Abstract
To overcome the limitations of the state‐of‐the‐art influenza surveillance systems in Europe, we established in 2008 a European‐wide consortium aimed at introducing an innovative information and communication technology approach for a web‐based surveillance system across different European countries, called Influenzanet. The system, based on earlier efforts in The Netherlands and Portugal, works with the participation of the population in each country to collect real‐time information on the distribution of influenza‐like illness cases through web surveys administered to volunteers reporting their symptoms (or lack of symptoms) every week during the influenza season. Such a large European‐wide web‐based monitoring infrastructure is intended to rapidly identify public health emergencies, contribute to understanding global trends, inform data‐driven forecast models to assess the impact on the population, optimize the allocation of resources, and help in devising mitigation and containment measures. In this article, we describe the scientific and technological issues faced during the development and deployment of a flexible and readily deployable web tool capable of coping with the requirements of different countries for data collection, during either a public health emergency or an ordinary influenza season. Even though the system is based on previous successful experience, the implementation in each new country represented a separate scientific challenge. Only after more than 5 years of development are the existing platforms based on a plug‐and‐play tool that can be promptly deployed in any country wishing to be part of the Influenzanet network, now composed of The Netherlands, Belgium, Portugal, Italy, the UK, France, Sweden, Spain, Ireland, and Denmark.
Keywords: Europe, infectious disease, influenza‐like illness, Internet, participatory surveillance
Introduction
Infectious diseases remain a serious medical burden all around the world, with 15 million deaths per year estimated to be directly related to infectious diseases 1. The emergence of new diseases such as human immunodeficiency virus/AIDS and SARS, and the rise of the new influenza strains H1N1, H5N1, and H7N9, as well as other respiratory pathogens such as the novel coronavirus Middle East respiratory syndrome coronavirus, represent a few examples of the global problems facing public health and medical science researchers. Even though pathogens such as the SARS coronavirus and H5N1 had a limited impact in terms of mortality, their sudden appearance has shown how abruptly health emergencies on a global scale can arise.
In recent times, our ability to control epidemic outbreaks has been, to a high degree, facilitated not only by advances in modern science (new cures, new drugs, and cooperative infrastructures for disease control and surveillance), but also by the worldwide spread of new technologies such as computers and smartphones, which allow more than 2 billion persons worldwide to have access to the Internet. This represents an unprecedented opportunity to collect and communicate health‐related information in real time and with high geographical resolution, including individuals who do not have contact with the healthcare system. Real‐time surveillance data are crucial for rapidly identifying public health emergencies, optimizing the allocation of resources to respond to them, and devising mitigation and containment measures.
Although existing disease surveillance systems (predominantly general practitioner (GP)‐based but also involving laboratory‐based reporting, mandatory notifications, etc.) have a fundamental role in monitoring and understanding the spread of communicable diseases, they also have several important limitations. One of the major issues is that, for diseases such as influenza‐like illness (ILI), only an unknown proportion of all infected individuals see a doctor. In addition, consultations frequently occur with a considerable delay, taking place only when a complication has occurred or a doctor's certificate is required (for example, in Sweden such a certificate is not required until after 1 week of absence from work). Other issues concern: the time delay in data reporting and aggregation; the lack of information on the patterns of household transmission; the lack of uniform standards for clinical definitions, which may vary considerably between countries and even between reporters (European Influenza Surveillance Network). Furthermore, age‐stratified rates of physician consultation may vary widely with different healthcare and health insurance systems. Healthcare‐seeking behaviour can change unpredictably during an epidemic, making extrapolations of those statistics to the general population uncertain.
To overcome the above limitations in the existing infectious diseases surveillance systems, with a focus on ILI, we proposed an innovative information and communication technology approach based on Web2.0 tools. Starting from the pivotal and successful experiences with Internet‐based monitoring systems in The Netherlands and Portugal 2, 3, 4, the Influenzanet consortium undertook the challenge of deploying an innovative real‐time and interlinked surveillance system across multiple European countries.
