Highlights
-
•
We reviewed the discovery and development process of broad-spectrum antiviral agents.
-
•
We summarized the information on 120 safe-in-man agents in a freely accessible database.
-
•
Further studies will increase the number of broad-spectrum antivirals, expand the spectrum of their indications, and identify drug combinations for treatment of emerging and re-emerging viral infections.
Keywords: Virus, Antiviral drug, Drug discovery and development, Broad-spectrum antiviral agents, BSAAs
Abstract
Viral diseases are one of the leading causes of morbidity and mortality in the world. Virus-specific vaccines and antiviral drugs are the most powerful tools to combat viral diseases. However, broad-spectrum antiviral agents (BSAAs, i.e. compounds targeting viruses belonging to two or more viral families) could provide additional protection of the general population from emerging and re-emerging viral diseases, reinforcing the arsenal of available antiviral options. Here, we review discovery and development of BSAAs and summarize the information on 120 safe-in-man agents in a freely accessible database (https://drugvirus.info/). Future and ongoing pre-clinical and clinical studies will increase the number of BSAAs, expand the spectrum of their indications, and identify drug combinations for treatment of emerging and re-emerging viral infections as well as co-infections.
Introduction
Viruses are one of the major causes of morbidity and mortality in the world (DALYs and Collaborators, 2018; Disease et al., 2018; Howard and Fletcher, 2012; WHO, 2015). Antiviral drugs and vaccines are used to fight viral infections in human (De Clercq and Li, 2016, Marston et al., 2014). Previously, there has been a focus on “one drug, one virus” dogma, which relied on targeting virus-specific factors. A counterpoint to this is the “one drug, multiple viruses” paradigm, which came with the discovery of broad-spectrum antiviral agents (BSAAs), small-molecules that inhibit a wide range of human viruses (Bekerman and Einav, 2015, de Clercq and Montgomery, 1983, Debing et al., 2015, Ianevski et al., 2019, Rada and Dragun, 1977, Sidwell et al., 1972). This paradigm was based on the observation that different viruses utilize similar pathways and host factors to replicate inside a cell (Bosl et al., 2019). Although the concept of BSAAs has been around for almost 50 years, the field received a new impetus with recent outbreaks of Ebola, Zika, Dengue, influenza and other viral infections, the discovery of novel host-directed agents, as well as development of drug repositioning methodology.
Drug repurposing, also called repositioning, redirecting, reprofiling, is a strategy for generating additional value from an existing drug by targeting disease other than that for which it was originally intended (Nishimura and Hara, 2018, Pushpakom et al., 2019). This has significant advantages over new drug discovery since chemical synthesis steps, manufacturing processes, reliable safety, and pharmacokinetic properties in pre-clinical (animal model) and early clinical developmental phases (phase 0, I and IIa) are already available (Figure 1 ). Therefore, repositioning of launched or even failed drugs to viral diseases provides unique translational opportunities, including a substantially higher probability of success to market as compared with developing new virus-specific drugs and vaccines, and a significantly reduced cost and timeline to clinical availability (Ianevski et al., 2019, Pizzorno et al., 2019, Zheng et al., 2018).
Figure 1.
Discovery of novel activities and follow-up development of broad-spectrum antiviral agents (BSAAs). Yellow shading indicates a process of discovery and development of safe-in-man BSAAs, for which pharmacokinetic (PK) properties in pre-clinical (animal model) and early clinical developmental phases (phase 0-IIa trials) are already available. Abbreviations: ESCs, human embryonic stem cells; iPSCs, human induced pluripotent stem cells (iPSCs).
Here, we detail the steps of BSAA repurposing, from discovery of novel antiviral activities in cell culture to post-market studies. Moreover, we summarized currently available information on BSAAs in freely available database, focusing on those antivirals, which have been already tested in human as antivirals, antibacterials, antiprotozoals, anthelmintics, etc. Finally, we discuss future perspectives of using safe-in-man BSAAs for treatment of emerging and re-emerging viral infections as well as viral and bacterial co-infections.
Discovery and development of safe-in-man BSAAs
Discovery of novel BSAA activities in immortalized cell cultures and co-cultures
The discovery of novel activities of BSAAs starts with exposing cells to the candidate antiviral agent at different concentrations and infecting the cells with a virus or mock. Immortalized cancerous cell cultures and co-cultures, which express appropriate viral receptors, are most commonly used in this first step. The half-maximal cytotoxic concentrations (CC50) for a compound are calculated based on their dose-response curves obtained on mock-infected cells. The half-maximal effective concentrations (EC50) are calculated based on the analysis of curves obtained on infected cells. Statistical analyses can help to determine if the differences between CC50 and EC50 are significant, given the inherent variability of the experiment (Meneghini and Hamasaki, 1967). A relative effectiveness of a drug is defined as selectivity index (SI = CC50/EC50).
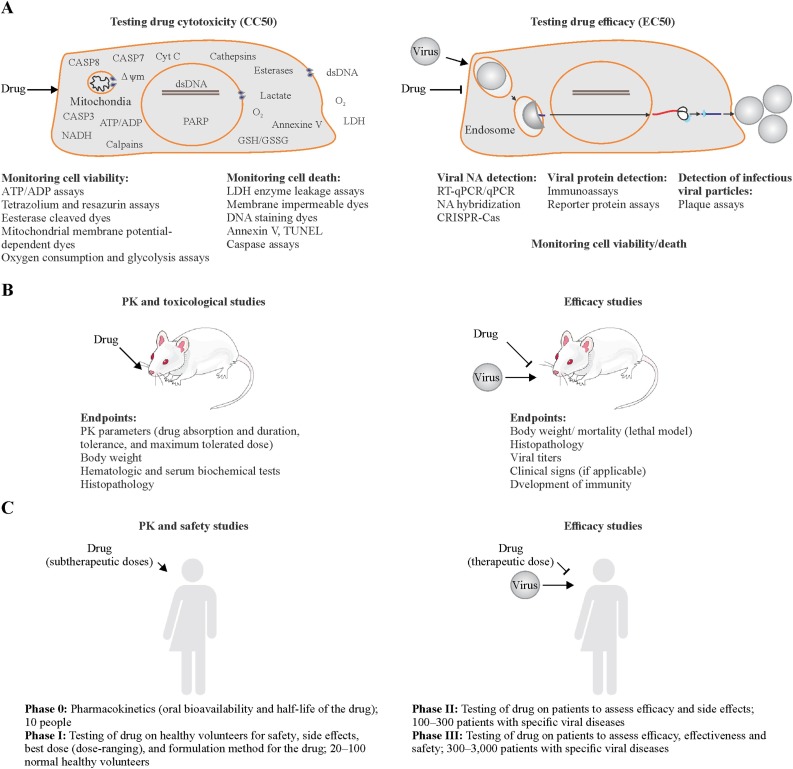
Cell viability assays and cell death assays are commonly used to assess the cytotoxicity and efficacy of BSAAs (Figure 2 A). Cell viability assays include MTT, MTS, resazurin or similar assays, mitochondrial membrane potential-dependent dyes-based assays, esterase cleaved dye-based assays, ATP-ADP assays, and assays that measure glycolytic flux and oxygen consumption. Other cell death assays include LDH enzyme leakage assays, membrane impermeable dye-based assays, and apoptosis assays, such as Annexin V, TUNEL, and caspase assays (Shen et al., 2019). For example, the Cell Titer Glo (CTG) assay quantifies ATP, an indicator of metabolically active living cells, whereas Cell Tox Green assay uses fluorescent asymmetric cyanine dye that stains the DNA of dead cells (Bosl et al., 2019, Bulanova et al., 2017, Ianevski et al., 2018, Muller et al., 2014).
Figure 2.
ABC of BSAA development process. (A) Testing BSAA toxicity (left panel) and efficacy (right panel) in immortalized cell cultures and co-cultures. (B) Testing BSAA toxicity (left panel) and efficacy (right panel) in animal models. If BSAA is repositioned from another disease (i.e. its PK/PD and toxicity profiles are available for the animal model) it could bypass the safety studies. (C) Clinical trials of BSAAs. (Left panel) Pharmacokinetics (PK) and safety studies. (Right panel) Efficacy studies. If the drug is repositioned from another disease (i.e. its safety profile in man is available) it could bypass the PK and safety studies in man.
