Summary
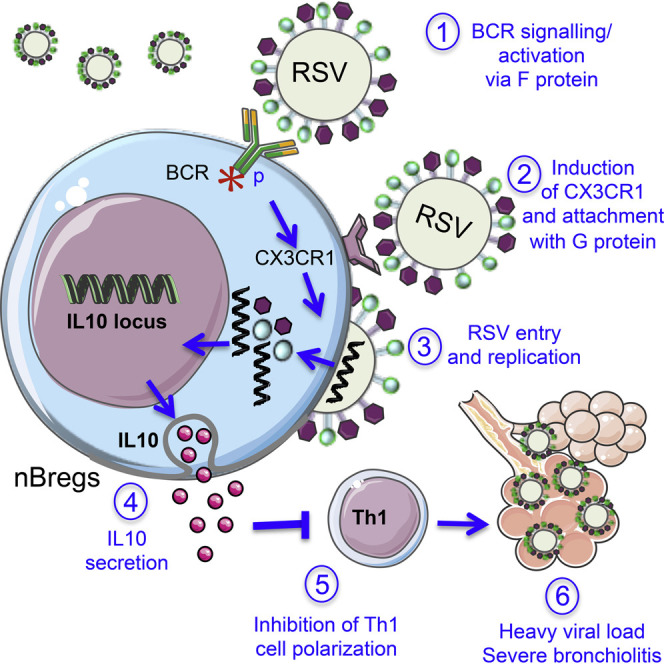
Respiratory syncytial virus (RSV) is the major cause of lower respiratory tract infections in infants and is characterized by pulmonary infiltration of B cells in fatal cases. We analyzed the B cell compartment in human newborns and identified a population of neonatal regulatory B lymphocytes (nBreg cells) that produced interleukin 10 (IL-10) in response to RSV infection. The polyreactive B cell receptor of nBreg cells interacted with RSV protein F and induced upregulation of chemokine receptor CX3CR1. CX3CR1 interacted with RSV glycoprotein G, leading to nBreg cell infection and IL-10 production that dampened T helper 1 (Th1) cytokine production. In the respiratory tract of neonates with severe RSV-induced acute bronchiolitis, RSV-infected nBreg cell frequencies correlated with increased viral load and decreased blood memory Th1 cell frequencies. Thus, the frequency of nBreg cells is predictive of the severity of acute bronchiolitis disease and nBreg cell activity may constitute an early-life host response that favors microbial pathogenesis.
Keywords: newborn, bronchiolitis, respiratory syncytial virus, Breg cell
Graphical Abstract
Highlights
-
•
Identified a neonatal-specific subset of regulatory B (nBreg) cells in the blood
-
•
Neonatal nBreg cells are infected by RSV via the BCR and CX3CR1
-
•
RSV-infected nBreg cells produce anti-inflammatory IL-10 that dowregulates Th1 cell responses
-
•
Blood nBreg cells are a biomarker of lung disease severity in RSV+ patients
RSV infection leads to respiratory distress in very young children whereas it is asymptomic later in life. Zhivaki et al. found a neonatal-specific regulatory B cell that is recruited in the respiratory tract, gets infected by the virus, and correlates with high viral load and disease severity.
Introduction
Human respiratory syncytial virus (RSV) is the major cause of lower respiratory tract infections in young infants leading to hospitalization and an increased risk factor for asthma development (Smyth and Openshaw, 2006). The immune system plays a critical role in the pathogenesis of RSV disease, and RSV is associated with the exacerbation of airway inflammation (Castro et al., 2008). In infants, fatal outcomes of primary RSV infection are associated with the pulmonary infiltration of B cells (Reed et al., 2009, Welliver et al., 2007), yet the role of these cells remains to be assessed.
Early-life susceptibility to infection contributes to high mortality rates in children under 5 years of age (PrabhuDas et al., 2011) who display biased T helper 2 (Th2) cell responses (Siegrist, 2001). We previously showed that B cells with regulatory properties (Breg cells) in murine neonates dampen innate inflammation and the functions of the dendritic cells (DCs) in a TLR-dependent manner, thereby inhibiting the Th1 cell responses (Sun et al., 2005, Zhang et al., 2007). Over the past decade, Breg cells, which act through IL-10 secretion, have attracted attention in the fields of autoimmunity, transplantation, and cancer (Mauri and Bosma, 2012). B cell-derived IL-10 can be induced through activation by CD40 (Mauri et al., 2003), the B cell receptor (BCR) (Fillatreau et al., 2002), or several Toll-like receptors (TLRs) (O’Garra et al., 1992, Zhang et al., 2007). The regulatory activity of IL-10-producing Breg cells is essential in the control of DCs (Sun et al., 2005) and inflammatory T cells (Matsushita et al., 2008) and in the induction of T regulatory (Treg) cells (Lemoine et al., 2011). Breg cell activity is not associated with a single B cell subset but rather develops under various conditions. In mice, regulatory activity has been extended to IL-35-producing B cells and to plasma cells in the context of inflammatory and bacterial diseases (Shen et al., 2014).
Here, we analyzed the B cell compartment in human newborns using high-content mass cytometry, transcriptomics, and functional studies. We identified a population of neonatal B lymphocytes with immunosuppressive activity (nBreg cells) through the production of IL-10. We showed that nBreg cells, which are a target for RSV, are highly permissive to infection because of BCR recognition of RSV F that drove nBreg cell activation and expression of the chemokine receptor CX3CR1. CX3CR1 interacted with RSV G glycoprotein-promoted infection of nBreg cells to induce IL-10 production. Analysis of clinical samples indicated that nBreg cells were associated with poor control of RSV infection in neonates. We propose the use of the frequency of nBreg cells as a prognostic tool to determine bronchiolitis severity and the use of both RSV F and G glycoprotein as targets for the development of interventions in the context of acute infection.
Results
RSV Infection Activates Neonatal Breg Cells, Resulting in IL-10 Production
To analyze the activity of putative Breg cell activity in healthy human newborns, we first examined neonatal B cells within the cord blood mononuclear cell (CBMC) population using a 35-parameter mass cytometric approach (CyTOF). Within the B cell population, unsupervised analysis using the viSNE algorythm that allows dimensionality reduction (Amir et al., 2013) revealed different B cell subsets with different phenotypes that clustered according to their high expression of CD5 or CD10 and that were associated with distinct sets of B cell markers (Figures 1 A, 1B, and S1). Mature naive (MN) CD19+ B cells were phenotypically defined as being CD5−CD10−CD1chiCD21hiCD45RAhiCD23loCD24intCD38intIgDhiIgMlo/hi, although there was some heterogeneity for some markers. Immature transitional B cells (IMT, N°2), basically corresponding to CD24hiCD38hi B cells, were phenotypically defined as being CD5loCD10+CD1c−CD21−CD45RAintCD23-CD24hiCD38hiIgDintIgMhi. We also found an additional, previously undescribed population of CD5hi B cells, phenotypically defined as CD5hiCD10−CD1cloCD21intCD45RAintCD23hiCD24loCD38loIgDloIgMlo (N°3) (Figure 1A). We compared the cytometry profiles for additional CD markers of MN B cells, IMT B cells, and the newly defined CD5hi population to bulk CD19+CD20+ B cells and further defined the CD5hi B cell phenotype as CD43+CD9−CD62L−CD40intDRintCD25+/−CD27+/−CD70− (Figure 1B). Adult blood IMT CD24hiCD38hi B cells have been shown to produce IL-10 in response to CD40 (Blair et al., 2010) or CpG and/or TLR9 activation (Menon et al., 2016). Therefore, the abundance of IMT B cells in cord blood suggested that these cells could be a major source of IL-10 in newborns. IMT B cells could be distinguished by CD10 expression with an intermediate cell surface expression of CD5 (Figure S1A), which accurately matched the high expression of CD24 and CD38 (Figure 1B). To analyze the functions of newborn B cells, the three cord blood B cell populations were sorted by fluorescence activated cell sorting (FACS) based on cell surface CD5 and CD10 expression (Figure S1B). Among these cord blood-derived cells, CD5hi B cells, but not IMT and MN B cells, produced IL-10 upon 48 hr of stimulation with A strain of human RSV (HRSV-A) (MOI = 2.5) (Figure 1C). Based on their IL-10 production, we thus named CD5hiB cells neonatal Breg (nBreg) cells. nBreg cells were abundant in neonatal blood from preterm and full-term babies and their frequency among total B cells quickly waned with age (Figure 1D). To determine whether these cells originated from the mothers of the babies or the babies themselves, we performed fluorescence in situ hybridization (FISH) on nBreg cells from male babies. FISH detected X and Y chromosomes in all of the nBreg cells isolated from the cord blood of male babies, indicating that the nBreg cells were derived from the babies rather than the mothers (Figure 1E). We then sought to determine the stimuli that can lead to IL-10 production from nBreg cells. While nBreg cells made IL-10 in response to HRSV (Figure 1C), the nBreg cells failed to produce IL-10 after infection with a large panel of RNA or DNA viruses, including influenza A virus (IAV), coronavirus 229E, HIV, HSV-1, HTLV, and MV (Figure S1C). To determine whether the induction of the IL-10 response by HRSV was specific to neonatal B cells, we tested the various adult blood B cell subsets and tested their responsiveness to HRSV. We first peformed CyTOF on adult IMT B cells, memory B cells, marginal zone B (MZB) cells, and naive B cells. Adult IMT cells displayed a phenotype similar to newborn IMT cells, whereas MZB cells and memory B cells were phenotypically distinct from nBreg cells (Figure S2A). FACS-sorted adult memory B cells, MZB cells, and IMT cells produced IL-10 in response to R848, a TLR7 agonist, but not in response to HRSV (MOI = 2.5) (Figure S2B). These data indicate that HRSV-activated nBreg cells may possess unique anti-inflammatory properties. The Th1-Th2 balance is critical at the time of the primary RSV infection, as impaired Th1 cell priming or a Th2 cell immunopathology have been shown to determine the outcome of secondary infection (Culley et al., 2002). We thus performed an inflammatory T cell suppression assay, which remains the gold standard for assessing Breg cell activity. Coculture of RSV-activated nBreg cells with activated CD4+ Th1 cells inhibited IFN-γ and IL-22, but not TNF-α, cytokine production from the CD4+ T cells (Figures 2 A–2C). Using IAV-activated neonatal plasmacytoid DCs (pDCs), we recently showed that type I IFN-dependent neonatal Th1 cell differentiation is induced by the allogeneic immune response (Zhang et al., 2014a). Similarly, RSV-activated neonatal pDCs induced a predominantly IFN-γ Th1 cell response that was associated with a mild IL-4 Th2 cell responses (Figures 2D–2F). To investigate whether nBreg cells could regulate Th1 cell response indirectly via pDCs, pDCs were cultured alone or with RSV-activated nBreg cells for 2 days and then these cells were co-cultured with naive T cells for 6 days after. We observed few pDCs infected by RSV when cultured alone or after coculture with nBreg cells (Figure S3A). RSV-activated nBreg cells were able to inhibit the ability of pDCs to prime an IFN-γ T cell response in an IL-10-dependent manner (Figures 2D–2F). This was associated with decreased APC functions of pDCs (HLA-DR and CD80) but not the IFN-α response (Figure S3B). Altogether, these data demonstrate that the nBreg cells can be specifically activated by RSV and may control the Th1 cell responses in an IL-10-dependent manner.
