Key Points
Free ISG15 causes tissue alert in the skin upon therapeutic vaccination in mice.
Free ISG15 is a potent adjuvant for the CTL response, acting, in part, via NK cells.
Free ISG15 function in mice requires amino acids defining ligand function in humans.
Abstract
Type I IFN is produced upon infection and tissue damage and induces the expression of many IFN-stimulated genes (ISGs) that encode host-protective proteins. ISG15 is a ubiquitin-like molecule that can be conjugated to proteins but is also released from cells in a free form. Free, extracellular ISG15 is suggested to have an immune-regulatory role, based on disease phenotypes of ISG15-deficient humans and mice. However, the underlying mechanisms by which free ISG15 would act as a “cytokine” are unclear and much debated. We, in this study, demonstrate in a clinically relevant mouse model of therapeutic vaccination that free ISG15 is an alarmin that induces tissue alert, characterized by extracellular matrix remodeling, myeloid cell infiltration, and inflammation. Moreover, free ISG15 is a potent adjuvant for the CTL response. ISG15 produced at the vaccination site promoted the vaccine-specific CTL response by enhancing expansion, short-lived effector and effector/memory differentiation of CD8+ T cells. The function of free ISG15 as an extracellular ligand was demonstrated, because the equivalents in murine ISG15 of 2 aa recently implicated in binding of human ISG15 to LFA-1 in vitro were required for its adjuvant effect in vivo. Moreover, in further agreement with the in vitro findings on human cells, free ISG15 boosted the CTL response in vivo via NK cells in the absence of CD4+ T cell help. Thus, free ISG15 is part of a newly recognized innate route to promote the CTL response.
Introduction
Infection and tissue damage lead to the production of type I IFNs (IFN-I). These cytokines induce the expression of many IFN-stimulated genes (ISGs), encoding proteins that protect the host in many different ways (1). This group of proteins includes ISG15 that has a diubiquitin-like structure (2). Isg15 is one of the genes most strongly upregulated in response to viral infection in a diversity of species, including humans (3, 4). ISG15 is also induced by bacterial infections (5, 6). Isg15-deficient mice and humans display phenotypes that indicate a role for ISG15 in the protection against infection, but the underlying mechanisms have not been fully elucidated (3, 7, 8). ISG15 can be conjugated to proteins but also exists in a free form and thus may act by different mechanisms either within or outside the cell.
Like ubiquitin, ISG15 can be covalently conjugated to lysine residues through a C-terminal diglycine motif (LRLRGG) (9). This process, termed ISGylation, relies on one E1-activating enzyme (UBE1L) (10), one E2 enzyme (UBC8) (11, 12), and one major E3 enzyme (HERC5 in humans and HERC6 in mice) (13, 14). ISGylation is reversible, and the major ISG15-deconjugating enzyme in vivo is USP18/UBP43 (15). All enzymes that regulate ISGylation are induced by type I IFN (9). It was shown that ISGylation can occur cotranslationally on newly synthesized proteins without apparent target specificity (16). Proteins are, in general, decorated with ISG15 monomers rather than polymeric chains, and ISG15 is not a signal for proteasomal targeting (17). Rather, by competition for ubiquitin, it can protect proteins from ubiquitination and ensuing proteasomal degradation (18). However, it was shown that ISG15 can also modify ubiquitin, thus forming ISG15–ubiquitin mixed chains (19). Various signal transduction proteins can be ISGylated, and this can affect signaling outcome, as shown in some specific cases (3). Several viruses have developed distinct strategies to counteract ISGylation (7), further indicating that ISGylation plays an important role in the arms race against viral infection.
Susceptibility to different viral infections has been studied in Isg15−/− mice, Ube1l−/− mice, and Usp18−/− mice (8). Response to a number of virus types, but not all, was found to be impaired in Isg15−/− mice. By comparing phenotypes of Isg15−/− mice with those of Ube1l−/− mice, in which only the conjugation to substrates, but not the function of free ISG15, is eliminated, the function of free ISG15 can be separated from that of ISGylation-dependent mechanisms. Irrespective of its protease function toward ISG15-modified substrates, USP18 represents a major negative regulator of the type I IFN response (20). Therefore, mice lacking USP18 protein expression exhibit phenotypical alterations not directly linked to ISG15. A knock-in mouse model with selective inactivation of only the protease function of USP18 exhibited enhanced ISGylation and increased viral resistance (21). From the collected work, it can be concluded that both ISGylation (22) and free ISG15 (23) can protect against certain viral infections in mice.
