (rI):poly(rC) transfection or EMCV infection [90]. Compared to mouse embryonic fibroblasts (MEFs), the levels of the different OASs (1–3 and L1) in mouse primary macrophages is overall much higher, possibly explaining the observed elevation in RNase L activity. Increased RNase L activation eventually leads to cell death, which may explain the reduced IFN-β production in macrophages compared to MEFs [90].
8. Role of RNase L in inflammasome signaling
Recently we reported that RNase L is essential to NLRP3 inflammasome activation and IL-1β secretion during IAV infections in LPS-primed bone marrow-derived primary dendritic cells (BMDCs), human monocytes, THP1 cells, and thioglycolate-elicited primary peritoneal macrophages [30]. Inflammasomes are macromolecular complexes that induce inflammatory signaling upon activation by various sensory signals, including pathogen-associated molecular patterns and danger-associated molecular patterns. The canonical inflammasome activates caspase 1, which leads to the cleavage of pro-IL-1β and production of mature IL-1β. Cleaved IL-1β is then secreted through an unconventional pathway.
Recently, both NLR (NLRP3, NLRP6) and non-NLR (AIM2) inflammasomes have been shown to be involved in viral infection events, further activating downstream antiviral responses [reviewed in [91]]. NLRP3 is a multi-domain protein consisting of a NT-PYD (pyrin domain), a central NATCH domain, and C-terminal-leucine-rich repeat (LRR). Two signals are required for NLRP3 inflammasome activation: (i) a priming signal, which involves the transcriptional activation of NFκβ target genes including NLRP3, caspase 1, and proIL-1β; and (ii) a second signal that is required for assembly of the macromolecular complex [92]. The NLRP3 inflammasome is activated during a wide range of RNA and DNA virus infections [reviewed in [91]]. We also found that RNase L increases IL-1β levels in bronchoalveolar lavage fluid and lung tissue after IAV infection [30]. We observed that RNase L activity had no effect on signal 1 (inflammasome priming) of LPS-primed mouse BMDCs, but did affect signal 2, activating the inflammasome.
Stable expression of wild-type human full-length RNase L, but not ribonuclease dead mutant (R667A), activates IL-1β and caspase 1 secretion in RNase L-deficient THP1 cells after virus infection or 2-5A transfection. Intracellular delivery of 2-5A activates IL-1β and caspase 1 secretion in culture supernatants of LPS-primed wild type mouse BMDCs but not in NLRP3, RNase L, and ASC-knockout BMDCs. RNase L-cleaved RNA products (both cellular total RNAs and IAV genomic RNAs) can activate IL-1β and caspase 1 in culture supernatants of LPS-primed BMDCs from wild-type mice and RNase L knockout mice but not in BMDCs of NLRP3 and ASC knockout mice [30]. Earlier studies showed that RNase L cleavage products are capable of activating the pathogen recognition receptors RIG-I and MDA5 [28], [29]. However, further investigation [30] showed that neither RIG-I nor MDA5 were involved in RNase L-mediated IL-1β production, but rather the Rig-I-like receptor (RLR) pathway adaptor, known as MAVS, played an essential role in IL-1β production during viral infection. Other studies have also implicated involvement of MAVS in NLRP3 inflammasome activation [93], [94]. In addition, our data suggested that the presence of 2′-3′ cyclic phosphate at the 3′ end of the cleaved RNA was essential for inflammasome activation.
The NLRP3 inflammasome pathways have been shown to be activated by the cytosolic delivery of different RNA molecules, including the dsRNA analogue poly(rI):poly(rC), bacterial RNA, and viral RNA molecules. DHX33 is a DExD/H-box RNA helicase with a helicase C-type domain like RIG-I and MDA5 but lacks a CARD domain to act as an upstream sensor of NLRP3 [95]. DHX33 binds to cytosolic RNAs and interacts with MAVS and NLRP3 [96]. We found that RNase L-cleaved RNAs directly interact with DHX33 to activate its association with RNA, MAVS, and NLRP3 [30]. We also found that the presence of 2′-3′ cyclic phosphate at the 3′ end of cleavage products is essential for the interaction between RNase L and DHX33 [30]. shRNA downregulation of DHX33 in THP1 cells inhibited caspase 1 activation and IL-1β secretion upon infection with Vesicular stomatitis virus (VSV), IAV, or transfection with poly(rI):poly(rC) or 2-5A.