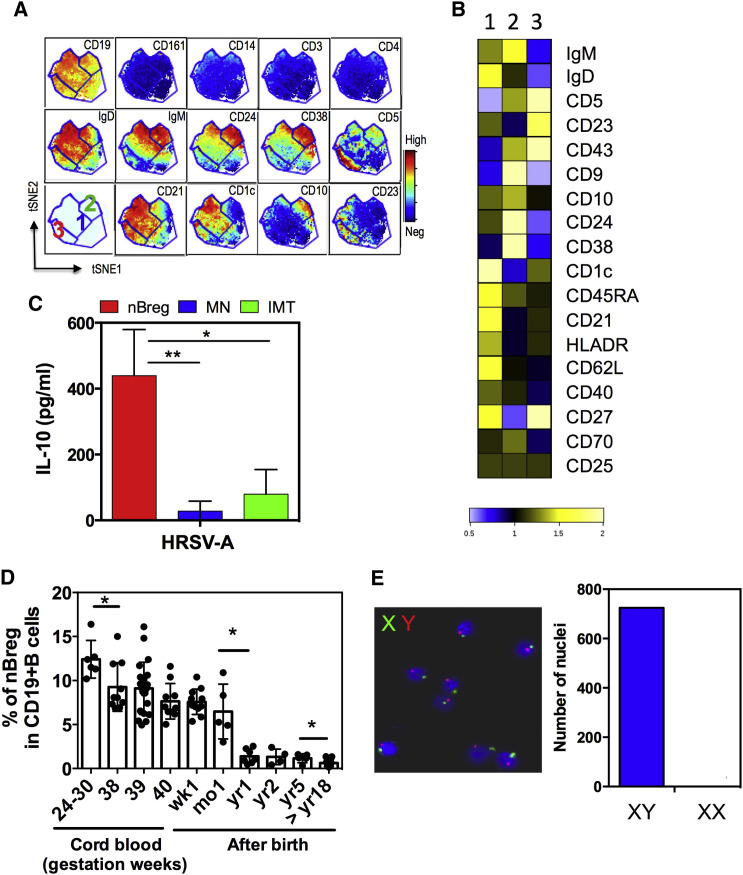
Figure 1.
Identification of Phenotype of New Population of Regulatory B Cells in the Human Neonate
(A and B) CyTOF analysis of cord blood CD19+ B cells within CMBCs for lineage and B cell markers.
(A) Data analysis using viSNE based on 19 markers delineating phenotype CD10−CD5− (1), CD10+CD5+ (2), and CD10−CD5+ (3).
(B) Heatmap analysis for data corresponding to the fold change expression of indicated markers for each subset as compared to the whole CD19+CD20+ B cell population.
(C) Cord blood B cell subsets were FACS sorted as CD10−CD5− (MN; blue), CD10+CD5+ (IMT; green), and CD10−CD5+ (nBreg; red). 105 cord blood B cell subsets were stimulated with HRSV-A (MOI = 2.5) and IL-10 production was measured at 48 hr by ELISA (n = 3). Results are expressed as the means ± SD.
(D) Percentage of the nBreg cell subset among the total B cells circulating in the blood during different developmental stages and ages.
∗p < 0.05, ∗∗p < 0.01.
(E) X and Y chromosome FISH staining of nBreg cells isolated from cord blood of a male baby. Bar chart indicates the number of nuclei associated with XY or XX staining.
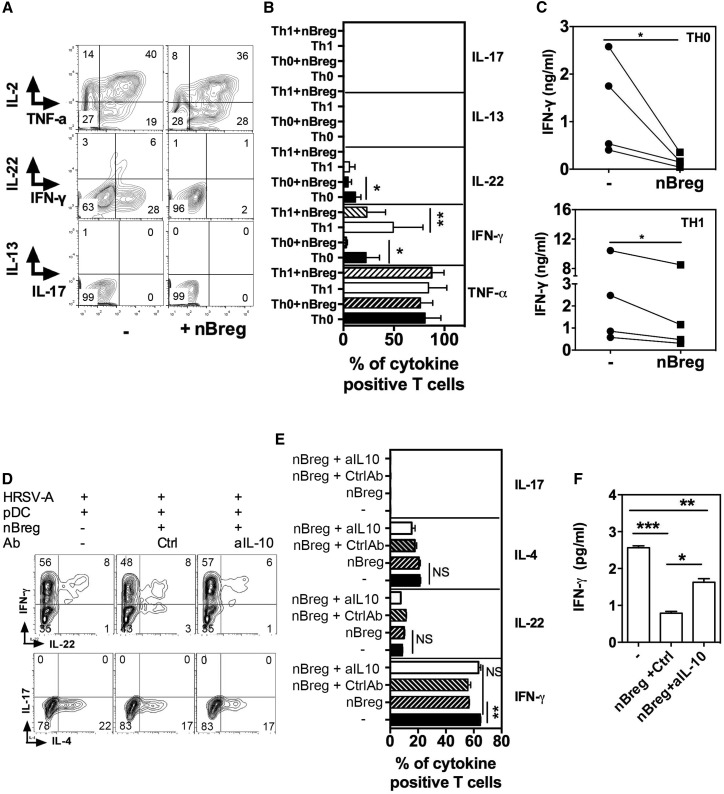
Figure 2.
nBreg Cells Regulate Th Cell Polarization Directly or Indirectly via pDCs
(A–C) Neonatal naive CD4+ T cells were activated by anti-CD3 + anti-CD28 and cultured with 10 ng/mL IL-12 (Th1) or without (Th0), alone or in co-culture with HRSV-activated nBreg cells for 6 days. FACS plots (A) and mean frequencies (B) of TNF-α-, IL-2-, IFN-γ-, IL-13-, IL-17-, or IL-22-secreting cells, as determined by intracellular staining for five donors (ANOVA test). Quantification of IFN-γ (C) in the supernatants of the same co-cultures, as determined by ELISA (n = 4 donors; paired t test was used for comparison).
(D–F) Neonatal pDCs were stimulated with HRSV-A either alone or in co-culture with nBreg cells in the presence of neutralizing anti-IL-10 or control antibody (Ctrl) for 48 hr. pDCs were FACS purified again before being used in Th cell differentiation assay. Intracellular IFN-γ, IL-4, IL-17, and IL-22 expression was analyzed by FACS (D and E) and secreted IFN-γ analyzed in the supernatants by ELISA (F).
Results are representatives of three experiments. Results are expressed as the means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗p < 0.001, and NS for non-significant.
Neonatal Breg Cells Are Highly Permissive to RSV Infection
We further investigated the capacity of RSV to directly activate the immunosuppressive properties of nBreg cells. RSV induced IL-10 transcription in sorted cord blood nBreg cells as soon as 6 hr after exposure (Figure 3 A). We used a rHRSV-expressing mCherry (rHRSV-Ch) construct (Rameix-Welti et al., 2014) to visualize the virus replication in cells by quantifying the red emitted fluorescence. We found that nBreg cells were preferentially infected by the virus (Figure 3B) compared to MN or IMT B cells isolated from cord blood. RSV infection did not affect the viability of nBreg cells (Figure S4A). Among adult B cell subsets, only memory B cells were able to be infected with RSV, but they showed much lower viral replication compared with nBreg cells (Figure S4B). nBreg cells harboring RSV produced more IL-10 compared to their RSV-negative counterparts (Figures 3C and 3D) and only live but not UV-treated RSV induced IL-10 production (Figure 3E), indicative of a role for viral infection in nBreg cell activation. Epithelial cells of the respiratory tract are the major targets of HRSV replication in vivo. We found that nBreg cells, but not MN or IMT B cells, produced IL-10 and were preferentially infected when cocultured with a RSV-infected human epithelial HEp-2 cell line (Figures 3F and 3G). Altogether, these data show that nBreg cells are specifically permissive to RSV infection and that IL-10 production by nBreg cells requires their viral infection.