Viral strains or cell lines expressing reporter proteins are also used to assess the efficacy of BSAAs in infected cells. For example, TZM-bl cells expressing firefly luciferase under control of HIV-1 LTR promoter allowed quantitation of BSAA action on HIV-1 infection (tat-protein expression by integrated HIV-1 provirus) using firefly luciferase assay (Sarzotti-Kelsoe et al., 2014, Xing et al., 2016). RFP-expressing RVFV, nanoLuc-expressing CHIKV and RRV, as well as GFP-expressing FLUAV, HCV and HMPV also allowed identification of novel activities of several BSAAs (Andersen et al., 2019b, Bosl et al., 2019, de Graaf et al., 2007, Habjan et al., 2008, Ianevski et al., 2018, Jupille et al., 2011, Kittel et al., 2004, Lee et al., 2017, Utt et al., 2016). In addition, qPCR/RT-qPCR, RNA/DNA sequencing, RNA/DNA hybridization, immuno- and plaque assays as well as CRISPR-Cas9 systems could be used for detection of inhibitory effects of BSAAs (Boonham et al., 2014, Fischer et al., 2017, Konig et al., 2019, Laamiri et al., 2018, Landry, 1990, Perez et al., 2013, Sashital, 2018, Zhou et al., 2018). Interestingly, CRISPR-Cas9, siRNA and shRNA approaches were used for identification of BSAA targets (Deans et al., 2016, Puschnik et al., 2017).
Novel anti-HSV-2 and anti-EV1 activities of emetine were discovered recently using CTG/plaque assays in human non-malignant RPE cells. Moreover, novel antiviral activities of the drug were identified using RFP-expressing RVFV, and GFP-expressing HMPV or FLUAV strains in RPE cells (Andersen et al., 2019a). Given that emetine also inhibits ZIKV, EBOV, RABV, CMV, HCoV-OC43 and HIV-1 infections (Chaves Valadao et al., 2015, MacGibeny et al., 2018, Mukhopadhyay et al., 2016, Shen et al., 2019, Yang et al., 2018), and that it is an FDA-approved anti-protozoal drug, it may represent a promising safe-in-man BSAA candidate.
Evaluation of BSAAs in human primary cell culture and co-cultures
Immortalized cell cultures/co-cultures and reporter viral strains represent excellent model systems for the discovery of novel activities of safe-in-man BSAAs. However, these genetically modified systems have certain limitations (attenuated or incomplete virus replication cycle, accumulation of mutations during repeated cell and virus passaging, defective innate immune responses and viral counter-responses, etc.) (Carter and Shieh, 2015). Thereby, novel antiviral activities of BSAAs should be further validated in primary human cells using different viral strains (including wild-type viruses), different viral loads, different times of compound addition, different endpoint measurements and compound concentration range. Primary cell cultures give more accurate images of drug responses (Alves et al., 2018, Denisova et al., 2012, Koban et al., 2018, Postnikova et al., 2018). They have a low population doubling level and therefore more closely recapitulate the physiological conditions observed in vivo.
Primary cells are cells isolated directly from tissues or blood using enzymatic or mechanical methods. The cells are characterized by their high degrees of specialization, are often fully differentiated and thus require defined culture conditions (serum-free media) in order to preserve their original phenotype. Peripheral blood mononuclear (PBMC), placental, amniotic and fetal primary cultures as well as vaginal/cervical epithelial and male germ cells have been used intensively to validate BSAA activity (Barrows et al., 2016, Denisova et al., 2012, Fink et al., 2018, Rausch et al., 2017, Robinson et al., 2018). Although primary cell cultures are relevant systems for validation of BSAAs, there are technical difficulties limiting their use, such as ethical issues, purity of population of primary cells, and limited shelf life of the cells. In addition, age, race, sex and other genetic and epigenetic factors of donor cells should be considered to determine common biological effect across a significant number of donors thereby avoiding minor variants (Lee et al., 2014, Zhang et al., 2013).
The obstacles associated with use of human primary cell cultures can be bypassed using human embryonic stem cells (ESCs) and human induced pluripotent stem cells (iPSCs). ESCs are isolated from surplus human embryos, whereas iPSCs are obtained by reprogramming somatic cells. These cells proliferate extensively and retain multi-lineage activity, which allows them to generate virtually any cell type of the body. The ESCs- and iPSC-derived cells have been used successfully to investigate the efficacy of several BSAAs against HBV, ZIKV, CHIKV and HSV-1 infections (Table S1) (Ferreira et al., 2019, Iwasawa et al., 2019, Lanko et al., 2017, Simonin et al., 2019, Xia et al., 2017, Zhou et al., 2017).
iPSCs, ESCs and primary tissue cells can be used to generate complex cultures termed organoids. Organoids are miniature and simplified version of organs. Establishing human airway, gut, skin, cerebral, liver, kidney, breast, retina and brain organoids allowed researchers to study toxicity and efficacy of several safe-in-man BSAAs against coronaviruses, influenza, enteroviruses, rotaviruses and flaviviruses (Li et al., 2017, Sacramento et al., 2017, Watanabe et al., 2017, Xu et al., 2016, Yin et al., 2015, Yin et al., 2018, Yin et al., 2016, Zhou et al., 2017). However, iPSCs, ESCs and iPSCs/ESCs-derived organoids have the same disadvantages as human primary cells (genetic differences, line-to-line and organoid batch-to-batch variability). On the other hand, these models allow researchers to predict the behavior of viruses in vivo and, therefore, to reduce animal use and in cases where animal models are unavailable to initiate clinical trials.
For example, novel anti-ZIKV activities of enoxacin, amodiaquine and niclosamide were discovered recently using human neural progenitor cells, human pluripotent stem cell-derived cortical neural progenitor cells, and human induced neural stem cells, respectively (Cairns et al., 2018, Xu et al., 2019, Zhou et al., 2017). Enoxacin is an oral broad-spectrum fluoroquinolone antibiotic, which also possesses anti-HCV and anti-HIV-1 activities in immortalized cell cultures (Kashiwase et al., 1999, Young et al., 2010). Amodiaquine is an anti-malaria drug, which also possesses antiviral activities against DENV, HCV, RRV, SINV, WNV, EFV, EBOV, LASV, RABV, VZV, and HSV-1 in immortalized cell cultures (Boonyasuppayakorn et al., 2014, Hulseberg et al., 2019, Mazzon et al., 2019). Niclosamide is an orally bioavailable anthelmintic drug which inhibits the broadest range of viruses in vitro and, in some cases, in vivo (Cairns et al., 2018, Fang et al., 2013, Huang et al., 2017, Hulseberg et al., 2019, Jurgeit et al., 2012, Kao et al., 2018, Mazzon et al., 2019, Stachulski et al., 2011, Wang et al., 2016, Wu et al., 2004). These safe-in-man BSAAs represent promising drug candidates.
Evaluation of BSAAs in animal models
In vitro and ex vivo models do not fully reflect the complexity and physiology of living organisms. Therefore, several in vivo models have been developed to test novel antiviral activities of BSAAs. These include immunocompetent and genetically or chemically immunocompromised mice, guinea pigs, hamsters, ferrets, pigs, macaques and other animals (Figure 2B) (Alves et al., 2018, Haese et al., 2016, Louz et al., 2013, Morrison and Diamond, 2017, Taylor, 2017, Thangavel and Bouvier, 2014). PK/PD studies determine drug absorption, dosage and half-life of BSAAs. Toxicological studies determine if the drugs have any adverse effects on the tissues and organs of the animals and define the dosage of adverse effects (Alabaster and In Vivo Pharmacology Training Group, 2002, Parasuraman, 2011, Rizk et al., 2017). Studying the efficacy of BSAAs is generally done by treating the animal with the drug or vehicle and infecting it with a virus of interest. Endpoints are usually body weight/ mortality (depending on the virus), histopathology, virus titers in organs, presence of clinical signs and development of immunity (Oh and Hurt, 2016, Smee and Barnard, 2013). Although animal models can give the initial characterization of BSAA, it is important to keep in mind that they differ significantly from humans, with respect to symptoms, disease manifestation, susceptibility, immune responses, pathogenesis, and pharmacokinetics (Barré-Sinoussi and Montagutelli, 2015, Shanks et al., 2009).