In the few ISG15-deficient human patients that have been reported, no evidence for increased susceptibility to viral infection has been detected so far (8). Initially, ISG15-deficient patients were discovered on basis of failed immunity to live attenuated mycobacteria (bacillus Calmette-Guérin) (5). A second group of ISG15-deficient patients that had not been vaccinated with bacillus Calmette-Guérin presented with a syndrome characterized by excessive type I IFN signaling (18), as did USP18-deficient patients (24). Mechanistic studies revealed that USP18 negatively regulates type I IFNR (IFNAR) signaling, independent of its de-ISGylation activity (18, 25). USP18 is subject to ubiquitin-dependent proteasomal degradation, which is inhibited by free intracellular ISG15. In this way, USP18 and ISG15 mediate negative feedback on IFNAR signaling, which explains the inflammatory phenotype in the ISG15-deficient patients (20). Remarkably, this stabilizing effect of free ISG15 on USP18 is found in humans, but not in mice (26), in which the affinity of the interaction is lower, likely because of the significant divergence in amino acid sequence of ISG15 between species (7).
The function of the free extracellular form of ISG15 has been an enigma. Free ISG15 does not have an N-terminal hydrophobic signal sequence (27, 28), so it is not secreted from cells in the classical way. However, ISG15 can be released from different cell types, including myeloid and lymphoid cells (5, 27–30) and is found in the serum of patients treated with type I IFN (27, 28, 31). The suggested immunomodulatory role of extracellular ISG15 is ill-defined and primarily based on cell culture experiments. ISG15 was shown to enhance IFN-γ secretion by NK cells and T cells (5, 31–33), which has been suggested to be its main antimycobacterial activity (5). A breakthrough has been the recent identification of the integrin LFA-1 (αLβ2) as cell surface receptor for extracellular ISG15. This work, performed on human cells in vitro, defined a signaling role for extracellular ISG15 by showing that 2 aa in ISG15 are critical for binding to LFA-1 and supporting IFN-γ secretion from IL-12–primed NK cells (33). One in vivo study indicates that ISG15 encoded by a DNA vaccine can promote the CTL response in mice (34), and another study reported that recombinant ISG15 protein can promote dendritic cell (DC) activation in mice (35).
We have addressed the potential role of extracellular ISG15 in supporting the CTL response in vivo and evaluated the underlying mechanisms in a clinically relevant mouse model of therapeutic vaccination. With this study, we demonstrate that extracellular ISG15 is an alarmin that promotes the CTL response via NK cells.
Materials and Methods
Mice
C57BL/6JRj mice were obtained from Janvier Laboratories (Le Genest-Saint-Isle, France). Isg15−/− mice (36) were provided by Dr. K.-P. Knobeloch (Freiburg, Germany). In all experiments, gender- and age-matched (8–12 wk) mice were used and maintained in individually ventilated cages (Innovive, San Diego, CA). Control and test mice were selected at random. Experiments were performed according to national and institutional guidelines and approved by the institutional committee for animal experimentation.
Cells
Bone marrow (BM) cells were isolated by flushing femurs of Isg15−/− mice with PBS supplemented with 2% FCS (Life Technologies BRL, Thermo Fisher Scientific). RBCs were lysed in 0.14 M NH4Cl and 0.017 M Tris-HCl (pH 7.2) for 1 min. DCs were generated by culturing 2 × 106 BM cells in IMDM supplemented with 8% FCS and rFlt3 ligand (homemade) for 8 d. HeLa cells were cultured in DMEM supplemented with 8% FCS. Phoenix-Eco packaging cells were cultured in IMDM supplemented with 5% FCS.
DNA constructs and gene expression
The E7SH DNA vaccine was generated as described (37, 38). Briefly, gene fragments of the human papilloma virus (HPV)–16 E7 gene were introduced into pVAX1 vector. The cDNA-encoding mouse ISG15 wild-type (WT) (NM_015783) or ISG15 ΔGG, synthesized as a gBlock gene fragment (Integrated DNA Technologies) was inserted into the pVAX1 plasmid (Invitrogen) using BamHI and NotI restriction sites. The mutant version of ISG15, which contains a leucine instead of a tyrosine residue at position 94 and an aspartate instead of a glutamine residue at position 100 (pVAX-ISG15-Y94L_Q100D), was generated using the QuikChange kit (StrataGene) in accordance with the manufacturer’s instructions. To express ISG15 variants in BM-derived DCs, ISG15 WT and ΔGG were subcloned into the pMX-IRES-GFP vector using BamHI and NotI restriction sites. BM-derived DCs expressing murine ISG15 WT or ΔGG were generated by retroviral transfection. For virus production, retroviral constructs were transfected using FuGENE HD Transfection Reagent (Promega) into Phoenix-Eco packaging cells, together with the pCL-Eco vector encoding the ecotropic retrovirus receptor. Medium that contained retrovirus was harvested from the Phoenix-Eco packaging cells 48 h later. For retroviral transduction, BM-derived DC precursors were cultured with Flt3 ligand for 3 d. Next, they were resuspended at 2 × 106 cells/ml retrovirus-containing medium plus Flt3 ligand and placed in nontissue culture–treated, 24-well plates (BD Biosciences) coated with 50 μg/ml RetroNectin (Takara Bio). Plates were spun for 90 min at 450 × g. Cells were cultured in this medium for 24 h. Cells were then transferred to BM-derived DC culture medium and maintained for 4 extra d before use. Transfections of cDNA in HeLa cells were performed using FuGENE HD transfection reagent (Promega) according to the manufacturer’s instructions.