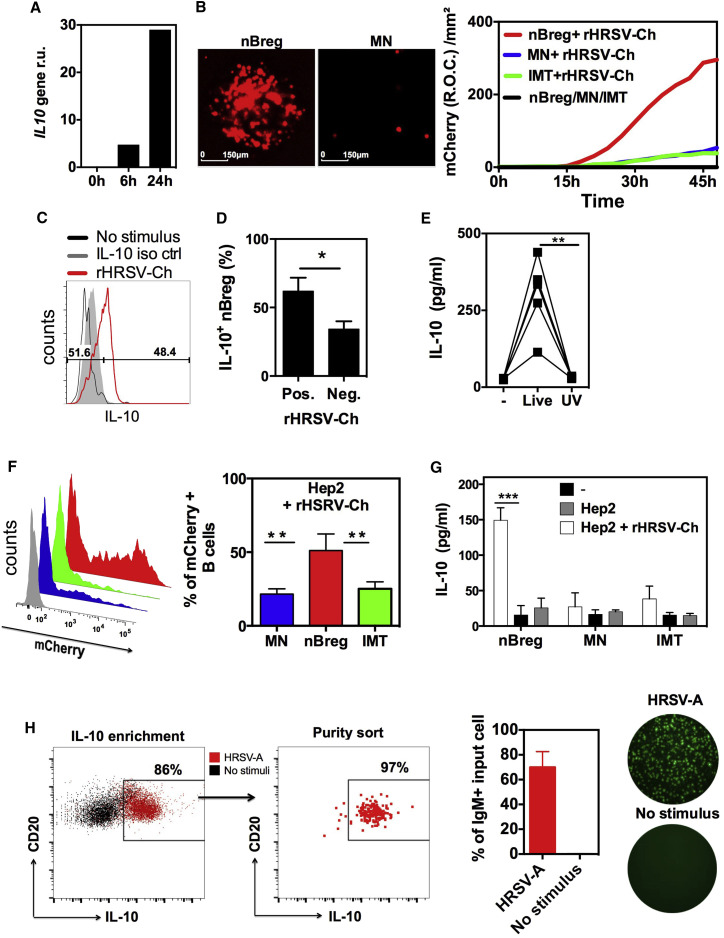
Figure 3.
nBreg Cells Are Preferentially Infected by RSV
(A–C) 105 cord blood nBreg cells were FACS sorted and left untreated (0 hr) or exposed to HRSV-A or to rHRSV-Ch (MOI = 2.5).
(A) Representative plot of IL10 gene expression, as measured by qRT-PCR at 0, 6, and 24 hr.
(B) 105 B cell subsets were FACS sorted as nBreg cells, MN, or IMT cells and stimulated with rHRSV-Ch. mCherry expression was assessed by fluorescent microscopy at 48 hr after infection (left) or by monitoring the red object count (R.O.C) through live imaging (right). Results are representitative of 3–5 independent experiments.
(C) Representative FACS plot for intra-cellular IL-10 expression at 48 hr after infection as compared to untreated cells (No stimulus).
(D) The frequency of IL-10+ nBreg cells among rHRSV-Ch-positive or -negative nBreg cells (n = 3). Unpaired t test was used for comparison.
(E) IL-10 production after nBreg cell exposure to live or UV-treated HRSV-mCherry was measured at 48 hr by ELISA (n = 5). Paired t test was also used to compare the three conditions.
(F and G) HEp-2 cells were infected with rHRSV-Ch (MOI = 0.1) then cocultured with B cell subsets.
(F) The percentage of rHRSV-Ch+ B cells in co-culture with HEp-2 cells is shown by FACS at 48 hr after coculture (n = 3). ANOVA test was used to compare the three groups.
(G) IL-10 production was measured by ELISA (n = 3) and unpaired t test was used for comparison.
(H) nBreg cells were stimulated or not with HRSV-A for 24 hr. IL-10+ nBreg cells were enriched using IL-10 enrichement beads, then FACS sorted IL-10+ nBreg cells were used for fluorescent IgM ELISPOT. Left panel is a representative FACS plot after IL-10 enrichement and sorting purity. Right panel indicates the frequency of IgM+ cells.
Results are expressed as the means ± SD of triplicates. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
RSV Is Recognized by IgM and Can Engage the BCR Pathway in nBreg Cells
During steady state or upon RSV-mediated activation, nBreg cells expressed immunoglobulin M (IgM) and IgD, but not IgA or IgG (Figures S1B and S1D). In addition, RSV-nBreg cell-specific interactions led to IgM secretion by the majority of IL-10-producing cells (Figure 3H), suggesting a role for the BCR. To investigate this hypothesis, we used a transcriptomic approach to compare nBreg cells after TLR (R848), BCR (anti-IgM), or viral (RSV or IAV) activation. Principal-component analysis (PCA) and hierarchical clustering showed the similarities and distinctness of the Breg cell response to different activators (Figures 4A and 4B). The transcriptional profiles of nBreg cells stimulated with anti-IgM (BCR) and RSV were closer than with TLR agonist, indicating that BCR activation could be involved in RSV infection. TLR7 or 8 activation by the R848 agonist did not recapitulate the transcriptional pattern of viral activation. Therefore, RSV RNA sensing by TLR7 or 8 may not be essential for the activation of nBreg cells and may explain why other RNA viruses did not induce IL-10. In addition, pathway analysis showed that RSV-activated nBreg cells significantly upregulated BCR-related pathways but not TLR-, RIG-I-, or CD40-related pathways (Figure 4C). RSV and BCR activation induced expression of common 534 genes in nBreg cells, as shown on the Venn diagram in Figure S5A. Gene set enrichment analysis (GSEA) of the transcriptome also indicated that BCR pathways were induced in nBreg cells by RSV whereas IAV infection did not specifically induce BCR pathways (Figure S5C). To confirm that RSV-infected nBreg cells were triggered through their BCRs, we performed an Igα (CD79a) phospho flow assay. Igα phosphorylation was detected in nBreg cells 30 min after treatment with anti-IgM or RSV, but not with R848 or IAV treatment (Figures 4D and 4E). All the stimuli activated nBreg cells, as shown by Erk phosphorylation (Figure S5D). In conclusion, the above experiments strongly indicate that RSV activation of nBreg cells is mediated in part by Ig recognition.
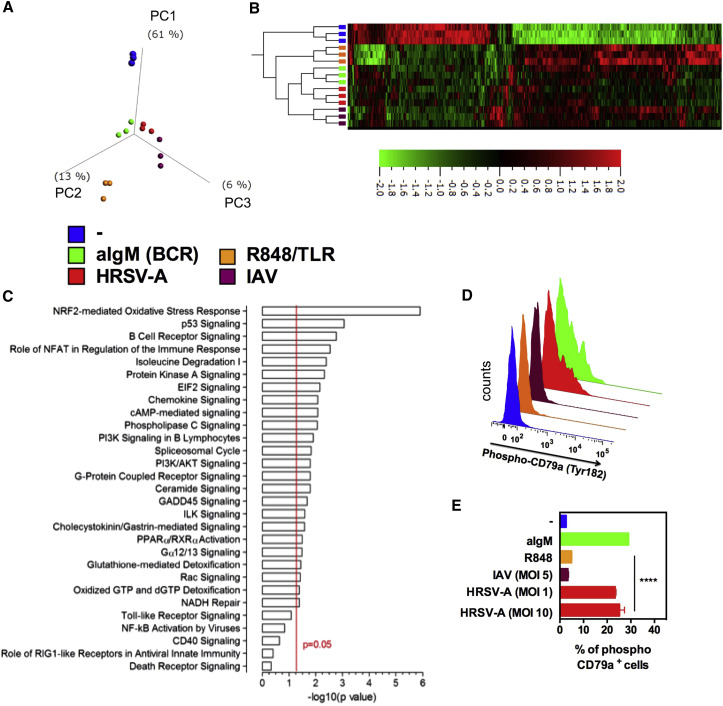
Figure 4.
RSV Activates the BCR Pathway
5 × 103 cord blood nBreg cells were FACS sorted as CD19+CD5+CD10− B cells and stimulated for 6 hr with aIgM, R848, HRSV-A, or IAV or left unstimulated. Gene expression profiles were compared by microarray analysis for three independent donors.
(A and B) PCA (A) and heatmap (B) of hierarchical clustering corresponding to the indicated stimulus (p = 0.0049 and q = 0.067; 745 genes).
(C) The list of differentially expressed genes (p < 0.05) was processed using the Ingenuity pathway analysis software. The list was then manually curated to remove pathways irrelevant to B cell biology. Canonical pathways were considered significant for p < 0.05 (red line).