9. The antibacterial role of RNase L
Although RNase L primarily acts in antiviral immunity, it has also been found to protect against bacterial infections, specifically Escherichia coli and Bacillus anthracis [97]. Infection of RNase L-deficient mice by these species increased bacterial load and mortality in comparison with wild-type mice [97]. Microarray profiles showed that RNase L deficiency enhanced the stabilization of cathepsin E mRNA (an endosomal aspartyl proteinase) in macrophages. Higher expression of cathepsin E was linked to decreased levels of LAMP1/2, which is required for phagosome maturation during which the late endosome fuses with the lysosome to eliminate bacteria through phagocytosis. Decreased LAMP1 and LAMP 2 levels in RNase L-deficient macrophages were associated with the accumulation of phagocytic vacuoles and reduced bacterial clearance. Another study reported that RNase L mediates protection against dextran sulfate sodium (DSS)-induced colitis and colitis-associated cancer in mice [98]. DSS-treated RNase-L−/− mice had significantly higher mortality and clinical morbidity score, delayed infiltration of leukocytes, and reduced expression of IFN-β and proinflammatory cytokines TNFα, IL-1β, and IL-18, as compared to DSS-treated wildtype mice. Moreover, DSS/azoxymethane-treated RNase L−/− mice displayed a higher tumor burden compared to wild-type mice. This finding indicates that the OAS-RNase L pathway contributes to maintenance of innate immune signaling within the intestinal environment. Studies have also shown that enteropathogenic E. coli (EPEC) infection inhibits RNase L activation through a Type III secretion system (T3SS), which helps EPEC to evade IFN-induced antibacterial activities [99]. Intestinal epithelial cell lines infected with a T3SS-deficient EPEC strain show higher RNase L activation and IFN-β production in an RNase L-dependent manner compared to infection with wild type EPEC; indicating novel roles for IFN-β and RNase L in the barrier functions of the intestinal epithelial cell, which is a target of EPEC secretion systems. This finding reinforces the potential of the OAS-RNase L system to serve as a critical therapeutic candidate for treating enteric infections and gastrointestinal tract diseases.
10. The role of RNase L in cell migration and adhesion
As evidenced by earlier genetic studies, RNase L has also been linked to hereditary prostate cancer. The RNase L gene (RNASE L) was identified as a candidate for the hereditary prostate cancer 1 (HPC1) locus on human chromosome 1q24-25 through a combined positional cloning and candidate gene approach [100]. Since then, however, several studies have found a positive association [101], [102], [103] or no association [104], [105], [106] between hereditary prostate cancer and mutations, and variants in the RNASE L gene. One possible explanation includes the influence of environmental factors, such as microbial populations, that may regulate the activation of RNase L. In addition to the association with prostate cancer, mutations in the RNASE L gene have been implicated to an increased risk for other cancers including non-melanoma skin cancer [107], head and neck, uterine, cervix, breast [108], pancreatic [109], and hereditary non-polyposis colorectal cancers [110]. Recently, RNase L activity has been linked to several cellular processes that may contribute to the development of cancer, including apoptosis, autophagy, inflammation, and the suppression of a mobile genetic element (LINE1) [111]. An earlier study demonstrated that the prostate cancer cell lines PC3, LNCaP, and DU145 express higher levels of cellular RNA molecules that are capable of binding and activating OASs than do normal prostate epithelial cells [112]. These activators of OAS were identified to be mRNAs for Raf kinase inhibitor protein, poly(rC)-binding protein 2 (PCBP2), and human endogenous retrovirus envelope RNAs. This study also showed that PCBP2 mRNA levels were elevated during prostate cancer progression.
RNase L was reported to influence the activity of filamin A by affecting actin cytoskeleton formation, thereby inhibiting virus entry [113]. During viral infection, however, RNase L was released from the filamin A complex, enabling the activation of its nuclease-dependent antiviral function, which may further be involved in processes such as cell adhesion and cell motility [113]. Recently, RNase L deficiency was found to enhance cell migration [85], [114]. The inactivated nuclease mutant RNase L (R667A), which lacks catalytic activity, failed to suppress cell migration in PC3 cells, a highly metastatic prostate cancer cell line [114]. The expression of a nuclease dead mutant (W460) mouse RNase L only partially restored the inhibition of cell migration in comparison to wild-type mouse RNase L, expressed in RNase L deficient MEF cells. These findings suggest that both the nuclease-dependent and -independent properties of RNase L may be involved in regulating cell migration in MEFs. Transfection of PC3 cells with the RNase L activator 2-5A also suppressed cell migration, providing further evidence that RNase L catalytic activity is necessary for this process in prostate cancer cell lines [114].