Western blotting
Cells were harvested, washed with PBS, and lysed in 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Nonidet P-40, and cOmplete Inhibitor Cocktail (Roche). Insoluble material was removed by centrifugation at 20,000 × g for 15 min. Protein concentration was determined by Bradford protein assay (Bio-Rad Laboratories). Equal amounts of lysate were separated on NuPAGE 4–12% Bis-Tris gels (Invitrogen), and proteins were transferred to nitrocellulose transfer packs (Bio-Rad Laboratories) using the Semi-dry Trans-Blot Turbo Transfer System (Bio-Rad Laboratories) according to manufacturer’s instructions. Membranes were blocked with Roche Western block solution (1:10) in TBS with 0.1% Tween 20 for 1 h at room temperature. Next, membranes were incubated overnight at 4°C with appropriate primary Abs in Roche Western block solution (1:20)/TBS with 0.1% Tween 20, washed with TBS with 0.1% Tween 20, and probed with the adequate secondary Abs (1:10,000) in Roche Western block solution/TBS with 0.1% Tween 20 for 1 h at room temperature. Primary Abs used were the following: rabbit anti-mouse ISG15 (1:5000, kindly provided by Dr. K.-P. Knobeloch), mouse anti-actin (1:10,000, clone C4; MAB1501R; MilliporeSigma), and anti-mouse GAPDH (1:2000, clone D4C6R; 97166S; Cell Signaling Technology). Secondary Abs used were the following: goat anti-mouse IRDye 682/800 (925-68070/926-32210) or goat anti-rabbit IRDye 682/800 (925-68071/925-32211) from LI-COR Biosciences. Immunoblots were developed with the aid of an Odyssey Imaging System (LI-COR Biosciences).
Intraepidermal DNA “tattoo” vaccination
On day 0, mice were anesthetized with isoflurane, and the hair on a hind leg was removed using depilating cream (Veet; Reckitt Benckiser). On days 0, 3, and 6, a 15-μl drop of a solution containing 2 mg/ml plasmid DNA (pDNA) mixture in 10 mM Tris-HCl and 1 mM EDTA (pH 8) was applied to the hairless skin of anesthetized animals and delivered into the epidermis with a Permanent Makeup Tattoo machine (MT.DERM) using a sterile disposable nine-needle bar with a needle depth of 1 mm and an oscillating frequency of 100 Hz for 45 s.
In vivo NK cell depletion
Mice were injected i.v. with 100 μl of anti-asialo GM1 (39) or control rabbit sera (Wako Chemicals) diluted 1∶10 in HBSS the day before the first DNA vaccination and on days 0 and 3.
Leukocyte isolation and flow cytometry
Blood was collected from tail bleeding using Microvette CB 300 LH tubes (Sarstedt). To isolate lymphocytes from spleen and inguinal draining lymph node (dLN), organs were passed through a 70-μm cell strainer (BD Falcon). RBCs were lysed in 0.14 M NH4Cl and 0.017 M Tris-HCl (pH 7.2) for 1 min at room temperature. Then, cell samples were centrifuged for 5 min at 400 × g and resuspended in FACS buffer (PBS with 2% FCS; Antibody Production Services). Surface staining with relevant mAbs and allophycocyanin–H-2Db/E749–57 tetramers was performed for 30 min on ice. Intracellular staining was performed after cell fixation and permeabilization using Foxp3 Transcription Factor Staining Buffer Set (eBioscience). Fluorochrome-labeled mAbs employed were as follows: anti-CD8α–V500 (1:200, clone 53-6.7) and anti–IFN-γ–eF450 (1:100, clone XMG1.2) from BD Biosciences; anti-CD127–BV421 (1:200, clone A7R34) and anti-CD3–Alexa Fluor 488 (1:200, clone 17A2) from BioLegend; anti-KLRG1–PEeF610 (1:200, clone 2F1), anti-CD44–PerCP-Cy5.5 (1:400, clone IM7), anti-CD49b–PE-Cy7 (1:200, clone DX5), anti-NK1.1–Alexa Fluor 700 (1:200, clone PK136), anti-CD4–eF450 (1:200, clone GK1.5), anti-Tbet–PE-Cy7, and anti-CD62L–FITC (1:100, MEL-14), from eBioscience; and anti–granzyme B (GZB)–PE (1:200, clone CLB-GB11) (Sanquin Reagents). To detect cytokine production by E749–57-specific CD8+ T cells in dLN and spleen, cells were incubated for 16 h with 1 μg/ml E749–57 or no peptide (negative control) in the presence of GolgiPlug (BD Biosciences) in IMDM supplemented with 8% FCS. Flow cytometry was performed using LSRFortessa (BD Biosciences), and data were analyzed with FlowJo software (Tree Star). Live cells were selected based on staining with LIVE/DEAD Near Infrared dye (Thermo Fisher Scientific).