(D and E) nBreg cells were activated for 30 min with aIgM, R848, HRSV-A, or IAV or left unstimulated. Phosphorylation of CD79a was assessed intracellularly by FACS. Representative FACS plots and means ± SD of 3 experiments are shown; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
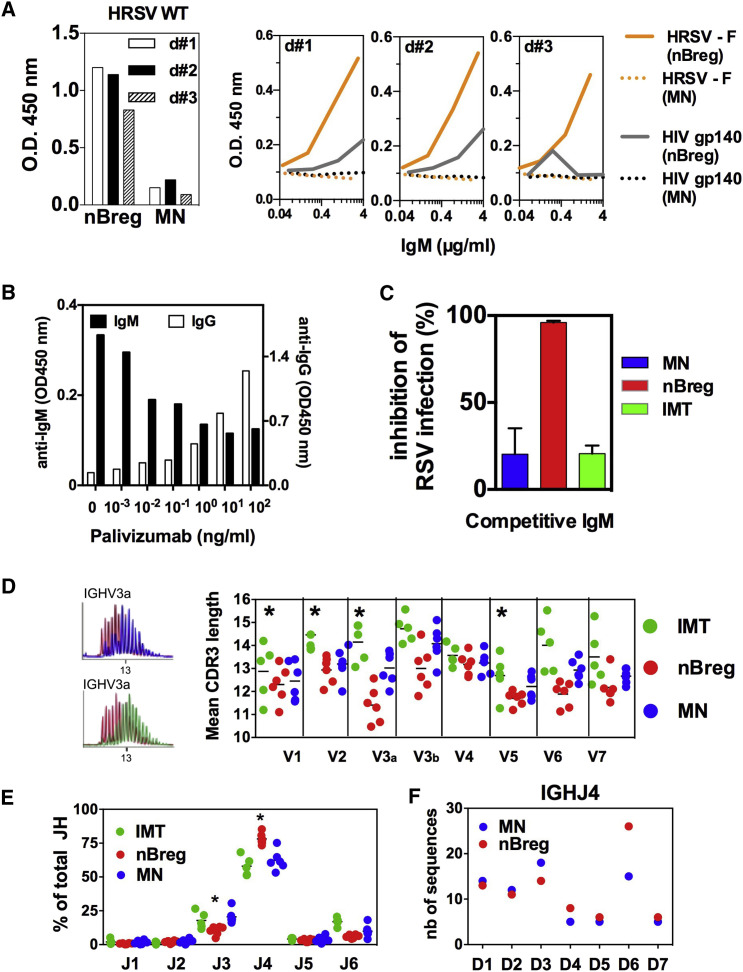
To further address the role of the BCR at the level of Ig recognition, we analyzed the IgM secreted by nBreg cells. nBreg cells produced 5-fold higher concentrations of IgM than MN cells and 50-fold higher concentrations than IMT cells (Figure S6A). This finding was confirmed using ELISPOT in which more than 80% were secreting IgM nBreg cells as compared to 10%–15% of MN B cells as assessed (Figure S6A). IgM produced by nBreg cells, but not MN or IMT B cells, showed specific binding to RSV particles (Figures 5 A and S6B). The mature HRSV envelope consists of glycoprotein (G), fusion (F) protein, and small hydrophobic (SH) protein. We found by ELISA that IgM from nBreg cells recognized the F fusion protein of HRSV but barely recognized the HIV-1 envelope glycoprotein gp140 (Figure 5A). These IgM still bound the rHRSV-ΔSH and the rHRSV-ΔG mutants showing that F protein, but not G or SH proteins, were recognized by the Ig (Figure S6B). Palivizumab, which is an IgG1 humanized monoclonal antibody that binds to F protein, outcompeted the binding of nBreg-cell-derived IgM to RSV in a dose-dependent manner (Figure 5B). In addition, IgM produced by nBreg cells, but not by MN or IMT B cells, competitively inhibited RSV infection of nBreg cells (Figure 5C). We concluded that the F protein, but not SH and G proteins on the virion, contributes to IgM-mediated recognition of RSV by nBreg cells. This further reinforced the results from transcriptomics and signaling analyses that indicated the engagement of the BCR in nBreg cell infection by RSV.
Figure 5.
Igs from nBreg Cells Recognize RSV and Display a Biased Repertoire
(A–C) IgMs from three donors (d#1, d#2, and d#3) were produced by nBreg cells and MN neonatal B cell subsets after their stimulation with CpG B (1826) for 4 days and then were tested by ELISA for recognition of WT HRSV-A (left, IgM 4 μg/mL), HRSV-F protein, or HIV-1 gp140 protein (right; serial dilutions of IgM). Results are expressed as optical density measured at 450 nm (OD 450 nm).
(B) IgMs (100 ng/mL) from nBreg cells were tested by ELISA for HRSV-A recognition in the presence of various doses of anti-F Palivizumab IgG Ab.
(C) rHRSV-Ch was pre-incubated with nBreg-, MN-, or IMT-derived IgMs (50 ng/mL) for 1 hr prior infection. RSV infection was assessed by monitoring mCherry expression by fluorescent live microscopy. Histogram plot shows the frequency of inhibition at 48 hr after infection with rHRSV-Ch/IgM mixes as compared to free rHRSV-Ch. Results are expressed as means ± SD of triplicates and are representative of two experiments.
(D–F) B cell subsets were subjected to Ig repertoire analysis of the IgM heavy chain (n = 5–6 donors).
(D) The mean CDR3 lengths was analyzed for the different IGHV and a profile comparison is shown for IGHV3.
(E) The frequencies of different IGHJ usage among V3a (V3-15, 49, 72, 73) subfamily were determined.
(F) For IGH3 subfamily 3a family, 165 clones from MN and 185 clones from nBreg cells were sequenced to determine IGHJ/IGHD junctions. Among IGHJ4, the number for the different D gene usage is indicated (n = 74 for MN, and n = 84 for nBreg cells).
Our results indicated that nBreg cell Igs recognized RSV in the absence of any previous exposure to the virus. One possible explanation of such a property may rely on polyreactivity of the nBreg cell IgM that may have developed in utero upon exposure to self-antigens. Compared to neonatal MN cell IgMs, nBreg cell IgMs displayed canonical features of polyreactive B cells, including self-antigen recognition (Figure S6A). To explore the molecular basis of this phenomenon, we compared the different neonatal B cell subsets at the level of their Ig repertoire. There was no bias in the usage of specific Ig heavy chain V genes (IGHV) in the IgMs derived from nBreg cells. However, nBreg cell IgMs exhibited a shorter complementarity determining region 3 (CDR3) for most of the IGHV genes, representing more than 90% of the BCR repertoire (Figures 5D, S6C, and S6D). A short CDR3 has been reported for MZB cells (Weller et al., 2008) that do not express CD23 and CD5 (Weill et al., 2009). CD27+ “B1-like cells” were described to have a 14-bp CDR3 (Griffin et al., 2011), whereas the CDR3 of nBreg cells was a 12.9 ± 0.2 bp in length (Figure 5D). We identified preferential usage of the IGHJ4 segment associated with IGHD6 in nBreg cells by analyzing the IGHV3 gene PCR products (Figures 5E, 5F, and S6E). Both CD27+ and CD27− nBreg cells showed similar repertoire characteristics and functional properties, such as equivalent susceptibility to RSV infection and similar concentrations of IL-10 production upon stimulation with RSV (Figures S6F and S6G). Therefore, the Ig repertoire analysis of nBreg cells showed that this population constituted a B cell subset with unique characteristics, presumably resulting from specific selection and/or maintenance processes. In summary, the repertoire traits, together with the viral particle recognition by nBreg cell IgMs, provide the molecular basis for the activation of the BCR pathway after exposure to RSV.
The RSV G Glycoprotein Interaction with CX3CR1 Is Critical to Infect nBreg Cells
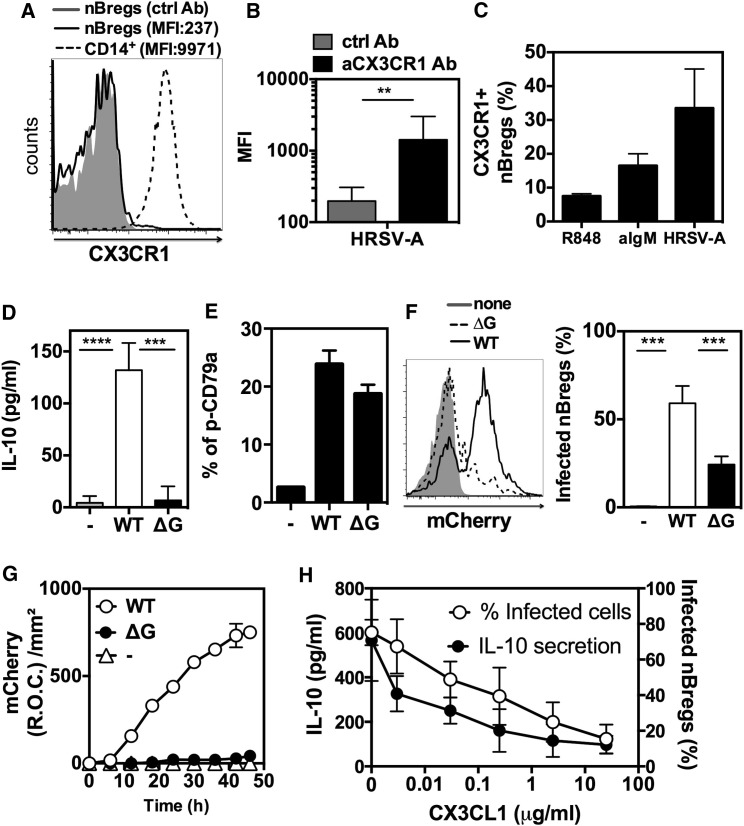
Because ultraviolet (UV) inactivation of RSV impaired IL-10 production, the polyreactive nature of nBreg IgMs was not sufficient to explain the triggering of nBreg cell activity by RSV. The G glycoprotein harbors a CX3C chemokine motif capable of chemokine mimicry when interacting with chemokine receptor CX3CR1 (Tripp et al., 2001). This interaction is reported as an important mechanism for RSV binding and infection in human lung epithelial cells (Jeong et al., 2015, Chirkova et al., 2015). Because nBreg cell exposure to RSV activated chemokine receptor pathways (Figure 4C), we analyzed CX3CR1 expression on cord blood B cells. CX3CR1 was expressed by monocytes, but not by B cells, including nBreg cells (Figure 6 A). However, after 48 hr of RSV exposure, CX3CR1 was induced on nBreg cells (Figure 6B), an effect that was mimicked by BCR activation but not by TLR activation (Figure 6C). Using rHRSV-Ch, we observed that viral infection of nBreg cells was associated with the highest frequencies of nBreg cell expression of cell surface CX3CR1 relative to other stimuli. We found that an RSV ΔG mutant poorly infected nBreg cells (Figures 6F and 6G) and was unable to induce IL-10 production (Figure 6D), despite its ability to trigger BCR Igα phosphorylation (Figure 6E). This indicates that G protein binding to CX3CR1 was necessary to induce IL-10 production. In the presence of the CX3CR1 ligand CX3CL1, RSV infection was strongly decreased, concomitant with the inhibition of the IL-10 secretion (Figure 6H), indicating that CX3CL1 is blocking the interaction of the RSV G protein with CX3CR1, which is therefore essential for the induction of nBreg cell activity. Our results show that RSV, upon binding of surface Ig on nBreg cells, induces the upregulation of CX3CR1 which, in turn, interacts with the G glycoprotein and favors viral entry and replication in Breg cells. This two-step mechanism can explain the permissiveness of nBreg cells to RSV infection and the specific induction of IL-10 that does not occur with the other viruses tested.