Materials and Methods
The first successful example of using the web for public health purposes dates back to 2003, when, in The Netherlands and Belgium, a Dutch scientific communication project was initiated by the small company Science in Action. The purpose was to inform the general population about influenza by means of a self‐reporting Internet platform in Dutch (http://www.degrotegriepmeting.nl), where volunteers in The Netherlands and Belgium (the Flemish part) could answer socio‐demographic, medical and behavioural questions, and report their influenza‐related symptoms each week. User participation was achieved through targeted communication and recruitment. A positive side effect of this project was the collection of almost real‐time data about influenza, including from individuals who did not visit a doctor when ill. Knowledge of volunteers' postal codes allowed the real‐time collection of data at the postal code level. Thanks to a vigorous communication campaign using both online and offline media, the project attracted 20 000 users during this first season.
The possibility of collecting such detailed data about influenza cases from such a wide audience attracted the interest of the Gulbenkian Institute of Science Epidemiology group in Portugal, who implemented their own platform in Portuguese, and deployed the system at the beginning of the 2005 influenza season (http://www.gripenet.pt). In Portugal, during the first 2 years of the project, approximately 5000 volunteers were attracted 3. On the basis of this success, the computational epidemiology group of the ISI Foundation in Turin, Italy decided to deploy a similar platform during the winter of 2007, to carry out influenza surveillance with the aim of informing computational models for studying the spread of ILI on a large scale. Data collected by these four platforms have been evaluated 3, 4 (Paolotti et al., 3rd International ICST Conference on Electronic Healthcare for the 21st Century, 2010, Abstract no. 30), and the estimated seasonal influenza incidence curves have been found to be in good agreement with those from the former European Influenza Surveillance Scheme.
Since 2009, the four Internet‐based monitoring systems have formed the foundation of a European‐wide network of platforms that became the key instrument for providing real‐time disease incidence for the epidemic forecast infrastructure developed by the EU‐funded research project EPIWORK (http://www.epiwork.eu). The proposed network of web platforms, called Influenzanet (http://www.influenzanet.eu), saw, for the first time, the collaboration of epidemiologists, public health practitioners and modellers with the aim of collecting epidemiological data in real time through the contributions of Internet volunteers self‐selected from among the general population. The epidemiology teams devised reference standard questionnaires for different diseases to collect unified data across European countries (presented in the supporting information). ILI has been the focus in the early deployment of the system, but the final goal is to considerably enlarge the portfolio of diseases and health conditions monitored. In June 2009, the plans to export the platform to other European countries were accelerated by the unfolding of the H1N1 pandemic in the northern hemisphere. A web system very similar to the Italian platform was rapidly deployed in the UK (https://flusurvey.org.uk/) to capture the whole progress of the H1N1 pandemic, with the involvement of the mathematical modelling group at the London School of Hygiene and Tropical Medicine. This work allowed researchers to detect the differences in healthcare‐seeking behaviour between different population groups and changes over time, with significant implications for estimates of total case numbers and the case‐fatality rate 5, 6.
The extension of the web platform in each country represented a separate scientific challenge and research problem, as the countries have different population stratifications and geographical distributions. The Influenzanet prototypical web platform implementation had to take into account these differences and design specific mapping/representation solutions, as well as find the optimal granularity for the data acquisition and statistical sampling in the different countries. It also had to provide easy management of content, news, and info‐graphics, and provide a user‐friendly interface for the volunteers to access the surveys aimed at collecting epidemiological data.
The data collection revolves around two main surveys: a background survey and a symptoms survey. The background questionnaire is completed at registration, and contains questions about the user's postal code, gender, birth date, household, level of education, employment, chronic health conditions, smoking habits, etc. The symptoms questionnaire is a weekly survey with a list of symptoms that the users employ to describe their health status.
The platform is equipped with a ‘Reminder’, i.e. a weekly email sent to users to remind them to fill in the symptoms survey. These features were included in a plug‐and‐play implementation to lessen the burden of deploying the platform in new countries, and to let the local teams concentrate on the communication and dissemination needed to attract participants, instead of having to deal with technical problems.