Often animals require higher concentration of an experimental antiviral as compared to effective in vitro concentrations. Moreover, it is relatively difficult to achieve micromolar EC50 in vivo. For example, aminoglycoside antibiotics, kasugamycin and neomycin were successfully tested against ZIKV, FLUAV, and HSV-2 infections in mice (Gopinath et al., 2018). Polyether antibiotic salinomycin also showed anti-FLUAV effect in mice (Jang et al., 2018). In addition, investigational anticancer agent, flavopiridol, was effective against FLUAV in mice (Soderholm et al., 2016). These findings support further development of these and other BSAAs.
Clinical trials and post-clinical studies of BSAAs
Clinical trials are the most critical and time-consuming step of a drug candidates’ journey to being approved (Figure 2C). However, safe-in-man BSAAs make this journey relatively short, because they have been already at phase 0, I and, sometime, at IIa of clinical trials as antibacterial, antiprotozoal, anticancer, etc. agent; i.e. they have been administered at sub-therapeutic doses to healthy volunteers to ensure the drugs are not harmful to the participants. Thus, safe-in-man BSAAs enter phase II and III, which assess the efficacy, effectiveness, safety and side effects of the drugs in clinic. It is important, however, to differentiate acute and chronic viral infections when repurposing BSAAs, given that drug concentrations and duration of the treatment could be different, and therefore, drug safety issues should be considered.
For phase II, patients with the viral disease in question are invited to join the study, where they are administered the BSAAs at the ideal therapeutic doses. Phase III is the longest of the phases, and includes multiple levels of securities to the studies, such as the use of placebos and double-blinded studies, to ensure the data is as unbiased as possible. Upon completing phase III, depending on its performance and efficacy, BSAAs may end either being approved or dropped. The U.S Food and Drug Administration (FDA) estimates that only 25–30% BSAA candidates that enter phase III are approved for use in the public (U.S Food and Drug Administration, 2018). After approval and marketing of the drug, phase IV may be initiated to follow up on the use of the drug in public, to surveil for rare effects (U.S Food and Drug Administration, 2018, Umscheid et al., 2011).
Forty-eight safe-in-man BSAAs undergo clinical studies as antivirals. There are currently 21 compounds in phase I, 34 agents in phase II and 11 compounds in phase III clinical trials. For example, nitazoxanide, remdesivir and brincidofovir are under clinical investigations against different viral infections (NCT03336619, NCT00302640, NCT03605862, NCT03719586, NCT01276756, NCT03905655, NCT01529073, NCT03395405, NCT03216967, NCT01431326, NCT02087306, NCT01769170).
Twenty-one BSAAs were approved by FDA, EMA or other agencies. These BSAAs altogether target 15 viruses. For example, favipiravir, also known as T-705, was approved against FLUAV in Japan; cidofovir is an injectable antiviral medication used as a treatment for CMV retinitis in people with AIDS; ribavirin, also known as tribavirin, is used for treatment of RSV and HCV infections; pleconaril is used against viruses in the picornaviridae family, including enterovirus and rhinovirus; and valacyclovir is used against CMV, EBV, HBV, HSV-1, HSV-2 and VZV infections.
Twenty BSAAs are undergoing surveillance studies (phase IV). Azithromycin, chloroquine, cyclosporine, ezetimibe, mycophenolic acid, nitazoxanide and rapamycin progressed to phase IV studies without approvals from national or international authorities (NCT01779570, NCT02058173, NCT02564471, NCT00821587, NCT03360682, NCT02328963, NCT02768545, NCT01624948, NCT01770483, NCT02683291, NCT01624948, NCT01469884, NCT03901001, NCT01412515, NCT02990312).
BSAA database
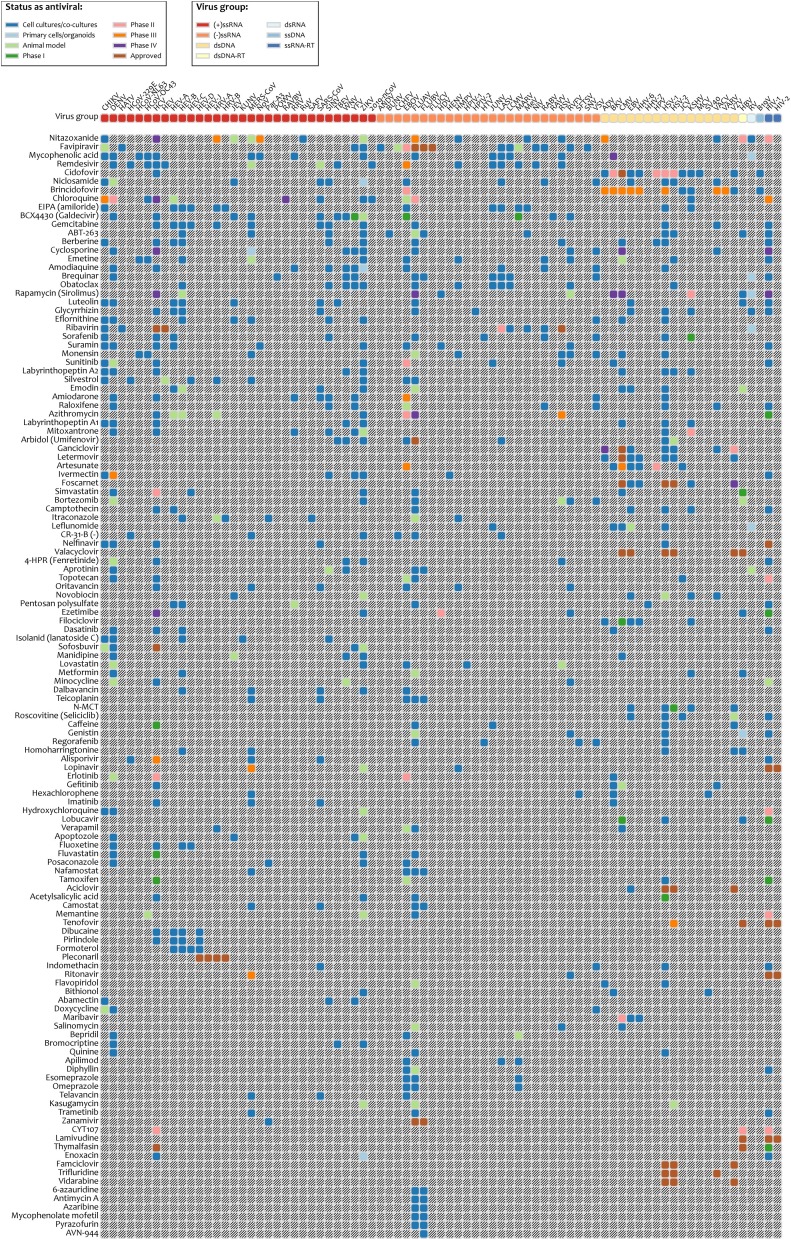
We have developed a database for safe-in-man BSAAs, which is available at https://drugvirus.info/ (Figure 3 ). The drug annotations were obtained from PubChem, DrugBank, DrugCentral, PubMed and clinicaltrials.gov databases (Table S1) (Kim et al., 2019, Ursu et al., 2019, Wishart et al., 2018). The information on virus families was exported from Virus Pathogen Database and Analysis Resource (Table S2) (Pickett et al., 2012). The database summarizes activities and developmental status of BSAAs. We decided to set no limits for EC50, SI and statistical significance because the studies were not harmonized (different cell lines, assays, end-point measurements, time of compound addition, etc.). The database allows interactive exploration of virus-BSAA interactions. It also includes information on BSA targets. A feedback form is available on the website. The database will be updated upon request or as soon as a new safe-in-man BSAA emerges or novel activity for an existing BSAA is reported.
Figure 3.