Figure 6.
HRSV Infects nBreg Cells via CX3CR1-G Protein Interaction
(A–C) FACS analysis of CX3CR1 expression in nBreg cells.
(A) Representative FACS plot of freshly isolated nBreg cells (MFI: 193 for Ab ctrl and 237 for CX3CR1 Ab) as compared to CD14+ monocytes (MFI: 9971).
(B) nBreg cells were infected with HRSV-A (MOI = 2.5) for 48 hr. MFI are shown for CX3CR1 Ab as compared to isotype control. Results are expressed as means ± SD of duplicates from three donors.
(C) nBreg cells were stimulated with R848, agM, or HRSV-A (MOI = 2.5) for 48 hr and their CX3CR1 expression was assessed by FACS. Results are expressed as means ± SD of duplicates from two independent experiments. nBreg cells were infected for 48 hr with WT or ΔG strains of rHRSV-Ch (MOI = 2.5).
(D) IL-10 production was measured by ELISA. Results are expressed as means ± SD of four independent experiments.
(E) Phopshorylation of CD79a was assessed intracellularly by FACS in nBreg cells after 30 min of exposure to ΔG RSV or WT RSV. Results are expressed as means ± SD of triplicates and are representative of three experiments.
(F) Representative FACS plot (left) of nBreg cell infection with ΔG RSV or WT RSV, as compared to non-infected cells (none). Histogram plot shows the percentage of infected nBreg cells, as measured by FACS as percent of mCherry+ cells (right), and is representative of three independent experiments.
(G) Viral replication measured through the detection of the mCherry fluorescence for 48 hr with ΔG as compared to WT counterpart. Results are expressed as means ± SD of triplicates and are representative of three independent experiments.
(H) nBreg cells were cultured on CX3CL1-coated plates and infected with rHRSV-Ch (MOI = 2.5). The percentage of infected nBreg cells and IL-10 secretion were measured at 48 hr. Results are expressed as means ± SD and represent three independent experiments. Unpaired t test was used for comparison. Results are expressed as the means ± SD.
∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
RSV Infects Infant nBreg Cells and nBreg Cells Are Predictive of Disease Severity
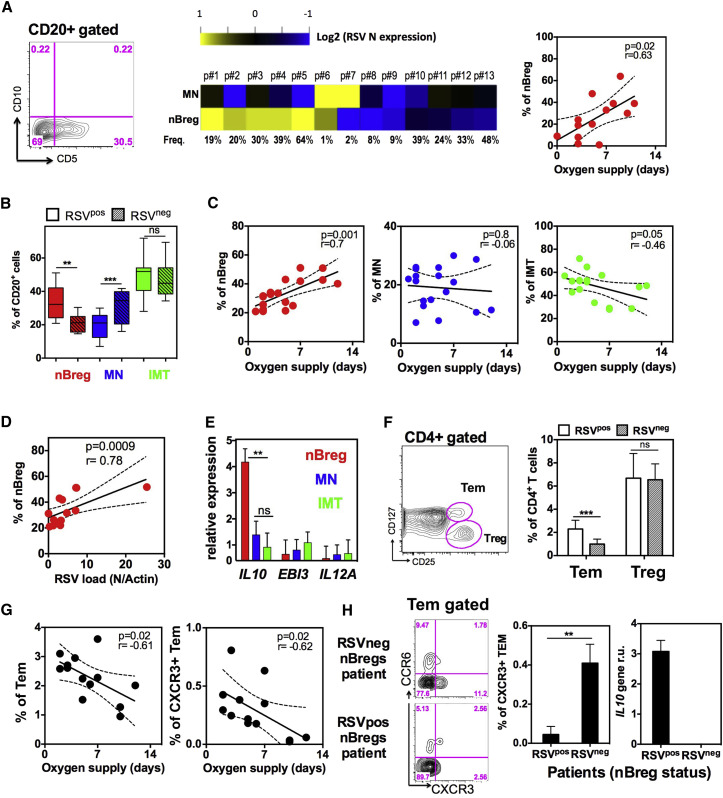
Infants under 3 months of age who develop acute severe bronchiolitis because of RSV infection may require ventilator support and are at a much higher risk to develop recurrent wheezing up through their teenage years (Stein, 2009). This is often thought to be associated with Th2 cell responses. However, post-mortem analysis in fatal cases reveals heavy pulmonary infiltration of B cells, but not T cells, in the lung upon RSV infection (Reed et al., 2009). RSV remains in the respiratory tract and does not spread to the blood. IL-10 can be detected in the nasopharyngeal aspirates (NPA) of RSV-infected children (Bont et al., 2001) and is associated with post-bronchiolitis wheeze (Schuurhof et al., 2011). We therefore looked for the presence of nBreg cells in the NPA of hospitalized RSV-infected children who required respiratory assistance. In 6 out of the 13 patients, RSV-infected nBreg cells were found in NPA swabs, whereas 2 patients showed RSV-infected MN B cells, highlighting the preferential infection of nBreg cells in vivo. The frequency of nBreg cells correlated with the severity of the disease, as assessed by the duration of oxygen support and hospitalization in the ICU (Figures 7 A and S7). We also found a higher frequency of nBreg cells in the blood of RSV-infected patients suffering from acute bronchiolitis compared to non-infected children, and a positive correlation between the percentage of nBreg cells with the disease severity and the viral load, but not with the age of the patient or the pregnancy term (Figures 7B–7D and S7). In contrast to MN and IMT B cells, nBreg cells purified from the blood of RSV-positive patients expressed IL10 mRNA upon RSV exposure, but not IL35, IL12A, and EBI3 subunits (Figure 7E), indicating the capacity of nBreg cell activity to be activated after their recruitment at the site of infection.
Figure 7.
nBreg Cells Are Infected by RSV in Patients and Predict the Severity of Acute Bronchiolitis
(A) CD20+ B cells in nasopharyngeal aspirates (NPA) collected from RSV-positive patients (n = 13) at the first day of hospital admission were analyzed and purified by FACS as nBreg cells and MN B cell subsets based on CD5 and CD10 expression. Heatmap shows sorted cell subsets analyzed for RSV nucleoprotein gene (RSV N) expression by qRT-PCR. Correlation plots (right) of nBreg cell frequency in the NPA with the duration of oxygen supply.
(B) Frequency of B cells subsets in the blood of RSV-positive (RSVpos; n = 18) and -negative (RSVneg; n = 10) patients.
(C) Correlation plot of B cell subset percentage in the blood of RSVpos patients (n = 18) with the duration of oxygen supply.
(D) Correlation plot of blood nBreg cell frequency with RSV load in the corresponding NPA of RSVpos patients (n = 13).
(E) nBreg, MN, and IMT B cells were sorted from the blood of RSVpos patients (n = 7) and stimulated or not with HRSV for 24 hr. IL10, EBI3, and IL12 gene expression was analyzed by qRT-PCR and normalized to housekeeping genes.
(F) CD4+ T cells were analyzed by FACS for Treg and Tem cells, as shown in the blood of RSV-positive and (RSVpos; n = 13) and -negative (RSVneg; n = 7) patients.
(G) Correlation plots of blood Tem and CXCR3+ Tem cell frequencies with the duration of oxygen supplementation (n = 13).
(H) Representative FACS analysis plot of blood CXCR3+ and CCR6+ Tem cells in patients whose nBreg cells were found infected or not in the NPA of (A). Histograms represent the frequency of CXCR3+ Tem cells in the blood (left histogram) and nBreg cells IL-10 gene expression in the NPA (right histogram) from patients corresponding to infected nBreg (RSVpos; n = 3) or non-infected nBreg (RSVneg; n = 3) cells.
Results are expressed as the means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. ns for non significant.
In the neonatal blood, we identified emerging CD4+ T cell effector memory (Tem) cells including CXCR3+ IFN-γ-producing Th1 cells (Zhang et al., 2014b). RSV-activated nBreg cells limited the development of neonatal Th1 cell responses in vitro (Figure 2). We found a higher frequency of Tem cells, but not Treg cells, in the blood of RSV-infected children compared to RSV un-infected children. The percentage of Tem cells and CXCR3+ Tem cells negatively correlated with bronchiolitis severity, but not with the age of the patient or the pregnancy term (Figures 7F, 7G, and S7). Among the RSV-infected patients, the frequency of CXCR3+ Tem cells was significantly lower when nBreg cells were infected with RSV, as measured in the NPA (Figure 7H). These findings indicate that RSV infection of nBreg cells may inhibit CXCR3+ Tem cell responses in patients to reduce viral clearance and to drive more severe lung disease.
Discussion
Using mass cytometry we identified nBreg cells, which are an age-dependent factor associated with the severity of RSV-induced acute bronchiolitis. RSV infects nBreg cells, through IgM recognition and induced CX3CR1 allowing viral interaction with the G glycoprotein. B cell interactions with pathogens without antigen specificity usually leads to B cell death and an impaired antibody responses (Nothelfer et al., 2015). The possibility of virus-mediated B cell subversion in an antigen-specific manner has been proposed recently in the context of influenza-specific B cells (Dougan et al., 2013). Salmonella spp. induce and/or activate Breg cells in a TLR-dependent manner (Neves et al., 2010). In inflammatory situations, the CD40- and TLR-mediated pathways are central in Breg cell activation (Mauri and Bosma, 2012). nBreg cells developed in utero cells and waned with age, likely reflecting a fetal-specific wave of B cell ontogeny and selection. The polyreactive nature of the Ig repertoire of nBreg cells suggests that other pathogens may target nBreg cells. However, the BCR was not sufficient to activate nBreg cells; a second receptor was required in the context of RSV. The mechanism we propose involves the combined role of RSV G and F glycoproteins in hijacking the newborn immune system to impair viral clearance. The pre-fusion form of the F protein appears to be the critical target for virus neutralization (McLellan et al., 2011, McLellan et al., 2013). Of note, TLR4 was reported to interact with the F protein of RSV in a CD14-dependent manner (Kurt-Jones et al., 2000). However, human B cells do not express TLR4. The F fusion protein-BCR interaction that initiates nBreg cell activation enables G-CX3CR1-mediated infection, and the nBreg cell IgMs outcompeted and decreased the initial viral interaction and further infection. The IL-10 production by nBreg cells was mainly associated with the infection of the cell, although additional mechanisms such as TLR activation might contribute to the amplification of the antiinflammatory response.