The new Influenzanet system started to be deployed in October 2010. The localization, translation, testing and fine‐tuning activities took place between October 2010 and February 2011. This powerful and flexible new platform has been successfully adopted by all of the ‘old’ countries (The Netherlands, Belgium, Portugal, the UK, and Italy), and, thanks to its plug‐and‐play features, it was deployed, in December 2011, in Sweden by the Swedish Institute for Infectious Disease Control, and by the Institut National de la Santé et de la Recherche Médicale in France. In both cases, in <2 months, the platform saw the enrolment of several thousands of participants from all over the country. The system has since been successfully deployed in Spain at the Instituto de Biocomputacion y Física de Sistemas Complejos, at the University of Saragoza, in September 2012, in Denmark at the Statens Serum Institut in Copenhagen, and in Ireland at the College of Engineering & Informatics, National University of Ireland, Galway in August 2013.
All of the national platforms upload their data to a centralized database, which provides uniform epidemiological real‐time data from >50 000 volunteers in ten European countries.
Results and Discussion
The Influenzanet network for influenza surveillance in Europe contains ten countries: The Netherlands, Belgium, Portugal, Italy, the UK, Sweden, France, Spain, Ireland, and Denmark. Identical questionnaires are implemented with questions about respiratory symptoms, access to and utilization of care and self‐medication, uptake of vaccines, attitudes to influenza vaccines, and absenteeism. The questionnaires can be used to monitor ILI in the community in real time, to track vaccine effectiveness 3, 4, 7, 8, and to estimate risk factors for ILI (not shown here).
In the ten countries, epidemiological data are collected with the participation of a self‐selected cohort followed over the influenza season. The success of the data collection strongly depends on the extent of participation of the volunteers involved in the projects. The determinants of participation and the representativeness of the self‐selected cohorts of participants in the various countries have been explored elsewhere 9. In order to assess how the web‐based systems for ILI surveillance can be a useful addition to the GP‐based system, we present the cross‐year and cross‐country incidence data, which can be readily compared and easily collected, once the platforms are in place. The national platforms collect data about the health status of the active cohort volunteers during the whole duration of the influenza season. Participants are considered to be active if they have completed, on average, at least one symptoms questionnaire every 3 weeks since the registration. To define ILI, the following ILI case definition is used, which resembles the WHO guidelines 10: a sudden onset of fever, namely, a measured body temperature of ≥38°C, accompanied by headache or muscle pain, and accompanied by cough or a sore throat. The date of fever onset is used as the date of ILI onset.
In Fig. 1, we show the weekly incidence calculated by dividing the number of active participants with ILI in a particular week by the total number of active participants in the same week. Data from 2002–2003 to 2012–2013 for eight countries (The Netherlands, Belgium, Portugal, Italy, the UK, Sweden, France, and Spain) are shown. Ireland and Denmark will start their data collection from 2013–2014. Data from the various countries have been previously published in separate papers 2, 3, 4, 5, 6, 7, 11 (Paolotti et al., 3rd International ICST Conference on Electronic Healthcare for the 21st Century, 2010).
Figure 1.
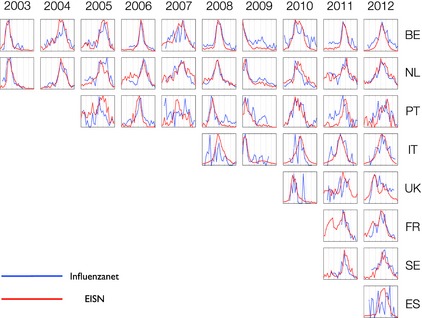
Weekly incidence data (influenza‐like illness cases/100 000) for eight countries from 2003–2004 to 2012–2013. All the curves are rescaled on the maximum. Vertical lines correspond to months from November to May for each year. EISN, European Influenza Surveillance Network; BE, Belgium; NL, The Netherlands; PT, Portugal; IT, Italy; FR, France; SE, Sweden; ES, Spain; UK, United Kingdom.
These incidence curves can be compared with the incidence rates as estimated from ILI consultations in the sentinel GP network (http://ecdc.europa.eu/en/activities/surveillance/EISN); this comparison shows very good agreement in the peak time. With different definitions of ILI cases and active participants, the incidence has the same trend, with a slight difference in the peak amplitude, but not in the timing (not shown). A more detailed figure for each country can be downloaded at https://dl.dropboxusercontent.com/u/8418871/influenzanet_supplementary.pdf.
In a few cases, such as Portugal in 2005 and Spain in 2012, the incidence curves are not perfectly congruent with the sentinel GP network data, because, for both countries, it was the first season, and the number of volunteers contributing to the data collection was very low at the beginning of the season, and rapidly increased during the unfolding of the influenza epidemics, leading to a noisy signal.