RNA preparation and sequencing
At day 4 postvaccination, total skin from the tattooed area was isolated, and total RNA was isolated using the RNeasy Mini Kit (catalog no. 74106; QIAGEN), including an on-column DNA digestion (catalog no. 79254; QIAGEN), according to the manufacturer’s instructions. Quality and quantity of the total RNA was assessed by the 2100 Bioanalyzer using an RNA Nano Chip and RNA Pico Chip (Agilent Technologies). Total RNA samples having RNA integrity number > 8 were subjected to library generation. Strand-specific cDNA libraries were generated using the TruSeq Stranded mRNA Sample Preparation Kit (Illumina) according to the manufacturer’s instructions. The libraries were analyzed on a 2100 Bioanalyzer using a DNA 7500 Chip (Agilent Technologies), diluted and pooled equimolar into a multiplex sequencing pool and stored at −20°C. The libraries were sequenced with 65 base single reads on a HiSeq 2500 using V4 chemistry (Illumina).
RNA sequence analysis
Differential expression of genes was assessed using the DESeq2 package (40) using default parameters in R v.3.5.3 (https://www.R-project.org/). Genes with adjusted p values ≤ 0.05 were deemed significantly differentially expressed. To find activated pathways, overexpressed in comparison with the control (according to log-fold change > 0, adjusted p value ≤ 0.1), were used as input for enrichment analysis using the Reactome enrichment analysis tool available at https://reactome.org/PathwayBrowser/#TOOL=AT. Additionally, we analyzed activated pathways using Ingenuity Pathway Analysis (IPA; QIAGEN). In this case, raw read counts were loaded into the software and analysis carried out using default settings. Data have been deposited in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE139469.
Statistical analysis
Statistical significance was determined with GraphPad Prism software as indicated in the figure legends.
Illustrations
Illustrations in Figs. 1A, 2A, and 6A were made with BioRender.
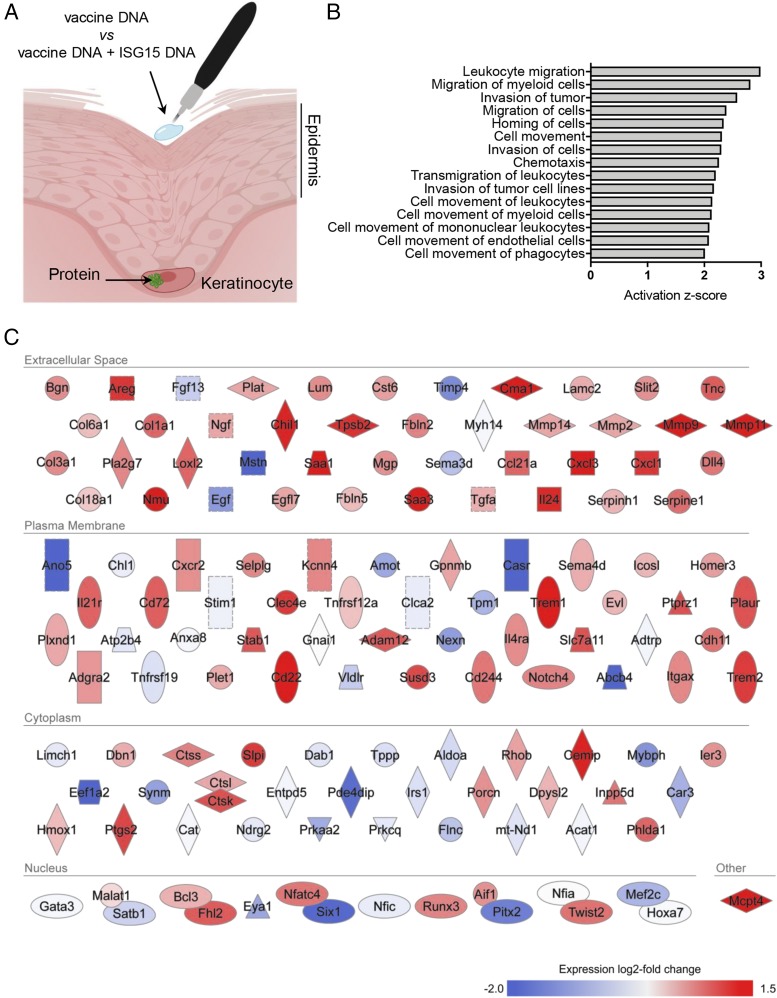
FIGURE 1.
