Human respiratory syncytial virus (HRSV), a member of the Paramyxoviridae family, is a nonsegmented, single-stranded RNA virus [1]. HRSV is the main viral cause of acute and severe lower respiratory tract infections in children worldwide. It has a single type, with two subgroups (A and B) that are further divided into several genotypes based on the variability of the G-protein gene. For HRSV A, 11 genotypes have been described, which are GA1–GA7, NA1, NA2, SAA1, and ON1 [2]. HRSV B has 17 genotypes, designated as GB1–GB4, SAB1–SAB3, and BA1–BA10 [3].
In Paraguay, we have previously found that children with severe acute respiratory infection required longer time of hospitalization (5–15 days) when HRSV was present alone or in co-infection with adenovirus, rhinovirus, or coronavirus [4]. The lack of genetic information about these Paraguayan HRSV isolates led us to carry out a molecular characterization of the circulating genotypes in the Central Department.
A total of 36HRSV were detected out of 280 nasopharyngeal aspirates, collected from children under 5 years of age with symptoms of severe acute respiratory infection, between May 2010 and December 2013, admitted to the Hospital General Pediátrico Niños de Acosta Ñu. HRSV detection was performed by real-time PCR (Fast-Track Respiratory Pathogens Plus Kit, FTD, Luxembourg). Genetic amplification of the attachment G-protein gene was carried out by Polymerase Chain Reaction. Phylogenetic relationships were reconstructed by the neighbor-joining method with Kimura’s two-parameters as the model of nucleotide substitution, as incorporated in MEGA v5 [5]. Partial nucleotide sequences obtained in this study were deposited in the GenBank database (accession numbers: KM508816–KM508827).
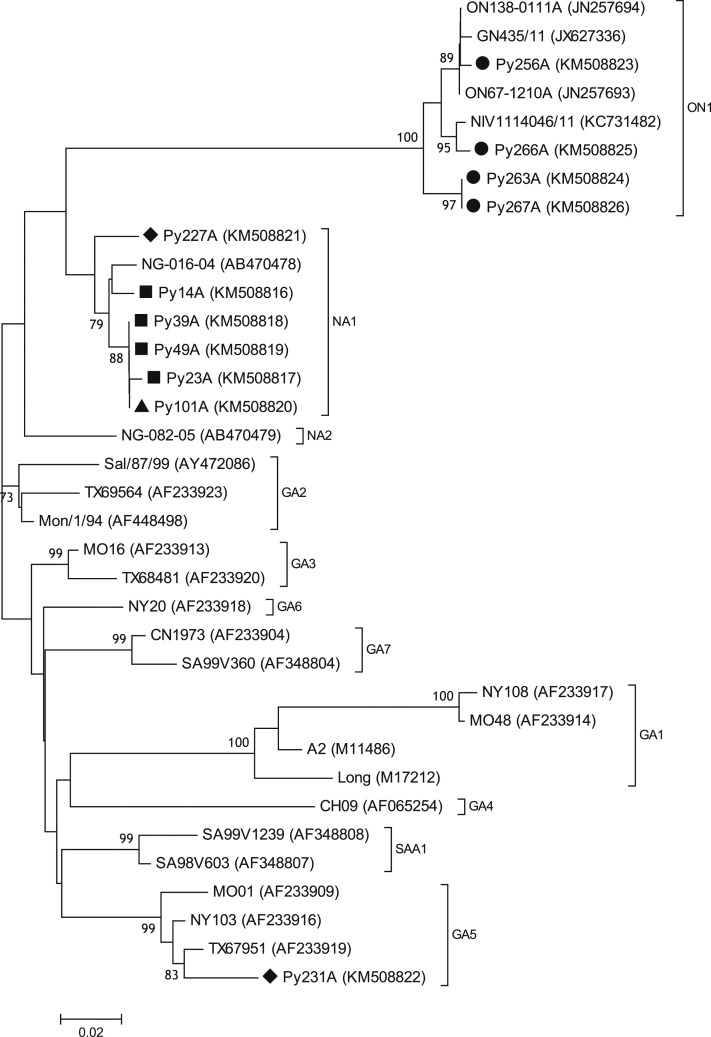
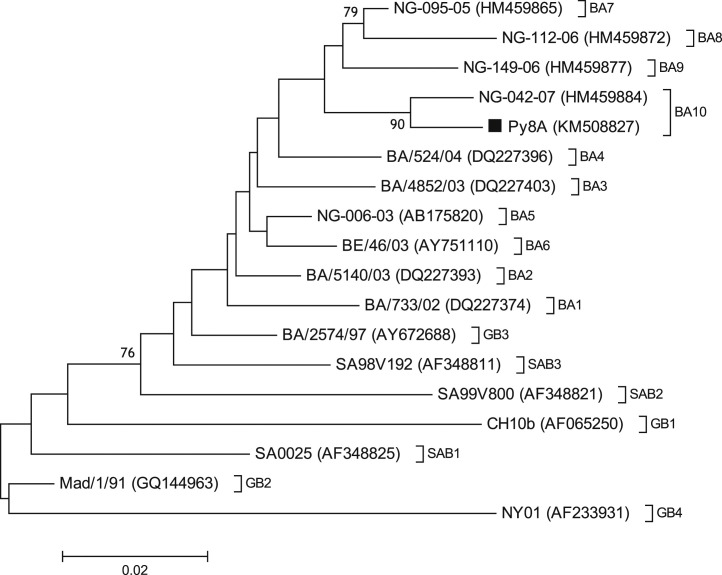