In contrast, another report showed that RNase L deficiency decreases migration of bone marrow-derived macrophages [115]. This may possibly be due to cell type differences. In MEFs, both catalytic and non-catalytic domains of RNase L played a role in inhibiting cell migration, whereas in PC3 cells, only the nuclease domain showed a prominent effect. The exact mechanism by which IFN regulates RNase L to suppress cell migration remains to be determined. However, certain findings have begun to reveal features of this process. RNA cleavage products generated by RNase L were previously shown to activate innate immune signaling mechanisms through RIG-I (RLR), MDA5, and MAVS-like receptors. In contrast, siRNA mediated down regulation of RIG-I or MDA5 or MAVS in PC3 cells did not affect cell migration. These results indicate that RNase L-mediated decay of mRNAs that encode for cell adhesion and migration proteins appears to be a key mechanism for RNase L-mediated inhibition of cell migration. RNase L-dependent mRNA decay selectively affects transcripts regulated by microRNAs (like miR17, miR-29, and miR-200 and other miRs) that negatively regulate mammalian cell adhesion and proliferation [85]. RNase L-mediated decay of miRNA targets may also potentially mimic the effects of some miRNAs that act as suppressors of proliferation and adhesion in mammalian cells. These studies extend the known biological roles and activities of RNase L to the inhibition of cell migration, adhesion, and proliferation, providing an explanation for the proposed link between mutations and variants in the RNASE L gene and increased cancer risk [101], [102], [103], [108], [109], [110].
11. Conclusion
RNase L is a unique IFN-regulated endoribonuclease that serves as an important mediator of antiviral innate immunity with possible roles in antibacterial defense and prostate cancer. The past few decades have witnessed remarkable advances in our understanding of RNase L and its biological activities. Probing studies on RNase L structure and function are providing insight into its mode of action and biological role. Also several cellular pathways are implicated that are directly or indirectly required to protect the host from various infectious agents, allowing for increased control of host inflammation and related diseases, including cancer. Targeting optimum RNase L activation may provide a potential platform to develop broad-spectrum anti-microbial therapies for inflammatory diseases and cancer.
Acknowledgement
The author S.B would like to thank the organizers of Cytokine 2015 and International Cytokine and Interferon Society (ICIS) and the Milstein Family for giving the “Young Investigator Award” in Cytokine 2015, Bamberg, Germany. In addition, the authors would like to thank Dr. Robert Silverman and Dr. Cassandra Talerico at the Cleveland Clinic for reading and critically reviewing the manuscript. Finally, the authors would like to apologize to all scientists whose efforts could not be cited because of the space constraints.
References
- 1.Silverman R.H. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 2007;81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silverman R.H. A scientific journey through the 2–5A/RNase L system. Cytokine Growth Factor Rev. 2007;18:381–388. doi: 10.1016/j.cytogfr.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hovanessian A.G., Brown R.E., Kerr I.M. Synthesis of low molecular weight inhibitor of protein synthesis with enzyme from interferon-treated cells. Nature. 1977;268:537–540. doi: 10.1038/268537a0. [DOI] [PubMed] [Google Scholar]
- 4.Roberts W.K., Clemens M.J., Kerr I.M. Interferon-induced inhibition of protein synthesis in L-cell extracts: an ATP-dependent step in the activation of an inhibitor by double-stranded RNA. Proc. Natl. Acad. Sci. USA. 1976;73:3136–3140. doi: 10.1073/pnas.73.9.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerr I.M., Brown R.E. PppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc. Natl. Acad. Sci. USA. 1978;75:256–260. doi: 10.1073/pnas.75.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clemens M.J., Williams B.R. Inhibition of cell-free protein synthesis by pppA2′p5′A2′p5′A: a novel oligonucleotide synthesized by interferon-treated L cell extracts. Cell. 1978;13:565–572. doi: 10.1016/0092-8674(78)90329-x. [DOI] [PubMed] [Google Scholar]
- 7.Ratner L., Wiegand R.C., Farrell P.J., Sen G.C., Cabrer B., Lengyel P. Interferon, double-stranded RNA and RNA degradation. Fractionation of the endonucleaseINT system into two macromolecular components; role of a small molecule in nuclease activation. Biochem. Biophys. Res. Commun. 1978;81:947–954. doi: 10.1016/0006-291x(78)91443-2. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt A., Zilberstein A., Shulman L., Federman P., Berissi H., Revel M. Interferon action: isolation of nuclease F, a translation inhibitor activated by interferon-induced (2′-5′) oligo-isoadenylate. FEBS Lett. 1978;95:257–264. doi: 10.1016/0014-5793(78)81006-0. [DOI] [PubMed] [Google Scholar]