The skin transcriptome reports ISG15 activity. (A) Illustration of the intraepidermal pDNA tattoo vaccination procedure and expression of pDNA-encoded protein (green) in transfected keratinocytes. Mice were vaccinated in comparative settings with pDNA encoding HPV-E7 (vaccine DNA) in combination with either pDNA-encoding ISG15 ΔGG (ISG15 DNA) or an equal amount of empty vector pDNA. (B) mRNA from total skin of mice (n = 3 per group) vaccinated with “vaccine DNA” versus “vaccine DNA + ISG15 DNA” was subjected to deep sequencing. Gene ontology analysis was performed by IPA of the 444 genes found to be differentially expressed between the comparative vaccination settings (p value ≤ 0.05). The functional categories connected to cell migration and with a predictive activation z-score ≥2 are depicted. (C) IPA-based rendering of differentially expressed molecules from the functional categories depicted in (B) and their subcellular localization. The total experiment was performed once.
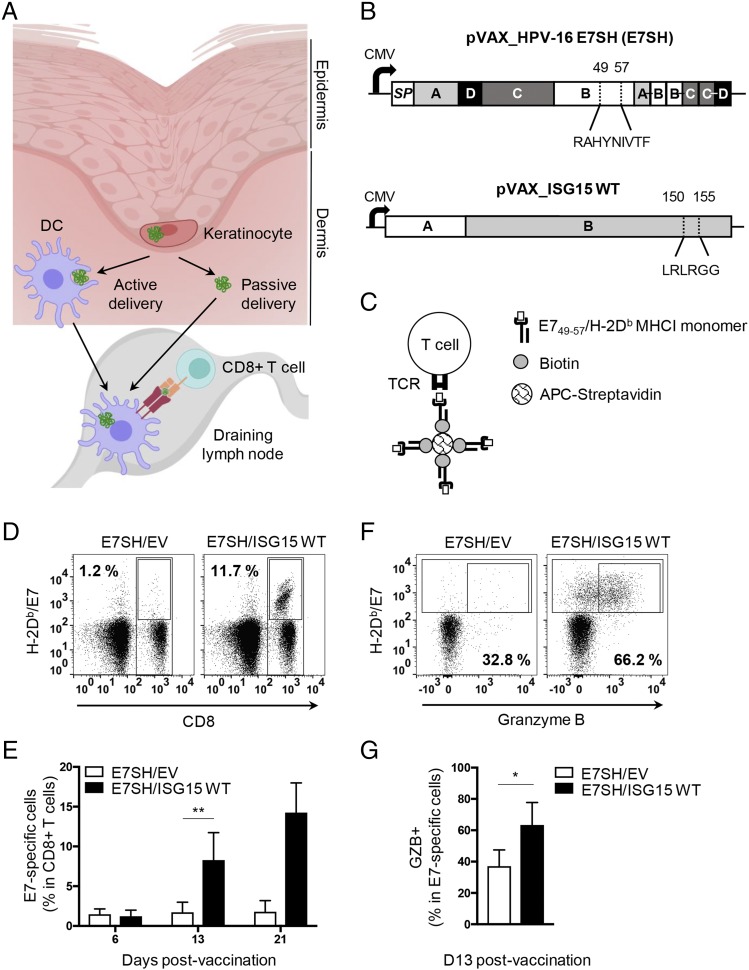
FIGURE 2.
WT ISG15 promotes the Ag-specific CTL response to vaccination. (A) Illustration of the cellular context of vaccine Ag delivery in the skin and the route of the Ag from keratinocytes to the dLN. (B) Scheme of the DNA sequence encoding the shuffled (SH) version of the HPV-16 E7 protein (E7SH vaccine) and ISG15 WT used for vaccination. For the E7 protein, the different exons (A–D) and the signal peptide (SP) are indicated. In exon B, the immunodominant H-2Db–restricted epitope RAHYNIVTF (corresponding to aa 49–57 from the original E7 protein) is depicted (37, 38). For ISG15, the two exons (A and B) are indicated (ENSMUST00000085425.5). In exon B, the conjugation site (LRLRGG, aa 150–155) is depicted. (C) MHC tetramer technology to identify by flow cytometry CD8+ T cells with a TCR that recognizes H-2Db/E749–57. Recombinant H-2Db MHC class I (MHCI) monomers are folded with E749–57 peptide, conjugated to biotin and multimerized with streptavidin conjugated to a fluorophore allophycocyanin. (D–G) Mice (n = 5 per group) were vaccinated with HPV-16 E7SH cDNA (E7SH) in combination with pVAX empty vector (EV) or pVAX-mouse ISG15 (ISG15 WT) on days 0, 3, and 6. The CD8+ T cell response was followed in time by flow cytometric analysis of peripheral blood using H-2Db/E749–57 tetramers. (D) Representative staining of cells with H-2Db/E749–57 tetramer and anti-CD8 Ab. Numbers indicate frequency of tetramer+ cells among total CD8+ T cells. (E) Quantification of the percentage of H-2Db/E749–57 tetramer+ cells among total CD8+ T cells over time postvaccination. (F) Representative staining of cells with H-2Db/E749–57 tetramer and Ab to GZB. Numbers indicate percentage of GZB+ cells among tetramer+ cells. (G) Quantification of the percentage of GZB+ cells among tetramer+ cells at day 13 postvaccination. Results are representative of at least three experiments. Statistical analysis was performed using two-tailed Student t test. *p < 0.05, **p < 0.01.