Sequencing/phylogenetic analyses of 12 samples showed the presence of HRSV subgroups A (11 samples) and B (1 sample) (Fig. 1, Fig. 2 ). Concerning HRSV subgroup A, genotype NA1 circulated between 2010 and 2012 (three seasons). In 2012 both genotypes NA1 and GA5 co-circulated. In 2013 the novel genotype ON1 circulated (Fig. 1); this genotype has a 72-nt duplication of G-protein gene, which was first described in Ontario during the 2010–2011 winter season [2]. Concerning HRSV subgroup B, genotype BA10 circulated in 2010 (Fig. 2); therefore, in this year both HRSV subgroups A and B co-circulated.
Fig. 1.
Phylogenetic tree of HRSV-A, constructed with a partial G-protein gene (ranging from nucleotide coding sequence 634–890 of reference strain Long, Genbank M17212), comparing 24 HRSV worldwide sequences and 11 Paraguayan isolates. Genotypes GA1–GA7, NA1, NA2, SAA1, and ON1 are grouped and indicated in parentheses. Each sequence is indicated with its corresponding name and GenBank accession number. Paraguayan isolates are labeled according to year of isolation: 2010 (■), 2011 (▲), 2012 (♦), and 2013 (●). Bootstrap values greater than 70% are shown at branch nodes. Branch distance is indicated by a scale bar at the bottom of the tree.
Fig. 2.
Phylogenetic tree of HRSV-B, constructed with a partial G-protein gene (ranging from nucleotide coding sequence 637–897 of reference strain CH10b, Genbank AF065250), comparing 17 HRSV worldwide sequences and 1 Paraguayan isolate. Genotypes GB1–GB4, SAB1–SAB3, and BA1–BA10 are grouped and indicated in parentheses. Each sequence is indicated with its corresponding name and GenBank accession number. Paraguayan isolate is labeled according to year of isolation: 2010 (■). Bootstrap values greater than 70% are shown at branch nodes. Branch distance is indicated by a scale bar at the bottom of the tree.
The 60-nt duplication of G-protein gene from HRSV-B was first described in Buenos Aires [6], from specimens collected at different times (1995–2001) and locations. The first report of BA10 circulation goes back to 2007 in Niigata, Japan [3]. The Paraguayan BA10 strain had two characteristic changes in its predicted G-protein, E226D and E292G, and conserved all its N-glycosylation sites, as compared with BA prototype strains.