- 9.Baglioni C., Minks M.A., Maroney P.A. Interferon action may be mediated by activation of a nuclease by pppA2′p5′A2′p5′A. Nature. 1978;273:684–687. doi: 10.1038/273684a0. [DOI] [PubMed] [Google Scholar]
- 10.Eppstein D.A., Samuel C.E. Mechanism of interferon action. Properties of an interferon-mediated ribonucleolytic activity from mouse L929 cells. Virology. 1978;89:240–251. doi: 10.1016/0042-6822(78)90056-9. [DOI] [PubMed] [Google Scholar]
- 11.Li Y., Banerjee S., Wang Y. Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc. Natl. Acad. Sci. USA. 2016;113(8):2241–2246. doi: 10.1073/pnas.1519657113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sadler A.J., Williams B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao L., Birdwell L.D., Wu A. Cell-type-specific activation of the oligoadenylate synthetase-RNase L pathway by a murine coronavirus. J. Virol. 2013;87:8408–8418. doi: 10.1128/JVI.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stark G.R., Kerr I.M., Williams B.R., Silverman R.H., Schreiber R.D. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 15.Der S.D., Zhou A., Williams B.R., Silverman R.H. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong B., Xu L., Zhou A. Intrinsic molecular activities of the interferon-induced 2–5A-dependent RNase. J. Biol. Chem. 1994;269:14153–14158. [PubMed] [Google Scholar]
- 17.Chakrabarti A., Jha B.K., Silverman R.H. New insights into the role of RNase L in innate immunity. J. Interferon Cytokine Res. 2011;31:49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper D.A., Jha B.K., Silverman R.H., Hesselberth J.R., Barton D.J. Ribonuclease L and metal-ion-independent endoribonuclease cleavage sites in host and viral RNAs. Nucl. Acids Res. 2014;42:5202–5216. doi: 10.1093/nar/gku118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wreschner D.H., McCauley J.W., Skehel J.J., Kerr I.M. Interferon action–sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature. 1981;289:414–417. doi: 10.1038/289414a0. [DOI] [PubMed] [Google Scholar]
- 20.Floyd-Smith G., Slattery E., Lengyel P. Interferon action: RNA cleavage pattern of a (2′-5′)oligoadenylate-dependent endonuclease. Science. 1981;212:1030–1032. doi: 10.1126/science.6165080. [DOI] [PubMed] [Google Scholar]
- 21.Siddiqui M.A., Malathi K. RNase L induces autophagy via c-Jun N-terminal kinase and double-stranded RNA-dependent protein kinase signaling pathways. J. Biol. Chem. 2012;287:43651–43664. doi: 10.1074/jbc.M112.399964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakrabarti A., Ghosh P.K., Banerjee S., Gaughan C., Silverman R.H. RNase L triggers autophagy in response to viral infections. J. Virol. 2012;86:11311–11321. doi: 10.1128/JVI.00270-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou A., Paranjape J., Brown T.L. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diaz-Guerra M., Rivas C., Esteban M. Activation of the IFN-inducible enzyme RNase L causes apoptosis of animal cells. Virology. 1997;236:354–363. doi: 10.1006/viro.1997.8719. [DOI] [PubMed] [Google Scholar]
- 25.Castelli J.C., Hassel B.A., Wood K.A. A study of the interferon antiviral mechanism: apoptosis activation by the 2–5A system. J. Exp. Med. 1997;186:967–972. doi: 10.1084/jem.186.6.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen J.B., Li X.L., Judge C.S. Role of 2–5A-dependent RNase-L in senescence and longevity. Oncogene. 2007;26:3081–3088. doi: 10.1038/sj.onc.1210111. [DOI] [PubMed] [Google Scholar]
- 27.Luthra P., Sun D., Silverman R.H., He B. Activation of IFN-β expression by a viral mRNA through RNase L and MDA5. Proc. Natl. Acad. Sci. USA. 2011;108:2118–2123. doi: 10.1073/pnas.1012409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malathi K., Saito T., Crochet N., Barton D.J., Gale M., Jr., Silverman R.H. RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA. 2010;16:2108–2119. doi: 10.1261/rna.2244210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malathi K., Dong B., Gale M., Jr., Silverman R.H. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakrabarti A., Banerjee S., Franchi L. RNase L activates the NLRP3 inflammasome during viral infections. Cell Host Microbe. 2015;17:466–477. doi: 10.1016/j.chom.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou A., Hassel B.A., Silverman R.H. Expression cloning of 2–5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell. 1993;72:753–765. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]
- 32.Zhou A., Paranjape J.M., Hassel B.A. Impact of RNase L overexpression on viral and cellular growth and death. J. Interferon Cytokine Res. 1998;18:953–961. doi: 10.1089/jir.1998.18.953. [DOI] [PubMed] [Google Scholar]