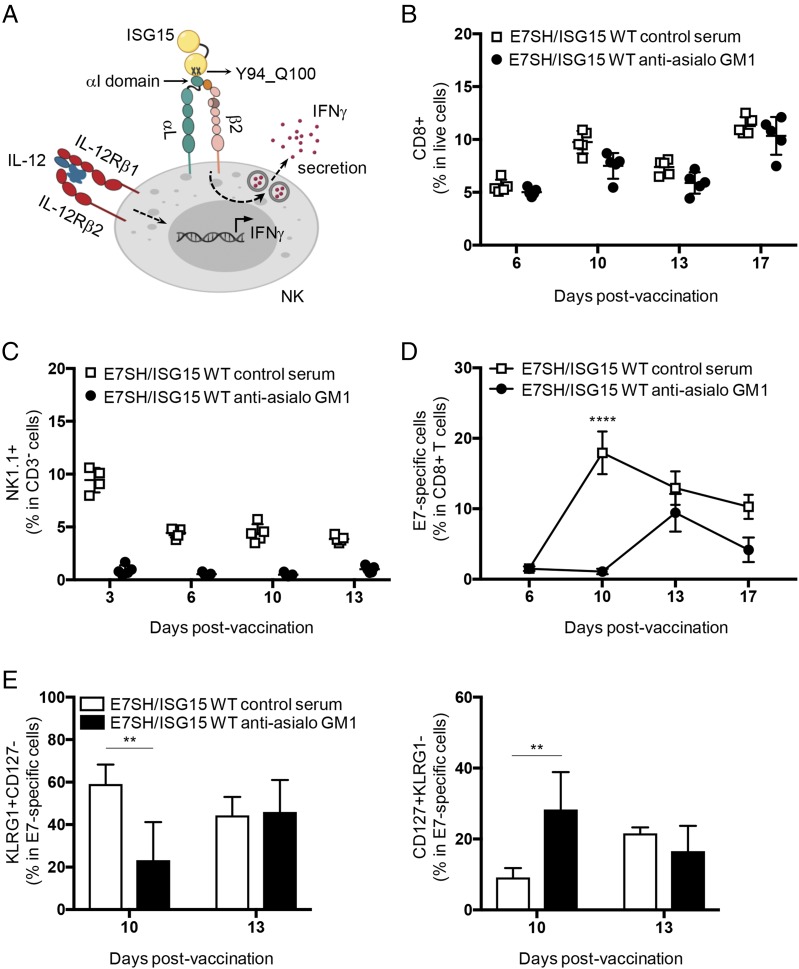
FIGURE 6.
NK cells contribute to relaying the effects of ISG15 to E7-specific CD8+ T cells. (A) Scheme showing ISG15 interaction with LFA-1 (αLβ2 integrin and CD11a/CD18) receptor in NK cells and its biological impact as described for human cells in vitro (33). (B–E) Mice (n = 4 per group) were vaccinated on days 0, 3, and 6 with plasmid encoding E7SH combined with ISG15 WT. On days −1, 0, 3, and 6, mice were injected i.v. with control serum or anti-asialo GM1 antiserum. (B) Frequency of CD8+ cells among live cells in blood at the indicated days postvaccination. (C) Frequency of NK1.1+ cells within CD3-negative cells in blood at the indicated days postvaccination to validate NK cell depletion. (D) E7-specific cells within CD8+ cells in blood at the indicated days postvaccination. (E) Percentage of SLECs (KLRG1+CD127−) (left) and MPECs (CD127+KLRG1−) (right) within E7-specific CD8+ T cells in blood at days 10 and 13. Results are representative of two experiments. Statistical evaluation was performed using one-way ANOVA evaluation and Tukey posttest and is indicated for day 10. **p < 0.01, ****p < 0.0001.
Results
ISG15 causes innate immune alert in the skin
We were intrigued by earlier findings that extracellular ISG15 may act as an adjuvant to support the CTL response (34). We aimed to corroborate this and to understand the mechanistic basis, making use of a versatile therapeutic DNA vaccination model in mice (37) that we have well characterized previously (38, 41, 42). In this model, pDNA is applied on the depilated skin and injected into the epidermis using a tattoo device (Fig. 1A). This results in transfection of keratinocytes with the pDNA of interest, which is subsequently transcribed and translated (42). To determine whether ISG15 protein expression had a local effect on the vaccinated skin, we performed the following experiment: two groups of mice were vaccinated with pDNA encoding specific Ag (to be described in Fig. 2) combined with pDNA encoding ISG15 or an equal dose of empty vector (Fig. 1A). At day 4 after vaccination, total skin from the area of vaccination was excised, and mRNA was isolated and subjected to deep sequencing.