In primary RSV infection in humans and mice, a type I immune response including NK cells, Th1 cells, and cytotoxic T lymphocytes (CTLs), which act as important sources of IFN-γ, is essential for viral clearance (Openshaw and Chiu, 2013). An unbalanced and dysregulated T cell response to HRSV limits viral clearance and is reported to cause immunopathology in the respiratory tract. Primary HRSV infection in newborn mice during the critical neonatal window led to the generation of a type II response, an enhanced airway inflammation, lymphocyte infiltration, and eosinophilia upon re-infection at adulthood, whereas delayed age priming led to enhanced IFN-γ production and less severe symptoms during reinfection (Culley et al., 2002). Th2 cell pathology occurs upon secondary RSV infection and is poorly associated with the primary infection. We found very few T cells in the NPA or effector memory Th1 cells in the blood and no detectable Th2 cell signature in the patients. Therefore, our in vivo and in vitro data support the role of RSV-activated nBreg cells in the control of IFN-γ Th1 cells and associated viral clearance.
It is unclear yet whether infected nBreg cells can reach the lymph nodes (LNs) and whether they directly influence Th1 cell priming. Human nBreg cells are related to neonatal B1a cells in terms of their regulatory properties (Sun et al., 2005, Zhang et al., 2007). Mouse B1a cells have been recently shown to be trans-infected by blood-borne retroviruses via LN macrophages (Sewald et al., 2015). In such a scenario, RSV might reach nBreg cells in the lung draining LN via myeloid cells. Such innate B cells produce natural antibodies with polyreactive properties. The hallmark of the human innate B cells is the expression of CD27, a marker corresponding to memory cells. Carsetti and colleagues defined “IgM memory cells” in the blood as cells that are IgM+IgD+CD22+CD27+ (Kruetzmann et al., 2003), splenic MZBs are defined as IgMhiIgDloCD23−CD21+CD1c+CD27+ (Weller et al., 2008), and B1 cell candidates are IgM+IgD+CD43+CD27+ B cells (Griffin et al., 2011). This IgM memory/MZB compartment develops after birth, possibly in response to the gut microbiota. The human B1 cells would represent a minority in neonatal blood but could account for 40% of all CD27+ memory B cells. This latter point raised some controversies about the phenotype of these cells, that would include T cell or monocyte contaminants (Descatoire et al., 2011, Perez-Andres et al., 2011). A small fraction of human B1 cell express CD5, which is the hallmark of nBreg cells, and do not have any repertoire bias in contrast to nBreg cells. The cyTOF approach we had clearly eliminated the possibility of other cell lineage contamination to CD5 and CD27 expression. The “human B1 cells” have been proposed to be pre-plasmablasts because they produce IgM, IgG, and IgA, and we showed nBreg cells to be free of any IgG- or IgA-positive cells. In addition, nBreg cells quickly wane with age whereas human B1 cell population would develop. This age depedency of nBreg cells might explain their contribution to RSV disease, which becomes asymptomatic later in life. We also showed that canonical adult memory B cells can be slightly infected by the RSV. Therefore it remains to be determined whether they correspond to a small fraction of RSV-specific B cells and whether their infection could play a role in the susecptibilty to the infection later in life, in the elderly poplulation.
In addition to lung epithelial cells, nBreg cells represent a newly described target cell for RSV and a biomarker for the severity of acute bronchiolitis. A recent study that defined biomarkers in bronchiolitis using whole blood RNA profiling highlighted an overexpression of neutrophil and interferon genes as well as suppression of B and T cell genes in children of less than 6 months (Mejias et al., 2013). The increased number of nBreg cells observed in the blood emphasizes how carefully B cell signatures need to be interpreted. Therefore, the appropriate complex signal deconvolution of whole blood signatures needs to take into account age-specific immune characteristics. If confirmed in the NPA, the “RSV-nBreg cell” signature that we defined may serve as a molecular biomarker of disease severity. Future work will determine whether the high frequency of nBreg cells is a consequence or a cause of the disease. In future investigations, large cohorts are needed to determine whether nBreg cells are a host risk factor that might predispose individuals to RSV-induced bronchiolitis. nBreg cell activity may constitute an early-life host response that favors microbial pathogenesis and may represent a target for the treatment of low respiratory tract viral infections and their pathological consequences later in life.
Experimental Procedures
Blood
Buffy coats were obtained from adult donors by the Etablissements Français du Sang (France). Heparinized cord blood samples from healthy neonates collected by the Thérapie Cellulaire of Hopital Saint-Louis (France). Written consent was obtained from the mothers. This study was conducted with the approval of the Ethics Committee of Institut Pasteur in agreement with the principles of the Declaration of Helsinki. For FACS analysis of infant B cells, we used blood samples from children from birth to 5 years of age that were admitted to the Hopital Erasme laboratory (Brussels, Belgium) for routine analysis of the common hematological parameters between March 2012 and June 2012. The final protocol of this study was approved by the Ethics Committee of Erasme Hospital, allowing us to test residual blood samples.
Cohort of Patients with Acute Bronchiolitis
46 infants admitted to the NICU of the Bicêtre Hospital for severe bronchiolitis were recruited. Infants being prophylactically treated with Palivizumab were excluded. After signed informed consent from the legal representatives of the children, 1 mL of blood in EDTA tubes (Sarstedt) and nasopharyngeal aspirates were obtained and stored at 4°C and at −80°C. Samples were processed within 12 hr. Diagnosis of viral bronchiolitis was confirmed using immunochromatography for RSV (Alere BinaxNow RSV, Alere) and respiratory virus Q-PCR techniques (either Simplexa Flu A/B & RSV Direct, Focus Diagnostics; or Argene Respiratory Multi Well System MWS r-gene range, Biomerieux). The study was approved by the local IRB (Pasteur Institute CoRC, no. 2013-14) and the French Ministry of Research (no. 13.644). Details of the demographic and diagnostic data are in the Supplemental Experimental Procedures.
Culture Medium and Reagents
Complete medium consisted of RPMI-1640 supplemented with 10% fetal calf serum (ICN Biomedicals), 5 × 10−5 M of 2-ME (Sigma), and antibiotics (GIBCO BRL). R848 and CpG1826 were purchased from InvivoGen. Human influenza virus A/PR/8/34 (IAV) was purchased from Charles River. Measles virus (MV, strain Schwarz) were amplified and titrated using Vero cells. Human coronavirus (HCoV-229E), herpes simplex virus 1 (HSV, strain KOS), and human immunodeficiency virus (HIV) were used. Human T-lymphotropic virus (HTLV-1) was produced with Mt2 cell supernatants (kindly provided by M.A. Thoulouze). Epstein-Barr virus (EBV) was generated using B95.8 cell line. Recombinant human BAFF, IL-2, and IL-12 were purchased from Peprotech. CX3CL1 was from R&D Systems.
HRSV Strains and Mutants
Human respiratory syncytial virus A (HRSV-A Long, kindly provided by F. Freymuth) was amplified and titrated on HEp-2 cells. Recombinant human RSV (rHRSV-A) and recombinant Human RSV-Cherry (rHRSV-Ch) were previously described (Rameix-Welti et al., 2014). The methods to generate the rHRSV-ΔG-Cherry and rHRSV-ΔSH are described in the Supplemental Experimental Procedures.
RSV-Specific ELISA
Maxisorp plates (NUNC) were coated with HRSV-A, or ΔSH and ΔG mutants or their WT counterparts described above at 4°C overnight. After blocking of the plates with 1% BSA in PBS at 37°C for 1 hr, IgMs from CpG-stimulated cord blood B cell subsets were added to the plates for 1 hr at room temperature. After washing, horseradish peroxidase (HRP) conjugated to goat anti-human IgM (Southern Biotech) with a TMB substrate was used. Optical densities (OD) were measured at 450 nm.
Mass Cytometry
The antibodies were labeled 100 μg at a time according to the manufacturer’s instruction with heavy metal-preloaded maleimide-coupled MAXPAR chelating polymers. Purified antibodies were purchased from Miltenyi. CBMCs were stained with these reagents, DNA content stained by an iridium-191/193 interchelator was used to identify individual cells, and by exclusion of a live-dead viability stain. Data were acquired using a CyTOF2 instrument (Fluidigm) and analyzed using vISNE algorithm on Cytobank (Fluidigm). Antibody clones used are detailed in the Supplemental Experimental Procedures.
Cell Purification and Culture
CBMCs or PBMCs from child patients or adults were isolated using Lymphoprep (Axis-Shield). B cells were positively enriched from CBMCs or PBMCs by using anti-CD19 magnetic beads with AutoMACS (Miltenyi Biotec). To recover the blood B cell subsets, the cells were sorted based on surface CD10 and CD5 markers to obtain CD10+CD5+ (IMT), CD10−CD5hi (nBreg cells), and CD10−CD5− (MN) B cell subsets using a FACS Aria 3 (BD). Cells sorted by AutoMACS and FACS were routinely >95% and 97%–99% pure, respectively. B cells were stimulated with indicated compounds or viruses for 48 hr (MOI = 2.5). For co-culture assay, mycoplasma-free HEp-2 cells were incubated for 2 hr at 37°C with rHRSV-Ch and supernatants were then discarded. 24 hr later, cells were washed twice with PBS (1×), and purified nBreg cells, MN, or IMT B cells were added to the infected HEp2 cells for 48 hr. Alternatively, cells were cultured with human BAFF (200 ng/mL), CpGB 1826 (5 μg/mL), and IL-2 (10 ng/mL). The supernatants were measured for IL-10 by ELISA (eBioscience). To monitor infection of cells, mCherry fluorescence was detected either by LSR Fortessa FACS (BD) or IncucyteZoom (Essen Bioscience) for live cell imaging. Antibodies used for FACS are described in the Supplemental Experimental Procedures.