- 33.Dong B., Niwa M., Walter P., Silverman R.H. Basis for regulated RNA cleavage by functional analysis of RNase L and Ire1p. RNA. 2001;7:361–373. doi: 10.1017/s1355838201002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka N., Nakanishi M., Kusakabe Y., Goto Y., Kitade Y., Nakamura K.T. Structural basis for recognition of 2′,5′-linked oligoadenylates by human ribonuclease L. EMBO J. 2004;23:3929–3938. doi: 10.1038/sj.emboj.7600420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang H., Zeqiraj E., Dong B. Dimeric structure of pseudokinase RNase L bound to 2–5A reveals a basis for interferon-induced antiviral activity. Mol. Cell. 2014;53:221–234. doi: 10.1016/j.molcel.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han Y., Donovan J., Rath S., Whitney G., Chitrakar A., Korennykh A. Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response. Science. 2014;343:1244–1248. doi: 10.1126/science.1249845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silverman R.H., Skehel J.J., James T.C., Wreschner D.H., Kerr I.M. RRNA cleavage as an index of ppp(A2′p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. J. Virol. 1983;46:1051–1055. doi: 10.1128/jvi.46.3.1051-1055.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wreschner D.H., James T.C., Silverman R.H., Kerr I.M. Ribosomal RNA cleavage, nuclease activation and 2–5A(ppp(A2′p)nA) in interferon-treated cells. Nucl. Acids Res. 1981;9:1571–1581. doi: 10.1093/nar/9.7.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Min J.Y., Krug R.M. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci USA. 2006;103:7100–7105. doi: 10.1073/pnas.0602184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engel D.A. The influenza virus NS1 protein as a therapeutic target. Antiviral Res. 2013;99:409–416. doi: 10.1016/j.antiviral.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krug R.M. Functions of the influenza A virus NS1 protein in antiviral defense. Curr. Opin. Virol. 2015;12:1–6. doi: 10.1016/j.coviro.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cooper D.A., Banerjee S., Chakrabarti A. RNase L targets distinct sites in influenza A virus RNAs. J. Virol. 2015;89:2764–2776. doi: 10.1128/JVI.02953-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez R., Mohr I. Inhibition of cellular 2′-5′ oligoadenylate synthetase by the herpes simplex virus type 1 Us11 protein. J. Virol. 2007;81:3455–3464. doi: 10.1128/JVI.02520-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang Y., Condit R.C., Vijaysri S., Jacobs B., Williams B.R., Silverman R.H. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J. Virol. 2002;76:5251–5259. doi: 10.1128/JVI.76.10.5251-5259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beattie E., Paoletti E., Tartaglia J. Distinct patterns of IFN sensitivity observed in cells infected with vaccinia K3L- and E3L- mutant viruses. Virology. 1995;210:254–263. doi: 10.1006/viro.1995.1342. [DOI] [PubMed] [Google Scholar]
- 46.Rivas C., Gil J., Melkova Z., Esteban M., Diaz-Guerra M. Vaccinia virus E3L protein is an inhibitor of the interferon (i.f.n.)-induced 2–5A synthetase enzyme. Virology. 1998;243:406–414. doi: 10.1006/viro.1998.9072. [DOI] [PubMed] [Google Scholar]
- 47.Liu S.W., Katsafanas G.C., Liu R., Wyatt L.S., Moss B. Poxvirus decapping enzymes enhance virulence by preventing the accumulation of dsRNA and the induction of innate antiviral responses. Cell Host Microbe. 2015;17:320–331. doi: 10.1016/j.chom.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burgess H.M., Mohr I. Cellular 5′-3′ mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell Host Microbe. 2015;17:332–344. doi: 10.1016/j.chom.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cruz J.L., Sola I., Becares M. Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog. 2011;7:e1002090. doi: 10.1371/journal.ppat.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sorgeloos F., Jha B.K., Silverman R.H., Michiels T. Evasion of antiviral innate immunity by Theiler’s virus L∗ protein through direct inhibition of RNase L. PLoS Pathog. 2013;9:e1003474. doi: 10.1371/journal.ppat.1003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Townsend H.L., Jha B.K., Silverman R.H., Barton D.J. A putative loop E motif and an H-H kissing loop interaction are conserved and functional features in a group C enterovirus RNA that inhibits ribonuclease L. RNA Biol. 2008;5:263–272. doi: 10.4161/rna.7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keel A.Y., Jha B.K., Kieft J.S. Structural architecture of an RNA that competitively inhibits RNase L. RNA. 2012;18:88–99. doi: 10.1261/rna.030007.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silverman R.H., Weiss S.R. Viral phosphodiesterases that antagonize double-stranded RNA signaling to RNase L by degrading 2–5A. J. Interferon Cytokine Res. 2014;34:455–463. doi: 10.1089/jir.2014.