Statistical analysis of normalized transcript read counts showed that 444 genes were differentially expressed in the skin as a result of ISG15 coexpression (Supplemental Table I; Gene Expression Omnibus submission GSE139469). Using IPA, we identified 29 functional categories predicted to be increased according to an activation z-score ≥2. Interestingly, 15 of these categories indicated that ISG15 stimulated cell migration, particularly of myeloid cells and endothelial cells (Fig. 1B). A zoom-in on the 140 specific molecules in the functional categories listed in Fig. 1B highlighted that ISG15 significantly upregulated the expression of various metalloproteases (Mmp2, Mmp9, Mmp11, Mmp14, and Adam12; downregulation of inhibitor Timp4) and collagens (Col1a1, Col3a1, Col6a1, and Col18a1) (Fig. 1C), suggesting remodeling of the extracellular matrix. This correlated with gene ontology analysis using Reactome, which more precisely specified metalloprotease activity, collagen degradation and formation, and extracellular matrix organization (Table I). The extracellular matrix remodeling and immune cell signature observed in the transcriptome strongly suggests that ISG15 is able to evoke tissue alarm.
Table I. Gene ontology analysis of ISG15-induced differential gene expression in skin.
| Pathway Name | No. Entities Found | No. Entities Total | p Value | False Discovery Rate |
|---|---|---|---|---|
| Collagen degradation | 15 | 57 | 6.35 × 10−13 | 1.13 × 10−10 |
| Extracellular matrix organization | 29 | 295 | 6.87 × 10−13 | 1.13 × 10−10 |
| Assembly of collagen fibrils and other multimeric structures | 14 | 61 | 2.28 × 10−11 | 2.49 × 10−9 |
| Degradation of extracellular matrix | 18 | 135 | 1.97 × 10−10 | 1.62 × 10−8 |
| Collagen formation | 15 | 88 | 2.57 × 10−10 | 1.67 × 10−8 |
| Collagen biosynthesis and modifying enzymes | 11 | 64 | 6.17 × 10−8 | 3.33 × 10−6 |
| Extracellular matrix proteoglycans | 10 | 51 | 7.50 × 10−8 | 3.53 × 10−6 |
| Collagen chain trimerization | 9 | 41 | 1.34 × 10−13 | 5.48 × 10−6 |
| Integrin cell surface interactions | 11 | 81 | 6.22 × 10−7 | 2.24 × 10−5 |
| NCAM1 interactions | 6 | 22 | 5.13 × 10−6 | 1.64 × 10−4 |
| Activation of matrix metalloproteases | 7 | 51 | 6.72 × 10−5 | 0.00195 |
| Axon guidance | 16 | 272 | 8.23 × 10−5 | 0.00222 |
| Signaling by PDGF | 7 | 57 | 1.33 × 10−4 | 0.00321 |
| NCAM signaling for neurite outgrowth | 6 | 40 | 1.39 × 10−4 | 0.00321 |
| Other semaphorin interactions | 4 | 14 | 1.71 × 10−4 | 0.00359 |
Pathway analysis using Reactome overrepresentation analysis of the 254 overexpressed genes (log fold change > 0 out of the total of 444 differentially expressed genes in Fig. 1B). The top 15 pathways are depicted.
WT ISG15 promotes the CD8+ T cell response to therapeutic vaccination
Using a fluorescent protein encoded by the vaccine, we have previously shown that vaccine protein expressed by keratinocytes can reach the proximal skin-draining dLN in two ways: the protein can be transported by migratory dermal DCs or drain passively via the lymph (42). In both cases, DCs can process the vaccine protein and cross-present relevant peptides in MHC class I, which can lead to the activation of Ag-specific CD8+ T cells (Fig. 2A). To examine CD8+ T cell activation, we used a DNA vaccine–encoding HPV-16 E7 protein in a shuffled configuration (E7SH) (37, 38). This protein contains an immunodominant epitope, E749–57, that is presented by H-2Db (Fig. 2B). To examine the effect of ISG15 on the T cell response, we vaccinated the mice with the E7SH vaccine in combination with empty vector or vector encoding WT ISG15. We followed the CD8+ T cell response by flow cytometry, using MHC tetramers (Fig. 2C). We knew that vaccination with E7SH elicits a weak CTL response (38, 41) and purposely used this vaccination setting to create a good window for testing the potential adjuvant effect of ISG15. The E7-specific CD8+ T cell response was followed in blood over time (Fig. 2D). This analysis revealed that the frequency of E7-specific cells within total CD8+ T cells was dramatically increased in the ISG15-adjuvanted setting. Analysis for expression of the CTL effector molecule GZB at day 13 emphasized that more CTLs were raised in the ISG15-adjuvanted setting (Fig. 2E).