Nasopharyngeal Aspirate Cells Isolation
Nasopharyngeal aspirates (NPA) were maintained on ice and processed within 24 hr. The samples were repeatedly washed with PBS with 5% FCS and centrifuged until no visible mucus clumps remained in the solution. The samples were then filtered using a Falcon 100-μm filter (Miltenyi Biotech). For nasal wash cell staining and isolation, filtered NPA cells were incubated with antibodies for 20 min.
B Cell Repertoire Analysis
We characterized the IgM repertoire at the molecular level in various B cell subsets from cord blood; the details can be found in the Supplemental Experimental Procedures.
Microarray Analysis
5 × 103 negatively enriched cord blood B cells were FACS sorted as nBreg cells (CD19+CD5+CD10−CD3−) and stimulated for 6 hr with 10 μg/mL F(ab’)2 goat anti-human IgM, R848 (1 μg/mL), HRSV-A (MOI = 5), or IAV (MOI = 4,000 HA/mL). The gene expression profiles were measured by Miltenyi Biotec using an Agilent DNA chip. We used the Agilent 60-mer Whole Human Genome Oligo Microarray containing approximately 44 K genes and gene candidates. Output data files were further analyzed using the Rosetta Resolver gene expression data analysis system. Microarrays Agilent files were processed, background corrected, and normalized using the quantile method with R and package Limma. Genes were averaged using ProbeID, and GeneName and transcripts were filtered using the RefSeq mRNA database. Principal component analyses on most differentially expressed genes, heatmaps, and hierarchical clustering were performed using Qlucore Omics Explorer 3.1.
Gene Expression
103 blood B cells stimulated or not with HRSV for 24 hr were then analyzed using appropriate primers for expression of EBI3 (5′-CCCTTCCCAGAGATCTTCTCAC-3′; 5′-CAGCCCTGAGGATGAAGGAC-3′), IL10 (5′-CCGTGGAGCAGGTGAAGAA-3′; 5′-GTCAAACTCACTCATGGCTTTGTA-3′), and IL12A (5′-CACAGTGGAGGCCTGTTTA-3′; 5′-TCTGGAATTTAGGCAACTCTCA-3′ RNA) on a Biomark System (Fluidigm).
Intracellular Staining Assay
CD19+ B cell fraction was stained with surface markers (CD20, CD10, CD5, and CD3) and live/dead-Blue to identify viable B cell subsets. Cells were stimulated for 30 min with 10 μg/mL F(ab’)2 goat anti-human IgM, R848 (1 μg/mL), HRSV-A (MOI = 1/10), or IAV (MOI = 4,000 HA/mL). Cells were then directly fixed and permeabilized using BD Cytofix/Cytoperm solution by following the manufacturer’s instructions (eBioscience) and then subjected to intracellular phospho-CD79a detection.
T Cell Differentiation In Vitro
5 × 104 purified cord blood naive CD4+ T cells activated with anti-CD3/CD28 beads in Th0 (no cytokine added) or Th1 (with 10 ng/mL of IL-12) cell conditions were co-cultured with 5 × 104 syngeneic B activated with HRSV. Alternatively, purified cord blood pDCs were stimulated with HRSV-A in the presence of nBreg cells for 48 hr. Activated pDCs were sorted again by gating on CD123hiCD20− cells on FACSAria II. 104 activated pDCs were co-cultured with 5 × 104 purified allogeneic cord blood naive CD4+ T cells. Five to six days later, differentiated T cells were restimulated with 50 ng/mL PMA, 1 μg/mL ionomycin, and GolgiPlug (BD) to detect intracellular cytokines (IL-2, IFN-γ, IL-13, IL-17, IL-22, and TNF-α), Alternatively, secreted IFN-γ, IL-13, and IL-17 were detected by ELISA with a specific kit (eBiosciences).
ELISPOT
nBreg cells were stimulated or not with HRSV-A for 24 hr. IL-10-secreting nBreg cells were enriched using IL-10 cytokine secretion assay according to the manufacturer’s protocol (Milteny Biotec). Enriched cells were then FACS sorted for IL-10-positive cells and used for human fluoroSpot IgM according to the manufacturer’s protocol (Mabtech). Alternatively, IgM-secreting cells were also analyzed with an HRP-based ELISPOT assay (Mabtech).
Statistical Analysis
Unpaired t tests were done in comparison of two groups (data are presented as the mean value ± SD). Paired t tests were also used to take into account donor to donor variation. ANOVA tests were used when comparing three groups or more. Spearman tests were used for correlations. p values < 0.05 were considered statistically significant.
Author Contributions
D.Z. designed and performed research. S.L. performed bioinformatics analysis of the transcriptomics data. A.L. completed the repertoire study. A.M., L.S., N.C., E.D., J.F., B.B., M.R.-L., J.M., B.L., and V.L. performed experiments and analyzed data. A.L., P.-O.V., M.-A.R.-W., P.-L.H., D.D., J.-F.E., O.S., F.P., F.M., H.M., and X.Z. provided reagents and contributed to experimental design. X.Z., P.T., and R.L.-M. designed and supervised the project. D.Z., P.-O.V., X.Z., S.R., J.-F.E., H.M., and P.T. critically revised the manuscript. R.L.-M. wrote the manuscript.
Acknowledgments
We are very grateful to T. Domet (Therapie Cellulaire, Hopital Saint-Louis) for the cord blood sample collection and M. Lucas-Hourani for some viral preparation. We thank F. Huetz for helpful discussion about B lymphocytes. This work was supported by an ANR grant (ANR 13-BSV3-0016) and by the Fondation pour la Recherche Médicale (grant no. DEQ20120323719). This study also received funding from the French Government’s Investissement d’Avenir program, Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” (grant no. ANR-10-LABX-62-IBEID). X.Z. and S.L. were supported by ANR and by the European Commission FP7 ADITEC program (HEALTH-F4-2011-280873). X.Z. was also partially supported by the Major Basic Research Project of Shanghai Science and Technology Commission (no. 13JC1405600); National Natural Science Foundation of China (31270961, 31470879); Interdisciplinary Innovation Team and External Cooperation Program (no. GJHZ201312); and Chinese Academy of Sciences. D.Z. was supported by DIM Malinf of Region IdF. Work in the O.S. lab is also supported by ANRS, Sidaction and Fondation Areva. We acknowledge the Center for Human Immunology at Institut Pasteur for support in conducting these studies.
Published: February 21, 2017
Footnotes
Supplemental Information includes seven figures and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2017.01.010.
Accession Numbers
Raw expression files and details have been deposited in GEO under accession number GSE78847.