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mazumder R., Iyer L.M., Vasudevan S., Aravind L. Detection of novel members, structure-function analysis and evolutionary classification of the 2H phosphoesterase superfamily. Nucl. Acids Res. 2002;30:5229–5243. doi: 10.1093/nar/gkf645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roth-Cross J.K., Stokes H., Chang G. Organ-specific attenuation of murine hepatitis virus strain A59 by replacement of catalytic residues in the putative viral cyclic phosphodiesterase ns2. J. Virol. 2009;83:3743–3753. doi: 10.1128/JVI.02203-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao L., Jha B.K., Wu A. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe. 2012;11:607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang R., Jha B.K., Ogden K.M. Homologous 2′,5′-phosphodiesterases from disparate RNA viruses antagonize antiviral innate immunity. Proc. Natl. Acad. Sci. USA. 2013;110:13114–13119. doi: 10.1073/pnas.1306917110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogden K.M., Hu L., Jha B.K. Structural basis for 2′-5′-oligoadenylate binding and enzyme activity of a viral RNase L antagonist. J. Virol. 2015;89:6633–6645. doi: 10.1128/JVI.00701-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thornbrough J.M., Jha B.K., Yount B. Middle East respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. MBio. 2016;7(2):e00258. doi: 10.1128/mBio.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gold M.G., Smith F.D., Scott J.D., Barford D. AKAP18 contains a phosphoesterase domain that binds AMP. J. Mol. Biol. 2008;375:1329–1343. doi: 10.1016/j.jmb.2007.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gusho E., Zhang R., Jha B.K. Murine AKAP7 has a 2′,5′-phosphodiesterase domain that can complement an inactive murine coronavirus ns2 gene. MBio. 2014;5:e01312–01314. doi: 10.1128/mBio.01312-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rorbach J., Nicholls T.J., Minczuk M. PDE12 removes mitochondrial RNA poly(A) tails and controls translation in human mitochondria. Nucl. Acids Res. 2011;39:7750–7763. doi: 10.1093/nar/gkr470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goldstrohm A.C., Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- 64.Kubota K., Nakahara K., Ohtsuka T. Identification of 2′-phosphodiesterase, which plays a role in the 2–5A system regulated by interferon. J. Biol. Chem. 2004;279:37832–37841. doi: 10.1074/jbc.M400089200. [DOI] [PubMed] [Google Scholar]
- 65.Wood E.R., Bledsoe R., Chai J. The role of phosphodiesterase 12 (PDE12) as a negative regulator of the innate immune response and the discovery of antiviral inhibitors. J. Biol. Chem. 2015;290:19681–19696. doi: 10.1074/jbc.M115.653113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poulsen J.B., Andersen K.R., Kjaer K.H., Vestergaard A.L., Justesen J., Martensen P.M. Characterization of human phosphodiesterase 12 and identification of a novel 2′-5′ oligoadenylate nuclease – the ectonucleotide pyrophosphatase/phosphodiesterase 1. Biochimie. 2012;94:1098–1107. doi: 10.1016/j.biochi.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 67.Bisbal C., Martinand C., Silhol M., Lebleu B., Salehzada T. Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2–5A pathway. J. Biol. Chem. 1995;270:13308–13317. doi: 10.1074/jbc.270.22.13308. [DOI] [PubMed] [Google Scholar]
- 68.Martinand C., Montavon C., Salehzada T., Silhol M., Lebleu B., Bisbal C. RNase L inhibitor is induced during human immunodeficiency virus type 1 infection and down regulates the 2–5A/RNase L pathway in human T cells. J. Virol. 1999;73:290–296. doi: 10.1128/jvi.73.1.290-296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martinand C., Salehzada T., Silhol M., Lebleu B., Bisbal C. The RNase L inhibitor (RLI) is induced by double-stranded RNA. J. Interferon Cytokine Res.: Off. J. Int. Soc. Interferon Cytokine Res. 1998;18:1031–1038. doi: 10.1089/jir.1998.18.1031. [DOI] [PubMed] [Google Scholar]
- 70.Martinand C., Salehzada T., Silhol M., Lebleu B., Bisbal C. RNase L inhibitor (RLI) antisense constructions block partially the down regulation of the 2–5A/RNase L pathway in encephalomyocarditis-virus-(EMCV)-infected cells. Eur. J. Biochem./FEBS. 1998;254:248–255. doi: 10.1046/j.1432-1327.1998.2540248.x. [DOI] [PubMed] [Google Scholar]
- 71.Thakur C.S., Jha B.K., Dong B. Small-molecule activators of RNase L with broad-spectrum antiviral activity. Proc. Natl. Acad. Sci. USA. 2007;104:9585–9590. doi: 10.1073/pnas.0700590104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kristiansen H., Gad H.H., Eskildsen-Larsen S., Despres P., Hartmann R. The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J. Interferon Cytokine Res. 2011;31:41–47. doi: 10.1089/jir.2010.0107. [DOI] [PubMed] [Google Scholar]