Safe-in-man broad-spectrum antiviral agents (BSAAs) and viruses they inhibit. A snapshot is taken from https://drugvirus.info/ website. Viruses are clustered by virus groups. BSAAs range from the highest to lowest number of targeted viruses. Different shadings indicate different development status of BSAAs. Gray shading indicates that the antiviral activity has not been either studied or reported. Abbreviations: ds, double-stranded; RT, reverse transcriptase; ss, single-stranded.
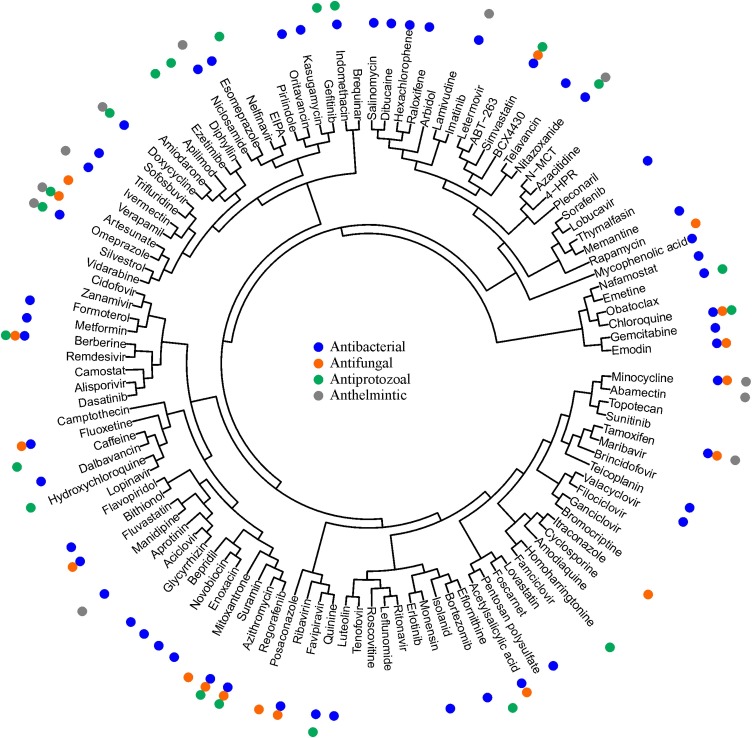
Altogether, the database contains 120 approved, investigational and experimental safe-in-man BSAAs, which inhibit 86 human viruses, belonging to 25 viral families. The BSAAs inhibit viral or host factors and block viral replication, reduce the viral burden to a level at which host immune responses can deal with it or facilitate apoptosis of infected cells (Table S1). Analysis of BSAA targets and structures (Figure 4 ) revealed that the most abundant are nucleotide and nucleoside analogues which inhibit viral RNA and DNA polymerases. Imatinib, erlotinib, gefitinib, and dasatinib, which inhibit tyrosine kinases, are the most abundant host-directed BSAAs. Most of the host targets (except Bcl-xL protein) are essential for viral replication but redundant for the cell, which is critical for reducing putative toxicities associated with blocking cellular pathways. The limited diversity of the targets and scaffolds could slow down the development of BSAA concept.
Figure 4.
Structure-activity relationship of safe-in-man BSAAs. Web-application serves C-SPADE was used to cluster BSAAs based on their structural similarities and visualize them as a dendrogram of compounds augmented with their functional annotations (https://cspade.fimm.fi/).
Emerging BSAAs, such as 5,6-dimethoxyindan-1-one, saliphenylhalamide, and GS-5734 (Denisova et al., 2012, Kuivanen et al., 2017, Muller et al., 2011, Muller et al., 2014, Patil et al., 2017, Sheahan et al., 2017), whose safety profiles in humans are not yet available, are not included in the database. However, they could serve as valuable antivirals in the future, pending the results of further pre-clinical and clinical investigations.
BSAAs as drug candidates for treatment of SARS-CoV-2 infections
SARS-CoV-2 is a novel strain of coronaviruses which is associated with a cluster of cases of pneumonia in China (Zhou et al., 2020). Coronaviruses (CoV) are a broad family of virusesthat includes SARS-CoV, MERS-CoV, HCoV-229E, HCoV-OC43, HCoV-NL63 and HCoV-HKU1 strains. HCoV-229E, HCoV-OC43, HCoV-NL63 and HCoV-HKU1 strains are usually associated with mild, self-limiting upper respiratory tract infections, such as the common cold. By contrast, people infected with MERS-CoV, SASRS-CoV or SARS-CoV-2 could develop severe respiratory illness, and many of the infected have died.
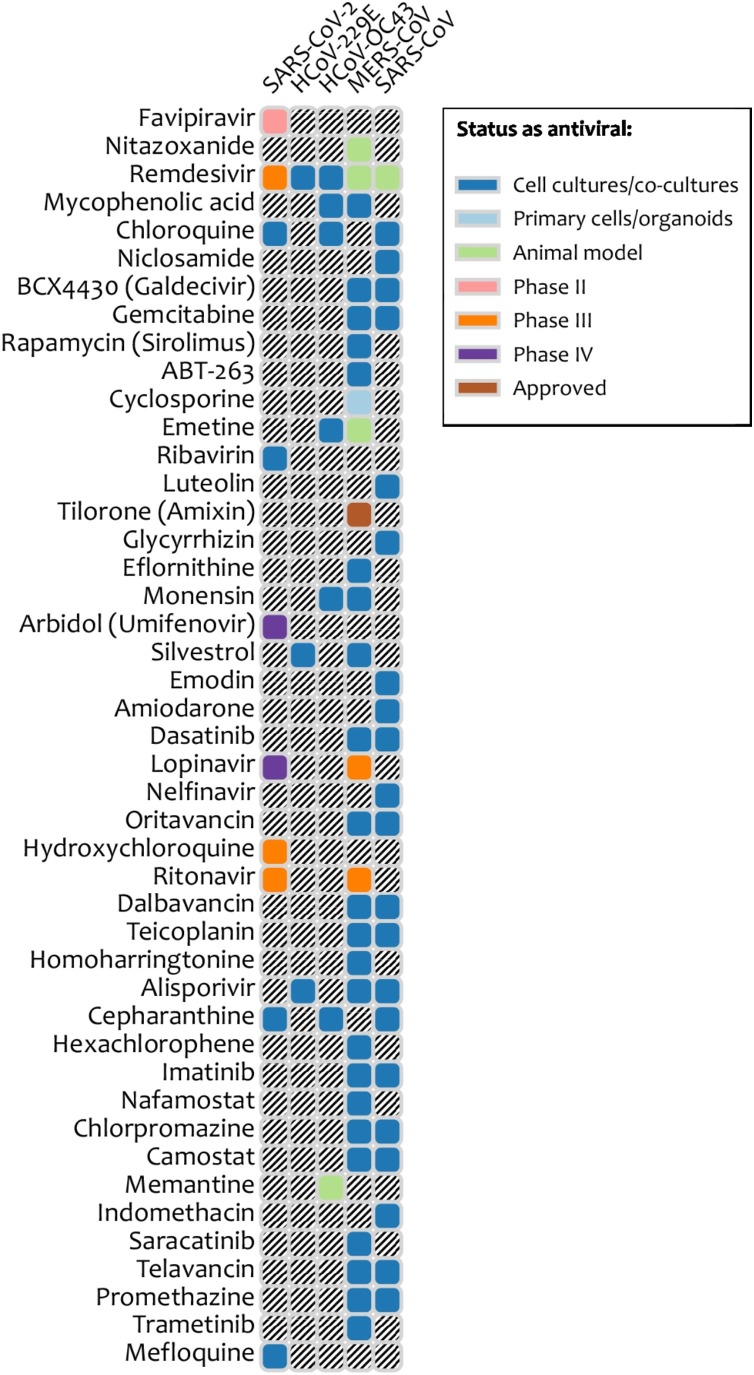
No vaccines and drugs are available for prevention, prophylaxis and treatment of coronavirus infections in humans (Eurosurveillance Editorial, 2020). However, safe-in-man BSAAs could be effective against SARS-CoV-2 and other coronaviruses (Figure 5 ). For example, teicoplanin, oritavancin, dalbavancin, monensin and emetine could be repurposed for treatment of COVID-19. Teicoplanin, ritavancin, dalbavancin and monensin are approved antibiotics, whereas emetine is an anti-protozoal drug. These drugs have been shown to inhibit several corona- as well as some other viral infections . Moreover, chloroquine and remdesivir were shown to effectively inhibit SARS-CoV-2 infection in vitro. In addition, clinical investigations into the effectiveness of lopinavir, ritonavir, remdesivir, hydroxychloroquine and arbidol against COVID-19 have started recently (NCT04252664; NCT0425487; NCT04255017; NCT04261517, NCT04260594).