It is worth mentioning that the answers of Influenzanet participants, when they were asked whether, during an ILI episode, they consulted a GP, showed wide variability 3, 4, 7 (Paolotti et al., 3rd International ICST Conference on Electronic Healthcare for the 21st Century, 2010) (Fig. 2) in the various countries, showing that, more often than not, the web platform can be of great advantage in collecting data about ILI cases that would otherwise go undetected by GP surveillance. The results show clearly the reliability and complementary relevance of the participatory platform data.
Figure 2.
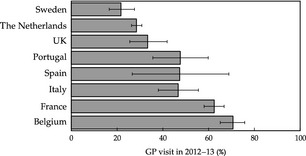
Visits to general practitioners (GPs) reported by Influenzanet users.
Conclusions
The Influenzanet network of web‐based platforms presented here highlights the benefit of using the Internet to carry out large‐scale real‐time surveillance for common diseases such as influenza. Influenzanet relies on a flexible and readily deployable web tool that is capable of coping with the requirements of different countries regarding the data collection method, either during a public health emergency or during an ordinary influenza season. Internet surveillance of healthcare usage can be used to complement traditional surveillance.
Transparency Declaration
This work was supported by the EC‐ICT IP EPIWORK grant agreement no. 231807. There are no conflicting interests.
Acknowledgements
The authors are grateful to all the Influenzanet consortium for all the data and the analysis.
Clin Microbiol Infect 2014; 20: 17–21 24350723
References
- 1. Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re‐emerging infectious diseases. Nature 2004; 430: 242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Marquet RL, Bartelds AIM, van Noort SP et al Internet‐based monitoring of influenza‐like illness (ILI) in the general population of the Netherlands during the 2003–2004 influenza season. BMC Public Health 2006; 6: 242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Noort SP, Muehlen M, Rebelo de Andrade H, Koppeschaar C, Lima Lourenço JM, Gomes MG. Gripenet: an internet‐based system to monitor influenza‐like illness uniformly across Europe. Euro Surveill 2007; 12: pii=722. [DOI] [PubMed]
- 4. Friesema IHM, Koppeschaar CE, Donker GA et al Internet‐based monitoring of influenza‐like illness in the general population: experience of five influenza seasons in the Netherlands. Vaccine 2009; 27: 6353–6357. [DOI] [PubMed] [Google Scholar]
- 5. Tilston NL, Eames KTD, Paolotti D, Ealden T, Edmunds WJ. Internet‐based surveillance of influenza‐like‐illness in the UK during the 2009 H1N1 influenza pandemic. BMC Public Health 2010; 10: 650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brooks‐Pollock E, Tilston N, Edmunds WJ, Eames KTD. Using an online survey of healthcare‐seeking behaviour to estimate the magnitude and severity of the 2009 H1N1v influenza epidemic in England. BMC Infect Dis 2011; 11: 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eames KT, Brooks‐Pollock E, Paolotti D, Perosa M, Gioannini C, Edmunds WJ. Rapid assessment of influenza vaccine effectiveness: analysis of an Internet‐based cohort. Epidemiol Infect, 2012; 140: 1309–1315 10.1017/S0950268811001804. [DOI] [PubMed] [Google Scholar]
- 8. Edmunds WJ, Funk S. Using the Internet to estimate influenza vaccine effectiveness. Expert Rev Vaccines 2012; 11: 1027–1029. [DOI] [PubMed] [Google Scholar]
- 9. Debin M, Turbelin C, Blanchon T et al Evaluating the feasibility, participants' representativeness of an online nationwide surveillance system for influenza in France. PLoS ONE 2013; 8: e73675 10.1371/journal.pone.0073675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. WHO/Europe influenza surveillance . Available at: http://www.euroflu.org/site/case_definitions.php (last accessed 15 August 2013)
- 11. Vandendijck Y, Faes C, Hens N. Eight years of the Great Influenza Survey to monitor influenza‐like illness in Flanders. PLoS ONE 2013; 8: e64156 10.1371/journal.pone.0064156. [DOI] [PMC free article] [PubMed] [Google Scholar]