- 73.Donovan J., Whitney G., Rath S., Korennykh A. Structural mechanism of sensing long dsRNA via a noncatalytic domain in human oligoadenylate synthetase 3. Proc. Natl. Acad. Sci. USA. 2015;112:3949–3954. doi: 10.1073/pnas.1419409112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Civril F., Deimling T., de Oliveira Mann C.C. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hartmann R., Justesen J., Sarkar S.N., Sen G.C., Yee V.C. Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′-5′-oligoadenylate synthetase. Mol. Cell. 2003;12:1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- 76.Zhu J., Ghosh A., Sarkar S.N. OASL – a new player in controlling antiviral innate immunity. Curr. Opin. Virol. 2015;12:15–19. doi: 10.1016/j.coviro.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu J., Zhang Y., Ghosh A. Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor. Immunity. 2014;40:936–948. doi: 10.1016/j.immuni.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ibsen M.S., Gad H.H., Thavachelvam K., Boesen T., Despres P., Hartmann R. The 2′-5′-oligoadenylate synthetase 3 enzyme potently synthesizes the 2′-5′-oligoadenylates required for RNase L activation. J. Virol. 2014;88:14222–14231. doi: 10.1128/JVI.01763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin R.J., Yu H.P., Chang B.L., Tang W.C., Liao C.L., Lin Y.L. Distinct antiviral roles for human 2′,5′-oligoadenylate synthetase family members against dengue virus infection. J. Immunol. 2009;183:8035–8043. doi: 10.4049/jimmunol.0902728. [DOI] [PubMed] [Google Scholar]
- 80.Kristiansen H., Scherer C.A., McVean M. Extracellular 2′-5′ oligoadenylate synthetase stimulates RNase L-independent antiviral activity: a novel mechanism of virus-induced innate immunity. J. Virol. 2010;84:11898–11904. doi: 10.1128/JVI.01003-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thavachelvam K., Gad H.H., Ibsen M.S. Rapid uptake and inhibition of viral propagation by extracellular OAS1. J. Interferon Cytokine Res. 2015;35:359–366. doi: 10.1089/jir.2014.0140. [DOI] [PubMed] [Google Scholar]
- 82.Dugan J.W., Albor A., David L. Nucleotide oligomerization domain-2 interacts with 2′-5′-oligoadenylate synthetase type 2 and enhances RNase-L function in THP-1 cells. Mol. Immunol. 2009;47:560–566. doi: 10.1016/j.molimm.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sabbah A., Chang T.H., Harnack R. Activation of innate immune antiviral responses by Nod2. Nat. Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Andersen J.B., Mazan-Mamczarz K., Zhan M., Gorospe M., Hassel B.A. Ribosomal protein mRNAs are primary targets of regulation in RNase-L-induced senescence. RNA Biol. 2009;6:305–315. doi: 10.4161/rna.6.3.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rath S., Donovan J., Whitney G., Chitrakar A., Wang W., Korennykh A. Human RNase L tunes gene expression by selectively destabilizing the microRNA-regulated transcriptome. Proc. Natl. Acad. Sci. USA. 2015;112:15916–15921. doi: 10.1073/pnas.1513034112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gog J.R., Afonso Edos S., Dalton R.M. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucl. Acids Res. 2007;35:1897–1907. doi: 10.1093/nar/gkm087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hutchinson E.C., von Kirchbach J.C., Gog J.R., Digard P. Genome packaging in influenza A virus. J. Gen. Virol. 2010;91:313–328. doi: 10.1099/vir.0.017608-0. [DOI] [PubMed] [Google Scholar]
- 88.Siddiqui M.A., Mukherjee S., Manivannan P., Malathi K. RNase L cleavage products promote switch from autophagy to apoptosis by caspase-mediated cleavage of beclin-1. Int. J. Mol. Sci. 2015;16:17611–17636. doi: 10.3390/ijms160817611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rasmussen S.B., Jensen S.B., Nielsen C. Herpes simplex virus infection is sensed by both Toll-like receptors and retinoic acid-inducible gene-like receptors, which synergize to induce type I interferon production. J. Gen. Virol. 2009;90:74–78. doi: 10.1099/vir.0.005389-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Banerjee S., Chakrabarti A., Jha B.K., Weiss S.R., Silverman R.H. Cell-type-specific effects of RNase L on viral induction of beta interferon. MBio. 2014;5:e00856–00814. doi: 10.1128/mBio.00856-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen I.Y., Ichinohe T. Response of host inflammasomes to viral infection. Trends Microbiol. 2015;23:55–63. doi: 10.1016/j.tim.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 92.Lamkanfi M., Dixit V.M. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 93.Franchi L., Eigenbrod T., Munoz-Planillo R. Cytosolic double-stranded RNA activates the NLRP3 inflammasome via MAVS-induced membrane permeabilization and K+ efflux. J. Immunol. 2014;193:4214–4222. doi: 10.4049/jimmunol.1400582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Subramanian N., Natarajan K., Clatworthy M.R., Wang Z., Germain R.N. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. 2013;153:348–361. doi: 10.1016/j.cell.2013.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mitoma H., Hanabuchi S., Kim T. The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity. 2013;39:123–135. doi: 10.1016/j.immuni.