Figure 5.
Safe-in-man broad-spectrum antiviral agents and coronaviruses they inhibit. A snapshot is taken from https://drugvirus.info/ website. Different shadings indicate different development status of BSAAs. Grey shading indicates that the antiviral activity has not been either studied or reported.
BSAA combinations
BSAAs could be combined with other antiviral agents to obtain synergistic or additive effects against certain viruses (Cheng et al., 2019, Zheng et al., 2018). Several combination therapies which include BSAAs (such as abacavir/dolutegravir/lamivudine (Triumeq), darunavir/cobicistat/emtricitabine/tenofovir (Symtuza), lopinavir/ritonavir (Kaletra), ledipasvir/sofosbuvir and sofosbuvir/velpatasvir) became a standard for the treatment of HIV or HCV infections. Several synergistic drug combinations, such as obatoclax/saliphenylhalamide and gemcitabine/pimodivir, could enter clinical studies and become effective treatment of ZIKV and FLUAV infections (Fu et al., 2016, Kuivanen et al., 2017).
By contrast to individual drugs, combinations of 2–3 BSAAs could be used to target an even broader range of viruses (Foucquier and Guedj, 2015, Zheng et al., 2018). Such combinations could serve as front line therapeutics against poorly characterized emerging viruses or re-emerging drug-resistant viral strains. For example, a cocktail of nitazoxanide, favipiravir, and niclosamide could be developed for the treatment of viruses belonging to 11 families.
Other activities of BSAAs
Fifty BSAAs possess not only antiviral but also antibacterial activity (Figure 4; Table S1) (Schor and Einav, 2018). Moreover, 13 of the 50 agents are approved as antibiotics (2 withdrawn). These agents with dual activity could be used for treatment of viral and bacterial co-infections or for the protection of patients from the secondary infections. For example, azithromycin could be used against FLUAV and Chlamydophila pneumoniae, Haemophilus influenzae, Mycoplasma pneumoniae or Streptococcus pneumoniae infections (NCT01779570) (Mandell et al., 2007).
In addition, BSAAs showed activity against a wide range of other medically important human pathogens, including fungi, protozoa and parasites (Table S1) (Montoya and Krysan, 2018), pointing out that some pathogens utilize common mechanisms to infect hosts. Moreover, structure-activity relationship analysis of BSAAs suggests that some agents, such as doxycycline, artesunate, omeprazole, nitazoxanide, suramin, azithromycin, minocycline and chloroquine, could have novel antibacterial, antiprotozoal, antifungal or anthelmintic activities (Figure 4). If confirmed, this could lead to development of broad-spectrum anti-infective drugs.
BSAAs could also serve as treatment of other co-morbidities, therefore, simplifying the therapy and lowering its cost (Table S1). For example, the concomitant actions of ezetimibe and statins could be beneficial for treatment of both hypertension and several viral infections in patients with these co-morbidities (NCT00908011, NCT00099684, NCT00843661, NCT03490097, NCT00994773, NCT00441493).
Conclusions and future perspectives
Here we reviewed a process of BSAA development and summarized information on 120 safe-in-man agents in freely available database. We hope that further pre-clinical and clinical studies on BSAAs will be harmonized, and data collection will be standardized. Furthermore, the follow-up studies as well as the results of on-going, finalized or terminated clinical trials will be made publicly available to allow prioritization and translation of emerging and existing BSAAs into clinical practice. This would allow BSAAs to play a pivotal role in the battle against emerging and re-emerging viral diseases.
Discovery of novel as well as repositioning existing safe-in-man BSAAs may shorten time and resources needed for development of virus-specific drugs and vaccines. In the future, BSAAs will have a global impact by decreasing morbidity and mortality from viral and other diseases, maximizing the number of healthy life years, improving the quality of life of infected patients and decreasing the costs of patient care.
Conflicts of interest
The authors declare no conflict of interest.
Funding sources
This study was supported by the European Regional Development Fund, the Mobilitas Pluss Project MOBTT39 (to D.K.).
Ethical approval
Approval was not required.
Acknowledgments
We thank Katarzyna Kolasa for illustrations. This manuscript has been released as a pre-print (doi: https://10.20944/preprints201910.0144.v46). We thank the European Regional Development Fund, the Mobilitas Pluss Project MOBTT39.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2020.02.018.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Alabaster V., In Vivo Pharmacology Training Group The fall and rise of in vivo pharmacology. Trends Pharmacol Sci. 2002;23:13–18. doi: 10.1016/s0165-6147(00)01882-4. [DOI] [PubMed] [Google Scholar]
- Alves M.P., Vielle N.J., Thiel V., Pfaender S. Research models and tools for the identification of antivirals and therapeutics against Zika virus infection. Viruses. 2018;10 doi: 10.3390/v10110593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen P.I., Krpina K., Ianevski A., Shtaida N., Jo E., Yang J. Novel antiviral activities of obatoclax, emetine, niclosamide, brequinar, and homoharringtonine. Viruses. 2019;11 doi: 10.3390/v11100964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen P.I., Krpina K., Ianevski A., Shtaida N., Jo E., Yang J. Novel antiviral activities of obatoclax, emetine, niclosamide, brequinar, and homoharringtonine. Viruses. 2019;11 doi: 10.3390/v11100964. pii: E964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barré-Sinoussi F., Montagutelli X. Animal models are essential to biological research: issues and perspectives. Future Sci OA. 2015;1 doi: 10.4155/fso.15.63. FSO63-FSO63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrows N.J., Campos R.K., Powell S.T., Prasanth K.R., Schott-Lerner G., Soto-Acosta R. A screen of FDA-approved drugs for inhibitors of Zika virus infection. Cell Host Microbe. 2016;20:259–270. doi: 10.1016/j.chom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekerman E., Einav S. Infectious disease. Combating emerging viral threats. Science. 2015;348:282–283. doi: 10.1126/science.aaa3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonham N., Kreuze J., Winter S., van der Vlugt R., Bergervoet J., Tomlinson J. Methods in virus diagnostics: from ELISA to next generation sequencing. Virus Res. 2014;186:20–31. doi: 10.1016/j.virusres.2013.12.007. [DOI] [PubMed] [Google Scholar]
- Boonyasuppayakorn S., Reichert E.D., Manzano M., Nagarajan K., Padmanabhan R. Amodiaquine, an antimalarial drug, inhibits dengue virus type 2 replication and infectivity. Antiviral Res. 2014;106:125–134. doi: 10.1016/j.antiviral.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosl K., Ianevski A., Than T.T., Andersen P.I., Kuivanen S., Teppor M. Common nodes of virus–host interaction revealed through an integrated network analysis. Front. Immunol. 2019;4:2186. doi: 10.3389/fimmu.2019.02186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulanova D., Ianevski A., Bugai A., Akimov Y., Kuivanen S., Paavilainen H. Antiviral properties of chemical inhibitors of cellular anti-apoptotic Bcl-2 proteins. Viruses. 2017;9 doi: 10.3390/v9100271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns D.M., Boorgu D., Levin M., Kaplan D.L. Niclosamide rescues microcephaly in a humanized in vivo model of Zika infection using human induced neural stem cells. Biol Open. 2018;7 doi: 10.1242/bio.031807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M., Shieh J. Chapter 14 — cell culture techniques. In: Carter M., Shieh J., editors. Guide to research techniques in neuroscience. Second edition. Academic Press; San Diego: 2015. pp. 295–310. [Google Scholar]