Supplemental Information
References
- Amir A.D., Davis K.L., Tadmor M.D., Simonds E.F., Levine J.H., Bendall S.C., Shenfeld D.K., Krishnaswamy S., Nolan G.P., Pe’er D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair P.A., Noreña L.Y., Flores-Borja F., Rawlings D.J., Isenberg D.A., Ehrenstein M.R., Mauri C. CD19(+)CD24(hi)CD38(hi) B cells exhibit regulatory capacity in healthy individuals but are functionally impaired in systemic lupus erythematosus patients. Immunity. 2010;32:129–140. doi: 10.1016/j.immuni.2009.11.009. [DOI] [PubMed] [Google Scholar]
- Bont L., Heijnen C.J., Kavelaars A., van Aalderen W.M., Brus F., Draaisma J.M., Pekelharing-Berghuis M., van Diemen-Steenvoorde R.A., Kimpen J.L. Local interferon-gamma levels during respiratory syncytial virus lower respiratory tract infection are associated with disease severity. J. Infect. Dis. 2001;184:355–358. doi: 10.1086/322035. [DOI] [PubMed] [Google Scholar]
- Castro M., Schweiger T., Yin-Declue H., Ramkumar T.P., Christie C., Zheng J., Cohen R., Schechtman K.B., Strunk R., Bacharier L.B. Cytokine response after severe respiratory syncytial virus bronchiolitis in early life. J. Allergy Clin. Immunol. 2008;122:726–733.e3. doi: 10.1016/j.jaci.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirkova T., Lin S., Oomens A.G.P., Gaston K.A., Boyoglu-Barnum S., Meng J., Stobart C.C., Cotton C.U., Hartert T.V., Moore M.L. CX3CR1 is an important surface molecule for respiratory syncytial virus infection in human airway epithelial cells. J. Gen. Virol. 2015;96:2543–2556. doi: 10.1099/vir.0.000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley F.J., Pollott J., Openshaw P.J. Age at first viral infection determines the pattern of T cell-mediated disease during reinfection in adulthood. J. Exp. Med. 2002;196:1381–1386. doi: 10.1084/jem.20020943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descatoire M., Weill J.C., Reynaud C.A., Weller S. A human equivalent of mouse B-1 cells? J. Exp. Med. 2011;208:2563–2564. doi: 10.1084/jem.20112232. author reply 2566–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan S.K., Ashour J., Karssemeijer R.A., Popp M.W., Avalos A.M., Barisa M., Altenburg A.F., Ingram J.R., Cragnolini J.J., Guo C. Antigen-specific B-cell receptor sensitizes B cells to infection by influenza virus. Nature. 2013;503:406–409. doi: 10.1038/nature12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillatreau S., Sweenie C.H., McGeachy M.J., Gray D., Anderton S.M. B cells regulate autoimmunity by provision of IL-10. Nat. Immunol. 2002;3:944–950. doi: 10.1038/ni833. [DOI] [PubMed] [Google Scholar]
- Griffin D.O., Holodick N.E., Rothstein T.L. Human B1 cells in umbilical cord and adult peripheral blood express the novel phenotype CD20+ CD27+ CD43+ CD70- J. Exp. Med. 2011;208:67–80. doi: 10.1084/jem.20101499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong K.-I., Piepenhagen P.A., Kishko M., DiNapoli J.M., Groppo R.P., Zhang L., Almond J., Kleanthous H., Delagrave S., Parrington M. CX3CR1 is expressed in differentiated human ciliated airway cells and co-localizes with respiratory syncytial virus on cilia in a G protein-dependent manner. PLoS ONE. 2015;10:e0130517. doi: 10.1371/journal.pone.0130517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruetzmann S., Rosado M.M., Weber H., Germing U., Tournilhac O., Peter H.H., Berner R., Peters A., Boehm T., Plebani A. Human immunoglobulin M memory B cells controlling Streptococcus pneumoniae infections are generated in the spleen. J. Exp. Med. 2003;197:939–945. doi: 10.1084/jem.20022020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt-Jones E.A., Popova L., Kwinn L., Haynes L.M., Jones L.P., Tripp R.A., Walsh E.E., Freeman M.W., Golenbock D.T., Anderson L.J., Finberg R.W. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 2000;1:398–401. doi: 10.1038/80833. [DOI] [PubMed] [Google Scholar]
- Lemoine S., Morva A., Youinou P., Jamin C. Human T cells induce their own regulation through activation of B cells. J. Autoimmun. 2011;36:228–238. doi: 10.1016/j.jaut.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Matsushita T., Yanaba K., Bouaziz J.-D., Fujimoto M., Tedder T.F. Regulatory B cells inhibit EAE initiation in mice while other B cells promote disease progression. J. Clin. Invest. 2008;118:3420–3430. doi: 10.1172/JCI36030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauri C., Bosma A. Immune regulatory function of B cells. Annu. Rev. Immunol. 2012;30:221–241. doi: 10.1146/annurev-immunol-020711-074934. [DOI] [PubMed] [Google Scholar]
- Mauri C., Gray D., Mushtaq N., Londei M. Prevention of arthritis by interleukin 10-producing B cells. J. Exp. Med. 2003;197:489–501. doi: 10.1084/jem.20021293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan J.S., Yang Y., Graham B.S., Kwong P.D. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J. Virol. 2011;85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan J.S., Chen M., Leung S., Graepel K.W., Du X., Yang Y., Zhou T., Baxa U., Yasuda E., Beaumont T. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejias A., Dimo B., Suarez N.M., Garcia C., Suarez-Arrabal M.C., Jartti T., Blankenship D., Jordan-Villegas A., Ardura M.I., Xu Z. Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLoS Med. 2013;10:e1001549. doi: 10.1371/journal.pmed.1001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon M., Blair P.A., Isenberg D.A., Mauri C. A regulatory feedback between plasmacytoid dendritic cells and regulatory B cells is aberrant in systemic lupus erythematosus. Immunity. 2016;44:683–697. doi: 10.1016/j.immuni.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves P., Lampropoulou V., Calderon-Gomez E., Roch T., Stervbo U., Shen P., Kühl A.A., Loddenkemper C., Haury M., Nedospasov S.A. Signaling via the MyD88 adaptor protein in B cells suppresses protective immunity during Salmonella typhimurium infection. Immunity. 2010;33:777–790. doi: 10.1016/j.immuni.2010.10.016. [DOI] [PubMed] [Google Scholar]
- Nothelfer K., Sansonetti P.J., Phalipon A. Pathogen manipulation of B cells: the best defence is a good offence. Nat. Rev. Microbiol. 2015;13:173–184. doi: 10.1038/nrmicro3415. [DOI] [PubMed] [Google Scholar]
- O’Garra A., Chang R., Go N., Hastings R., Haughton G., Howard M. Ly-1 B (B-1) cells are the main source of B cell-derived interleukin 10. Eur. J. Immunol. 1992;22:711–717. doi: 10.1002/eji.1830220314. [DOI] [PubMed] [Google Scholar]
- Openshaw P.J., Chiu C. Protective and dysregulated T cell immunity in RSV infection. Curr. Opin. Virol. 2013;3:468–474. doi: 10.1016/j.coviro.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Andres M., Grosserichter-Wagener C., Teodosio C., van Dongen J.J., Orfao A., van Zelm M.C. The nature of circulating CD27+CD43+ B cells. J. Exp. Med. 2011;208:2565–2566. doi: 10.1084/jem.20112203. author reply 2566–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PrabhuDas M., Adkins B., Gans H., King C., Levy O., Ramilo O., Siegrist C.-A. Challenges in infant immunity: implications for responses to infection and vaccines. Nat. Immunol. 2011;12:189–194. doi: 10.1038/ni0311-189. [DOI] [PubMed] [Google Scholar]
- Rameix-Welti M.A., Le Goffic R., Hervé P.L., Sourimant J., Rémot A., Riffault S., Yu Q., Galloux M., Gault E., Eléouët J.F. Visualizing the replication of respiratory syncytial virus in cells and in living mice. Nat. Commun. 2014;5:5104. doi: 10.1038/ncomms6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.L., Welliver T.P., Sims G.P., McKinney L., Velozo L., Avendano L., Hintz K., Luma J., Coyle A.J., Welliver R.C., Sr. Innate immune signals modulate antiviral and polyreactive antibody responses during severe respiratory syncytial virus infection. J. Infect. Dis. 2009;199:1128–1138. doi: 10.1086/597386. [DOI] [PubMed] [Google Scholar]
- Schuurhof A., Janssen R., de Groot H., Hodemaekers H.M., de Klerk A., Kimpen J.L., Bont L. Local interleukin-10 production during respiratory syncytial virus bronchiolitis is associated with post-bronchiolitis wheeze. Respir. Res. 2011;12:121. doi: 10.1186/1465-9921-12-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewald X., Ladinsky M.S., Uchil P.D., Beloor J., Pi R., Herrmann C., Motamedi N., Murooka T.T., Brehm M.A., Greiner D.L. Retroviruses use CD169-mediated trans-infection of permissive lymphocytes to establish infection. Science. 2015;350:563–567. doi: 10.1126/science.aab2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P., Roch T., Lampropoulou V., O’Connor R.A., Stervbo U., Hilgenberg E., Ries S., Dang V.D., Jaimes Y., Daridon C. IL-35-producing B cells are critical regulators of immunity during autoimmune and infectious diseases. Nature. 2014;507:366–370. doi: 10.1038/nature12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist C.A. Neonatal and early life vaccinology. Vaccine. 2001;19:3331–3346. doi: 10.1016/s0264-410x(01)00028-7. [DOI] [PubMed] [Google Scholar]
- Smyth R.L., Openshaw P.J. Bronchiolitis. Lancet. 2006;368:312–322. doi: 10.1016/S0140-6736(06)69077-6. [DOI] [PubMed] [Google Scholar]
- Stein R.T. Long-term airway morbidity following viral LRTI in early infancy: recurrent wheezing or asthma? Paediatr. Respir. Rev. 2009;10(Suppl 1):29–31. doi: 10.1016/S1526-0542(09)70013-2. [DOI] [PubMed] [Google Scholar]
- Sun C.M., Deriaud E., Leclerc C., Lo-Man R. Upon TLR9 signaling, CD5+ B cells control the IL-12-dependent Th1-priming capacity of neonatal DCs. Immunity. 2005;22:467–477. doi: 10.1016/j.immuni.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Tripp R.A., Jones L.P., Haynes L.M., Zheng H., Murphy P.M., Anderson L.J. CX3C chemokine mimicry by respiratory syncytial virus G glycoprotein. Nat. Immunol. 2001;2:732–738. doi: 10.1038/90675. [DOI] [PubMed] [Google Scholar]
- Weill J.-C., Weller S., Reynaud C.-A. Human marginal zone B cells. Annu. Rev. Immunol. 2009;27:267–285. doi: 10.1146/annurev.immunol.021908.132607. [DOI] [PubMed] [Google Scholar]
- Weller S., Mamani-Matsuda M., Picard C., Cordier C., Lecoeuche D., Gauthier F., Weill J.-C., Reynaud C.-A. Somatic diversification in the absence of antigen-driven responses is the hallmark of the IgM+ IgD+ CD27+ B cell repertoire in infants. J. Exp. Med. 2008;205:1331–1342. doi: 10.1084/jem.20071555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welliver T.P., Garofalo R.P., Hosakote Y., Hintz K.H., Avendano L., Sanchez K., Velozo L., Jafri H., Chavez-Bueno S., Ogra P.L. Severe human lower respiratory tract illness caused by respiratory syncytial virus and influenza virus is characterized by the absence of pulmonary cytotoxic lymphocyte responses. J. Infect. Dis. 2007;195:1126–1136. doi: 10.1086/512615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Deriaud E., Jiao X., Braun D., Leclerc C., Lo-Man R. Type I interferons protect neonates from acute inflammation through interleukin 10-producing B cells. J. Exp. Med. 2007;204:1107–1118. doi: 10.1084/jem.20062013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Casartelli N., Lemoine S., Mozeleski B., Azria E., Le Ray C., Schwartz O., Launay O., Leclerc C., Lo-Man R. Plasmacytoid dendritic cells engagement by influenza vaccine as a surrogate strategy for driving T-helper type 1 responses in human neonatal settings. J. Infect. Dis. 2014;210:424–434. doi: 10.1093/infdis/jiu103. [DOI] [PubMed] [Google Scholar]
- Zhang X., Mozeleski B., Lemoine S., Dériaud E., Lim A., Zhivaki D., Azria E., Le Ray C., Roguet G., Launay O. CD4 T cells with effector memory phenotype and function develop in the sterile environment of the fetus. Sci. Transl. Med. 2014;6:238ra72. doi: 10.1126/scitranslmed.3008748. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.