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu Y., Lu N., Yuan B. The interaction between the helicase DHX33 and IPS-1 as a novel pathway to sense double-stranded RNA and RNA viruses in myeloid dendritic cells. Cell. Mol. Immunol. 2014;11:49–57. doi: 10.1038/cmi.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li X.L., Ezelle H.J., Kang T.J. An essential role for the antiviral endoribonuclease, RNase-L, in antibacterial immunity. Proc. Natl. Acad. Sci. USA. 2008;105:20816–20821. doi: 10.1073/pnas.0807265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Long T.M., Chakrabarti A., Ezelle H.J. RNase-L deficiency exacerbates experimental colitis and colitis-associated cancer. Inflamm. Bowel Dis. 2013;19:1295–1305. doi: 10.1097/MIB.0b013e318281f2fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Long T.M., Nisa S., Donnenberg M.S., Hassel B.A. Enteropathogenic Escherichia coli inhibits type I interferon- and RNase L-mediated host defense to disrupt intestinal epithelial cell barrier function. Infect. Immun. 2014;82:2802–2814. doi: 10.1128/IAI.00105-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Carpten J., Nupponen N., Isaacs S. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat. Genet. 2002;30:181–184. doi: 10.1038/ng823. [DOI] [PubMed] [Google Scholar]
- 101.Casey G., Neville P.J., Plummer S.J. RNASEL Arg462Gln variant is implicated in up to 13% of prostate cancer cases. Nat. Genet. 2002;32:581–583. doi: 10.1038/ng1021. [DOI] [PubMed] [Google Scholar]
- 102.Rokman A., Ikonen T., Seppala E.H. Germline alterations of the RNASEL gene, a candidate HPC1 gene at 1q25, in patients and families with prostate cancer. Am. J. Hum. Genet. 2002;70:1299–1304. doi: 10.1086/340450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fesinmeyer M.D., Kwon E.M., Fu R., Ostrander E.A., Stanford J.L. Genetic variation in RNASEL and risk for prostate cancer in a population-based case-control study. Prostate. 2011;71:1538–1547. doi: 10.1002/pros.21370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maier C., Haeusler J., Herkommer K. Mutation screening and association study of RNASEL as a prostate cancer susceptibility gene. Br. J. Cancer. 2005;92:1159–1164. doi: 10.1038/sj.bjc.6602401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wiklund F., Jonsson B.A., Brookes A.J. Genetic analysis of the RNASEL gene in hereditary, familial, and sporadic prostate cancer. Clin. Cancer Res. 2004;10:7150–7156. doi: 10.1158/1078-0432.CCR-04-0982. [DOI] [PubMed] [Google Scholar]
- 106.Orr-Urtreger A., Bar-Shira A., Bercovich D. RNASEL mutation screening and association study in Ashkenazi and non-Ashkenazi prostate cancer patients. Cancer Epidemiol. Biomarkers Prev. 2006;15:474–479. doi: 10.1158/1055-9965.EPI-05-0606. [DOI] [PubMed] [Google Scholar]
- 107.Farzan S.F., Karagas M.R., Christensen B.C. RNASEL and MIR146A SNP-SNP interaction as a susceptibility factor for non-melanoma skin cancer. PLoS ONE. 2014;9:e93602. doi: 10.1371/journal.pone.0093602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Madsen B.E., Ramos E.M., Boulard M. Germline mutation in RNASEL predicts increased risk of head and neck, uterine cervix and breast cancer. PLoS ONE. 2008;3:e2492. doi: 10.1371/journal.pone.0002492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bartsch D.K., Fendrich V., Slater E.P. RNASEL germline variants are associated with pancreatic cancer. Int. J. Cancer. 2005;117:718–722. doi: 10.1002/ijc.21254. [DOI] [PubMed] [Google Scholar]
- 110.Kruger S., Silber A.S., Engel C. Arg462Gln sequence variation in the prostate-cancer-susceptibility gene RNASEL and age of onset of hereditary non-polyposis colorectal cancer: a case-control study. Lancet Oncol. 2005;6:566–572. doi: 10.1016/S1470-2045(05)70253-9. [DOI] [PubMed] [Google Scholar]
- 111.Zhang A., Dong B., Doucet A.J., Moldovan J.B., Moran J.V., Silverman R.H. RNase L restricts the mobility of engineered retrotransposons in cultured human cells. Nucl. Acids Res. 2014;42:3803–3820. doi: 10.1093/nar/gkt1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Molinaro R.J., Jha B.K., Malathi K., Varambally S., Chinnaiyan A.M., Silverman R.H. Selection and cloning of poly(rC)-binding protein 2 and Raf kinase inhibitor protein RNA activators of 2′,5′-oligoadenylate synthetase from prostate cancer cells. Nucl. Acids Res. 2006;34:6684–6695. doi: 10.1093/nar/gkl968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Malathi K., Siddiqui M.A., Dayal S. RNase L interacts with Filamin A to regulate actin dynamics and barrier function for viral entry. MBio. 2014;5:e02012. doi: 10.1128/mBio.02012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Banerjee S., Li G., Li Y. RNase L is a negative regulator of cell migration. Oncotarget. 2015;6:44360–44372. doi: 10.18632/oncotarget.6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yi X., Zeng C., Liu H. Lack of RNase L attenuates macrophage functions. PLoS ONE. 2013;8:e81269. doi: 10.1371/journal.pone.0081269. [DOI] [PMC free article] [PubMed] [Google Scholar]