In the ON1 genotype, the duplication of the G-protein gene is at one-third of the C-terminal region. In our study, the Paraguayan ON1 strains retained two unique amino acid substitutions (E232G, and T252K) in this region [2]. It is known that ON1 genotype has rapidly disseminated across the world and replaced other genotypes of HRSV-A, but up to date this replacement has not been associated with changes in the severity of illness or number of intensive care admissions [7], [8], [9].
Conflict of interests
None.
Funding
This study was partially funded by the Universidad Nacional de Asunción, Paraguay.
Ethical approval
Written informed consent was obtained from parents or guardians prior to study participation. This study was approved by the Ethics Committee at Instituto de Investigaciones en Ciencias de la Salud, Universidad Nacional de Asunción (IICS-UNA) and the Hospital General Pediátrico Niños de Acosta Ñu.
Acknowledgements
We would like to thank Dr. Viviana Pavlicich and Dr. Carolina Aquino, from Hospital General Pediátrico Niños de Acosta Ñu (Paraguay), for their professional support throughout the study, and Fondation Mérieux—GABRIEL Network (France), for their technical support with the multiplex PCR.
References
- 1.Collins P.L., Karron R.A. Respiratory syncytial virus and metapneumovirus. In: Knipe D.M., Howley P.M., Cohen J.I., Griffin D.E., Lamb R.A., Martin M.A., editors. 6th ed. vol. 1. Wolters Kluwer/Lippincott Williams & Wilkins Health; Philadelphia, PA: 2013. pp. 1086–1123. (Fields Virology). [Google Scholar]
- 2.Eshaghi A., Duvvuri V.R., Lai R., Nadarajah J.T., Li A., Patel S.N. Genetic variability of human respiratory syncytial virus A strains circulating in Ontario: a novel genotype with a 72 nucleotide G gene duplication. PLoS One. 2012;7:e32807. doi: 10.1371/journal.pone.0032807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dapat I.C., Shobugawa Y., Sano Y., Saito R., Sasaki A., Suzuki Y. New genotypes within respiratory syncytial virus group B genotype BA in Niigata, Japan. J. Clin. Microbiol. 2010;48:3423–3427. doi: 10.1128/JCM.00646-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espinola E.E., Basualdo W., Guillen R.M., Pavlicich V., Maldonado L., Aquino C. High incidence of viral co-infections and atypical bacterial detection in acute respiratory infections among hospitalized children in the Central Department of Paraguay, 2010–2011. J. Infect. 2013;66:196–198. doi: 10.1016/j.jinf.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trento A., Galiano M., Videla C., Carballal G., Garcia-Barreno B., Melero J.A. Major changes in the G protein of human respiratory syncytial virus isolates introduced by a duplication of 60 nucleotides. J. Gen. Virol. 2003;84:3115–3120. doi: 10.1099/vir.0.19357-0. [DOI] [PubMed] [Google Scholar]
- 7.Ren L., Xia Q., Xiao Q., Zhou L., Zang N., Long X. The genetic variability of glycoproteins among respiratory syncytial virus subtype A in China between 2009 and 2013. Infect. Genet. Evol. 2013;27:339–347. doi: 10.1016/j.meegid.2014.07.030. [DOI] [PubMed] [Google Scholar]
- 8.Tabatabai J., Prifert C., Pfeil J., Grulich-Henn J., Schnitzler P. Novel respiratory syncytial virus (RSV) genotype ON1 predominates in Germany during winter season 2012–13. PLoS One. 2014;9:e109191. doi: 10.1371/journal.pone.0109191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pierangeli A., Trotta D., Scagnolari C., Ferreri M.L., Nicolai A., Midulla F. Rapid spread of the novel respiratory syncytial virus A ON1 genotype, central Italy, 2011 to 2013. Euro Surveill. 2014:19. doi: 10.2807/1560-7917.es2014.19.26.20843. [DOI] [PubMed] [Google Scholar]