- Chaves Valadao A.L., Abreu C.M., Dias J.Z., Arantes P., Verli H., Tanuri A. Natural plant alkaloid (emetine) inhibits HIV-1 replication by interfering with reverse transcriptase activity. Molecules. 2015;20:11474–11489. doi: 10.3390/molecules200611474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.S., Williamson P.R., Zheng W. Improving therapy of severe infections through drug repurposing of synergistic combinations. Curr Opin Pharmacol. 2019;48:92–98. doi: 10.1016/j.coph.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DALYs, G.B.D., Collaborators, H Global, regional, and national disability-adjusted life-years (DALYs) for 359 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1859–1922. doi: 10.1016/S0140-6736(18)32335-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E., Li G. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev. 2016;29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Clercq E., Montgomery J.A. Broad-spectrum antiviral activity of the carbocyclic analog of 3-deazaadenosine. Antiviral Res. 1983;3:17–24. doi: 10.1016/0166-3542(83)90011-6. [DOI] [PubMed] [Google Scholar]
- de Graaf M., Herfst S., Schrauwen E.J., van den Hoogen B.G., Osterhaus A.D., Fouchier R.A. An improved plaque reduction virus neutralization assay for human metapneumovirus. J Virol Methods. 2007;143:169–174. doi: 10.1016/j.jviromet.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Deans R.M., Morgens D.W., Okesli A., Pillay S., Horlbeck M.A., Kampmann M. Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nat Chem Biol. 2016;12:361–366. doi: 10.1038/nchembio.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debing Y., Neyts J., Delang L. The future of antivirals: broad-spectrum inhibitors. Curr Opin Infect Dis. 2015;28:596–602. doi: 10.1097/QCO.0000000000000212. [DOI] [PubMed] [Google Scholar]
- Denisova O.V., Kakkola L., Feng L., Stenman J., Nagaraj A., Lampe J. Obatoclax, saliphenylhalamide, and gemcitabine inhibit influenza a virus infection. J Biol Chem. 2012;287:35324–35332. doi: 10.1074/jbc.M112.392142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disease G.B.D., Injury I., Prevalence C. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1789–1858. doi: 10.1016/S0140-6736(18)32279-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eurosurveillance Editorial T. Note from the editors: World Health Organization declares novel coronavirus (2019-nCoV) sixth public health emergency of international concern. Euro Surveill. 2020 doi: 10.2807/1560-7917.ES.2020.25.5.200131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J., Sun L., Peng G., Xu J., Zhou R., Cao S. Identification of three antiviral inhibitors against Japanese encephalitis virus from library of pharmacologically active compounds 1280. PLoS One. 2013;8 doi: 10.1371/journal.pone.0078425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira A.C., Reis P.A., de Freitas C.S., Sacramento C.Q., Villas Boas Hoelz L., Bastos M.M. Beyond members of the flaviviridae family, sofosbuvir also inhibits chikungunya virus replication. Antimicrob Agents Chemother. 2019;63 doi: 10.1128/AAC.01389-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink S.L., Vojtech L., Wagoner J., Slivinski N.S.J., Jackson K.J., Wang R. The antiviral drug arbidol inhibits Zika virus. Sci Rep. 2018;8:8989. doi: 10.1038/s41598-018-27224-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer C., Torres M.C., Patel P., Moreira-Soto A., Gould E.A., Charrel R.N. Lineage-specific real-time RT-PCR for yellow fever virus outbreak surveillance, Brazil. Emerg Infect Dis. 2017;23:1867–1871. doi: 10.3201/eid2311.171131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foucquier J., Guedj M. Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect. 2015;3 doi: 10.1002/prp2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Gaelings L., Soderholm S., Belanov S., Nandania J., Nyman T.A. JNJ872 inhibits influenza A virus replication without altering cellular antiviral responses. Antiviral Res. 2016;133:23–31. doi: 10.1016/j.antiviral.2016.07.008. [DOI] [PubMed] [Google Scholar]
- Gopinath S., Kim M.V., Rakib T., Wong P.W., van Zandt M., Barry N.A. Topical application of aminoglycoside antibiotics enhances host resistance to viral infections in a microbiota-independent manner. Nat Microbiol. 2018;3:611–621. doi: 10.1038/s41564-018-0138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M., Penski N., Spiegel M., Weber F. T7 RNA polymerase-dependent and -independent systems for cDNA-based rescue of Rift Valley fever virus. J Gen Virol. 2008;89:2157–2166. doi: 10.1099/vir.0.2008/002097-0. [DOI] [PubMed] [Google Scholar]
- Haese N.N., Broeckel R.M., Hawman D.W., Heise M.T., Morrison T.E., Streblow D.N. Animal models of Chikungunya virus infection and disease. J Infect Dis. 2016;214:S482–S487. doi: 10.1093/infdis/jiw284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard C.R., Fletcher N.F. Emerging virus diseases: can we ever expect the unexpected? Emerg Microbes Infect. 2012;1:e46. doi: 10.1038/emi.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Yang M., Yuan Y., Li X., Kuang E. Niclosamide inhibits lytic replication of Epstein-Barr virus by disrupting mTOR activation. Antiviral Res. 2017;138:68–78. doi: 10.1016/j.antiviral.2016.12.002. [DOI] [PubMed] [Google Scholar]
- Hulseberg C.E., Feneant L., Szymanska-de Wijs K.M., Kessler N.P., Nelson E.A., Shoemaker C.J. Arbidol and other low-molecular-weight drugs that inhibit lassa and Ebola viruses. J Virol. 2019;93 doi: 10.1128/JVI.02185-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianevski A., Andersen P.I., Merits A., Bjoras M., Kainov D. Expanding the activity spectrum of antiviral agents. Drug Discov Today. 2019;24:1224–1228. doi: 10.1016/j.drudis.2019.04.006. [DOI] [PubMed] [Google Scholar]
- Ianevski A., Zusinaite E., Kuivanen S., Strand M., Lysvand H., Teppor M. Novel activities of safe-in-human broad-spectrum antiviral agents. Antiviral Res. 2018;154:174–182. doi: 10.1016/j.antiviral.2018.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasawa C., Tamura R., Sugiura Y., Suzuki S., Kuzumaki N., Narita M. Increased cytotoxicity of herpes simplex virus thymidine kinase expression in human induced pluripotent stem cells. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20040810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang Y., Shin J.S., Yoon Y.S., Go Y.Y., Lee H.W., Kwon O.S. Salinomycin inhibits influenza virus infection by disrupting endosomal acidification and viral matrix protein 2 function. J Virol. 2018;92 doi: 10.1128/JVI.01441-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupille H.J., Oko L., Stoermer K.A., Heise M.T., Mahalingam S., Gunn B.M. Mutations in nsP1 and PE2 are critical determinants of Ross River virus-induced musculoskeletal inflammatory disease in a mouse model. Virology. 2011;410:216–227. doi: 10.1016/j.virol.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgeit A., McDowell R., Moese S., Meldrum E., Schwendener R., Greber U.F. Niclosamide is a proton carrier and targets acidic endosomes with broad antiviral effects. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao J.C., HuangFu W.C., Tsai T.T., Ho M.R., Jhan M.K., Shen T.J. The antiparasitic drug niclosamide inhibits dengue virus infection by interfering with endosomal acidification independent of mTOR. PLoS Negl Trop Dis. 2018;12 doi: 10.1371/journal.pntd.0006715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwase H., Momota K., Ohmine T., Komai T., Kimura T., Katsube T. A new fluoroquinolone derivative exhibits inhibitory activity against human immunodeficiency virus type 1 replication. Chemotherapy. 1999;45:48–55. doi: 10.1159/000007164. [DOI] [PubMed] [Google Scholar]
- Kim S., Chen J., Cheng T., Gindulyte A., He J., He S. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47:D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittel C., Sereinig S., Ferko B., Stasakova J., Romanova J., Wolkerstorfer A. Rescue of influenza virus expressing GFP from the NS1 reading frame. Virology. 2004;324:67–73. doi: 10.1016/j.virol.2004.03.035. [DOI] [PubMed] [Google Scholar]
- Koban R., Neumann M., Daugs A., Bloch O., Nitsche A., Langhammer S. A novel three-dimensional cell culture method enhances antiviral drug screening in primary human cells. Antiviral Res. 2018;150:20–29. doi: 10.1016/j.antiviral.2017.12.005. [DOI] [PubMed] [Google Scholar]
- Konig A., Yang J., Jo E., Park K.H.P., Kim H., Than T.T. Efficient long-term amplification of hepatitis B virus isolates after infection of slow proliferating HepG2-NTCP cells. J Hepatol. 2019;71:289–300. doi: 10.1016/j.jhep.2019.04.010. [DOI] [PubMed] [Google Scholar]
- Kuivanen S., Bespalov M.M., Nandania J., Ianevski A., Velagapudi V., De Brabander J.K. Obatoclax, saliphenylhalamide and gemcitabine inhibit Zika virus infection in vitro and differentially affect cellular signaling, transcription and metabolism. Antiviral Res. 2017;139:117–128. doi: 10.1016/j.antiviral.2016.12.022. [DOI] [PubMed] [Google Scholar]
- Laamiri N., Aouini R., Marnissi B., Ghram A., Hmila I. A multiplex real-time RT-PCR for simultaneous detection of four most common avian respiratory viruses. Virology. 2018;515:29–37. doi: 10.1016/j.virol.2017.11.021. [DOI] [PubMed] [Google Scholar]
- Landry M.L. Nucleic acid hybridization in viral diagnosis. Clin Biochem. 1990;23:267–277. doi: 10.1016/0009-9120(90)80053-l. [DOI] [PubMed] [Google Scholar]
- Lanko K., Eggermont K., Patel A., Kaptein S., Delang L., Verfaillie C.M. Replication of the Zika virus in different iPSC-derived neuronal cells and implications to assess efficacy of antivirals. Antiviral Res. 2017;145:82–86. doi: 10.1016/j.antiviral.2017.07.010. [DOI] [PubMed] [Google Scholar]
- Lee M., Yang J., Jo E., Lee J.Y., Kim H.Y., Bartenschlager R. A novel inhibitor IDPP interferes with entry and egress of HCV by targeting glycoprotein E1 in a genotype-specific manner. Sci Rep. 2017;7:44676. doi: 10.1038/srep44676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.N., Ye C., Villani A.C., Raj T., Li W., Eisenhaure T.M. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343 doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Deng Y.Q., Wang S., Ma F., Aliyari R., Huang X.Y. 25-Hydroxycholesterol protects host against Zika virus infection and its associated microcephaly in a mouse model. Immunity. 2017;46:446–456. doi: 10.1016/j.immuni.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louz D., Bergmans H.E., Loos B.P., Hoeben R.C. Animal models in virus research: their utility and limitations. Crit Rev Microbiol. 2013;39:325–361. doi: 10.3109/1040841X.2012.711740. [DOI] [PubMed] [Google Scholar]
- MacGibeny M.A., Koyuncu O.O., Wirblich C., Schnell M.J., Enquist L.W. Retrograde axonal transport of rabies virus is unaffected by interferon treatment but blocked by emetine locally in axons. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell L.A., Wunderink R.G., Anzueto A., Bartlett J.G., Campbell G.D., Dean N.C. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44(Suppl 2):S27–72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston H.D., Folkers G.K., Morens D.M., Fauci A.S. Emerging viral diseases: confronting threats with new technologies. Sci Transl Med. 2014;6 doi: 10.1126/scitranslmed.3009872. [DOI] [PubMed] [Google Scholar]
- Mazzon M., Ortega-Prieto A.M., Imrie D., Luft C., Hess L., Czieso S. Identification of broad-spectrum antiviral compounds by targeting viral entry. Viruses. 2019;11 doi: 10.3390/v11020176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini K.A., Hamasaki D.I. The electroretinogram of the iguana and Tokay gecko. Vision Res. 1967;7:243–251. doi: 10.1016/0042-6989(67)90088-0. [DOI] [PubMed] [Google Scholar]
- Montoya M.C., Krysan D.J. Repurposing estrogen receptor antagonists for the treatment of infectious disease. MBio. 2018;9 doi: 10.1128/mBio.02272-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison T.E., Diamond M.S. Animal models of Zika virus infection, pathogenesis, and immunity. J Virol. 2017:91. doi: 10.1128/JVI.00009-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R., Roy S., Venkatadri R., Su Y.P., Ye W., Barnaeva E. Efficacy and mechanism of action of low dose emetine against human cytomegalovirus. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller K.H., Kainov D.E., El Bakkouri K., Saelens X., De Brabander J.K., Kittel C. The proton translocation domain of cellular vacuolar ATPase provides a target for the treatment of influenza A virus infections. Br J Pharmacol. 2011;164:344–357. doi: 10.1111/j.1476-5381.2011.01346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller K.H., Spoden G.A., Scheffer K.D., Brunnhofer R., De Brabander J.K., Maier M.E. Inhibition by cellular vacuolar ATPase impairs human papillomavirus uncoating and infection. Antimicrob Agents Chemother. 2014;58:2905–2911. doi: 10.1128/AAC.02284-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y., Hara H. Editorial: drug repositioning: current advances and future perspectives. Front Pharmacol. 2018;9:1068. doi: 10.3389/fphar.2018.01068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D.Y., Hurt A.C. Using the ferret as an animal model for investigating influenza antiviral effectiveness. Front Microbiol. 2016;7 doi: 10.3389/fmicb.2016.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasuraman S. Toxicological screening. J Pharmacol Pharmacother. 2011;2:74–79. doi: 10.4103/0976-500X.81895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil S.A., Patil V., Patil R., Beaman K., Patil S.A. Identification of novel 5,6-dimethoxyindan-1-one derivatives as antiviral agents. Med Chem (Shariqah (United Arab Emirates)) 2017;13:787–795. doi: 10.2174/1573406413666170330094822. [DOI] [PubMed] [Google Scholar]
- Perez J.T., Garcia-Sastre A., Manicassamy B. Insertion of a GFP reporter gene in influenza virus. Curr Protocols in microbiology. 2013 doi: 10.1002/9780471729259.mc15g04s29. Chapter 15, Unit 15G.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett B.E., Greer D.S., Zhang Y., Stewart L., Zhou L., Sun G. Virus pathogen database and analysis resource (ViPR): a comprehensive bioinformatics database and analysis resource for the coronavirus research community. Viruses. 2012;4:3209–3226. doi: 10.3390/v4113209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzorno, A., Padey, B., Terrier, O., Rosa-Calatrava, M., 2019. Drug repurposing approaches for the treatment of influenza viral infection: reviving old drugs to fight against a long-lived enemy. 10. [DOI] [PMC free article] [PubMed]
- Postnikova E., Cong Y., DeWald L.E., Dyall J., Yu S., Hart B.J. Testing therapeutics in cell-based assays: factors that influence the apparent potency of drugs. PLoS One. 2018;13 doi: 10.1371/journal.pone.0194880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puschnik A.S., Majzoub K., Ooi Y.S., Carette J.E. A CRISPR toolbox to study virus-host interactions. Nat Rev Microbiol. 2017;15:351–364. doi: 10.1038/nrmicro.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- Rada B., Dragun M. Antiviral action and selectivity of 6-azauridine. Ann N Y Acad Sci. 1977;284:410–417. doi: 10.1111/j.1749-6632.1977.tb21977.x. [DOI] [PubMed] [Google Scholar]
- Rausch K., Hackett B.A., Weinbren N.L., Reeder S.M., Sadovsky Y., Hunter C.A. Screening bioactives reveals nanchangmycin as a broad spectrum antiviral active against Zika virus. Cell Rep. 2017;18:804–815. doi: 10.1016/j.celrep.2016.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk M.L., Zou L., Savic R.M., Dooley K.E. Importance of drug pharmacokinetics at the site of action. Clin Transl Sci. 2017;10:133–142. doi: 10.1111/cts.12448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C.L., Chong A.C.N., Ashbrook A.W., Jeng G., Jin J., Chen H. Male germ cells support long-term propagation of Zika virus. Nat Commun. 2018;9:2090. doi: 10.1038/s41467-018-04444-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacramento C.Q., de Melo G.R., de Freitas C.S., Rocha N., Hoelz L.V., Miranda M. The clinically approved antiviral drug sofosbuvir inhibits Zika virus replication. Sci Rep. 2017;7:40920. doi: 10.1038/srep40920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarzotti-Kelsoe M., Bailer R.T., Turk E., Lin C.L., Bilska M., Greene K.M. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods. 2014;409:131–146. doi: 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital D.G. Pathogen detection in the CRISPR-Cas era. Genome Med. 2018;10:32. doi: 10.1186/s13073-018-0543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schor S., Einav S. Combating intracellular pathogens with repurposed host-targeted drugs. ACS Infect Dis. 2018;4:88–92. doi: 10.1021/acsinfecdis.7b00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks N., Greek R., Greek J. Are animal models predictive for humans? Philos Ethics Humanit Med. 2009;4:2. doi: 10.1186/1747-5341-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Niu J., Wang C., Huang B., Wang W., Zhu N. High-throughput screening and identification of potent broad-spectrum inhibitors of coronaviruses. J Virol. 2019;93 doi: 10.1128/JVI.00023-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidwell R.W., Huffman J.H., Khare G.P., Allen L.B., Witkowski J.T., Robins R.K. Broad-spectrum antiviral activity of Virazole: 1-beta-D-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science. 1972;177:705–706. doi: 10.1126/science.177.4050.705. [DOI] [PubMed] [Google Scholar]
- Simonin Y., Erkilic N., Damodar K., Cle M., Desmetz C., Bollore K. Zika virus induces strong inflammatory responses and impairs homeostasis and function of the human retinal pigment epithelium. EBioMedicine. 2019;39:315–331. doi: 10.1016/j.ebiom.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smee D.F., Barnard D.L. Methods for evaluation of antiviral efficacy against influenza virus infections in animal models. Methods Mol Biol (Clifton, N.J.) 2013;1030:407–425. doi: 10.1007/978-1-62703-484-5_31. [DOI] [PubMed] [Google Scholar]
- Soderholm S., Kainov D.E., Ohman T., Denisova O.V., Schepens B., Kulesskiy E. Phosphoproteomics to characterize host response during influenza a virus infection of human macrophages. Mol Cell Proteomics. 2016;15:3203–3219. doi: 10.1074/mcp.M116.057984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachulski A.V., Pidathala C., Row E.C., Sharma R., Berry N.G., Lawrenson A.S. Thiazolides as novel antiviral agents. 2. Inhibition of hepatitis C virus replication. J Med Chem. 2011;54:8670–8680. doi: 10.1021/jm201264t. [DOI] [PubMed] [Google Scholar]
- Taylor G. Animal models of respiratory syncytial virus infection. Vaccine. 2017;35:469–480. doi: 10.1016/j.vaccine.2016.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangavel R.R., Bouvier N.M. Animal models for influenza virus pathogenesis, transmission, and immunology. J Immunol Methods. 2014;410:60–79. doi: 10.1016/j.jim.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S Food and Drug Administration . 2018. The Drug Development Process: Step 3. [Google Scholar]
- Umscheid C.A., Margolis D.J., Grossman C.E. Key concepts of clinical trials: a narrative review. Postgrad Med. 2011;123:194–204. doi: 10.3810/pgm.2011.09.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursu O., Holmes J., Bologa C.G., Yang J.J., Mathias S.L., Stathias V. DrugCentral 2018: an update. Nucleic Acids Res. 2019;47:D963–D970. doi: 10.1093/nar/gky963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utt A., Quirin T., Saul S., Hellstrom K., Ahola T., Merits A. Versatile trans-replication systems for chikungunya virus allow functional analysis and tagging of every replicase protein. PLoS One. 2016;11 doi: 10.1371/journal.pone.0151616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.M., Lu J.W., Lin C.C., Chin Y.F., Wu T.Y., Lin L.I. Antiviral activities of niclosamide and nitazoxanide against chikungunya virus entry and transmission. Antiviral Res. 2016;135:81–90. doi: 10.1016/j.antiviral.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Buth J.E., Vishlaghi N., de la Torre-Ubieta L., Taxidis J., Khakh B.S. Self-organized cerebral organoids with human-specific features predict effective drugs to combat Zika virus infection. Cell Rep. 2017;21:517–532. doi: 10.1016/j.celrep.2017.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2015. WHO publishes list of top emerging diseases likely to cause major epidemics.www.who.int/medicines/ebola-treatment/WHO-list-of-top-emerging-diseases/en/ [Google Scholar]
- Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.J., Jan J.T., Chen C.M., Hsieh H.P., Hwang D.R., Liu H.W. Inhibition of severe acute respiratory syndrome coronavirus replication by niclosamide. Antimicrob Agents Chemother. 2004;48:2693–2696. doi: 10.1128/AAC.48.7.2693-2696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Carpentier A., Cheng X., Block P.D., Zhao Y., Zhang Z. Human stem cell-derived hepatocytes as a model for hepatitis B virus infection, spreading and virus-host interactions. J Hepatol. 2017;66:494–503. doi: 10.1016/j.jhep.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L., Wang S., Hu Q., Li J., Zeng Y. Comparison of three quantification methods for the TZM-bl pseudovirus assay for screening of anti-HIV-1 agents. J Virol Methods. 2016;233:56–61. doi: 10.1016/j.jviromet.2016.03.008. [DOI] [PubMed] [Google Scholar]
- Xu M., Lee E.M., Wen Z., Cheng Y., Huang W.K., Qian X. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med. 2016;22:1101–1107. doi: 10.1038/nm.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.P., Qiu Y., Zhang B., Chen G., Chen Q., Wang M. Zika virus infection induces RNAi-mediated antiviral immunity in human neural progenitors and brain organoids. Cell Res. 2019;29:265–273. doi: 10.1038/s41422-019-0152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Xu M., Lee E.M., Gorshkov K., Shiryaev S.A., He S. Emetine inhibits Zika and Ebola virus infections through two molecular mechanisms: inhibiting viral replication and decreasing viral entry. Cell Discov. 2018;4:31. doi: 10.1038/s41421-018-0034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Bijvelds M., Dang W., Xu L., van der Eijk A.A., Knipping K. Modeling rotavirus infection and antiviral therapy using primary intestinal organoids. Antiviral Res. 2015;123:120–131. doi: 10.1016/j.antiviral.2015.09.010. [DOI] [PubMed] [Google Scholar]
- Yin Y., Chen S., Hakim M.S., Wang W., Xu L., Dang W. 6-Thioguanine inhibits rotavirus replication through suppression of Rac1 GDP/GTP cycling. Antiviral Res. 2018;156:92–101. doi: 10.1016/j.antiviral.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Wang Y., Dang W., Xu L., Su J., Zhou X. Mycophenolic acid potently inhibits rotavirus infection with a high barrier to resistance development. Antiviral Res. 2016;133:41–49. doi: 10.1016/j.antiviral.2016.07.017. [DOI] [PubMed] [Google Scholar]
- Young D.D., Connelly C.M., Grohmann C., Deiters A. Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis C virus infection and hepatocellular carcinoma. J Am Chem Soc. 2010;132:7976–7981. doi: 10.1021/ja910275u. [DOI] [PubMed] [Google Scholar]
- Zhang Y.H., Zhao Y., Li N., Peng Y.C., Giannoulatou E., Jin R.H. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat Commun. 2013;4:1418. doi: 10.1038/ncomms2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Sun W., Simeonov A. Drug repurposing screens and synergistic drug-combinations for infectious diseases. Br J Pharmacol. 2018;175:181–191. doi: 10.1111/bph.13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Peng R., Zhang R., Li J. The applications of CRISPR/Cas system in molecular detection. J Cell Mol Med. 2018;22:5807–5815. doi: 10.1111/jcmm.13925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Tan L., Cederquist G.Y., Fan Y., Hartley B.J., Mukherjee S. High-content screening in hPSC-neural progenitors identifies drug candidates that inhibit Zika virus infection in fetal-like organoids and adult brain. Cell Stem Cell. 2017;21 doi: 10.1016/j.stem.2017.06.017. 274–283.e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.