Free ISG15 enhances the generation of vaccine Ag-specific CTLs
We next examined whether it was free or conjugated ISG15 that promoted the CTL response. For this purpose, we compared the activity of ISG15 WT with the activity of the ISG15 ΔGG mutant that lacks the C-terminal glycine residues that are required for substrate conjugation (Fig. 3A). Lack of ISG15 conjugation was validated by expression of ISG15 WT and the ISG15 ΔGG mutant in Isg15−/− DCs. As expected, transduction of a vector encoding ISG15 WT led to expression of the free form as well as ISG15 conjugation to multiple proteins, whereas transduction of the vector encoding ISG15 ΔGG led to expression of the free form only (Fig. 3B).
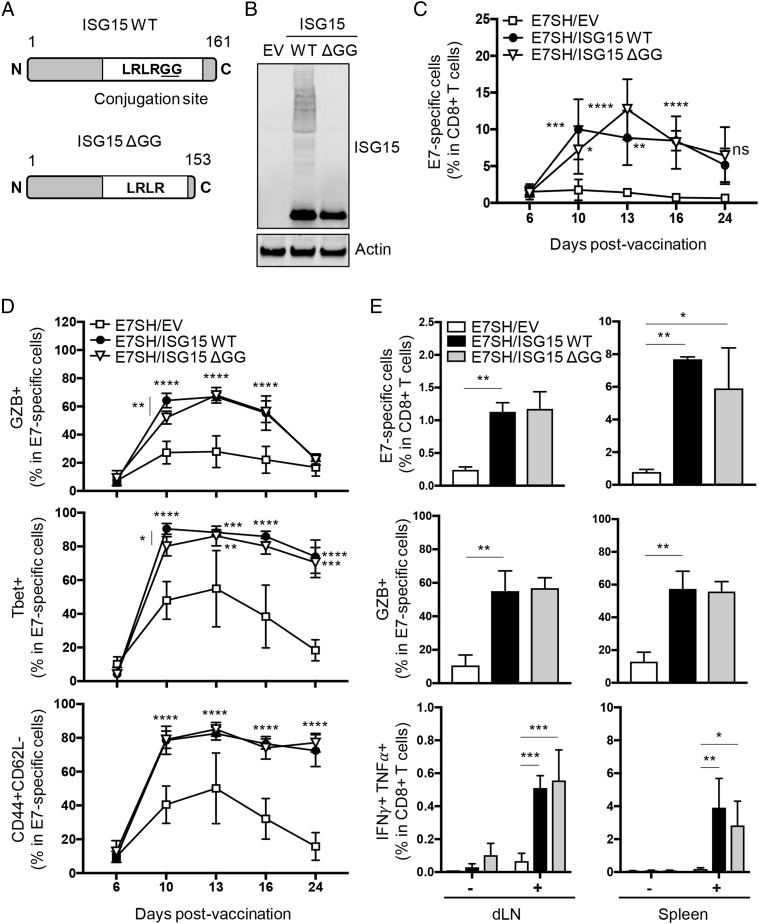
FIGURE 3.
The free form of ISG15 promotes CTL priming in response to vaccination. (A) Scheme of mouse ISG15 WT protein (aa 1–161) and its C-terminal truncated version, ISG15 ΔGG (aa 1–153). The minimal sequence (LRLRGG) required for its conjugation to intracellular proteins is depicted in ISG15 WT. (B) Validation of ISG15 constructs by assessment of ISGylation of intracellular proteins. pDNA encoding ISG15 WT or ISG15 ΔGG and empty control vector (EV) were expressed in Isg15−/− BM-derived DCs, and ISGylation was assessed by Western blotting on total cell lysates. Actin served as loading control. Results are representative of multiple independent analyses with individual samples. (C–E) Mice (n = 7 per group) were vaccinated with plasmid encoding E7SH, either combined with EV, ISG15 WT, or ISG15 ΔGG as outlined in Fig. 2. (C and D) The E7-specific CTL response was followed over time in peripheral blood by flow cytometric analysis using H-2Db/E749–57 tetramers and Abs to CD8, GZB, Tbet, CD44, and CD62L. (C) Quantification of the percentage of H-2Db/E749–57 tetramer+ cells among total CD8+ T cells. (D) Quantification of the percentage of GZB+, Tbet+, and CD44+CD62L− cells within H-2Db/E749–57 tetramer+ cells. (E) On day 16, three mice per group were sacrificed, and the T cell response was assessed in dLN and spleen. Depicted are percentage of H-2Db/E749–57 tetramer+ cells within total CD8+ T cells, percentage of GZB+ cells within tetramer+ cells ex vivo, and percentage of IFN-γ+TNF-α+ cells among total CD8+T cells after in vitro stimulation. Results are representative of two experiments. Statistical analysis was performed using one-way ANOVA (C and D) or two-way ANOVA (E) and Tukey posttest. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
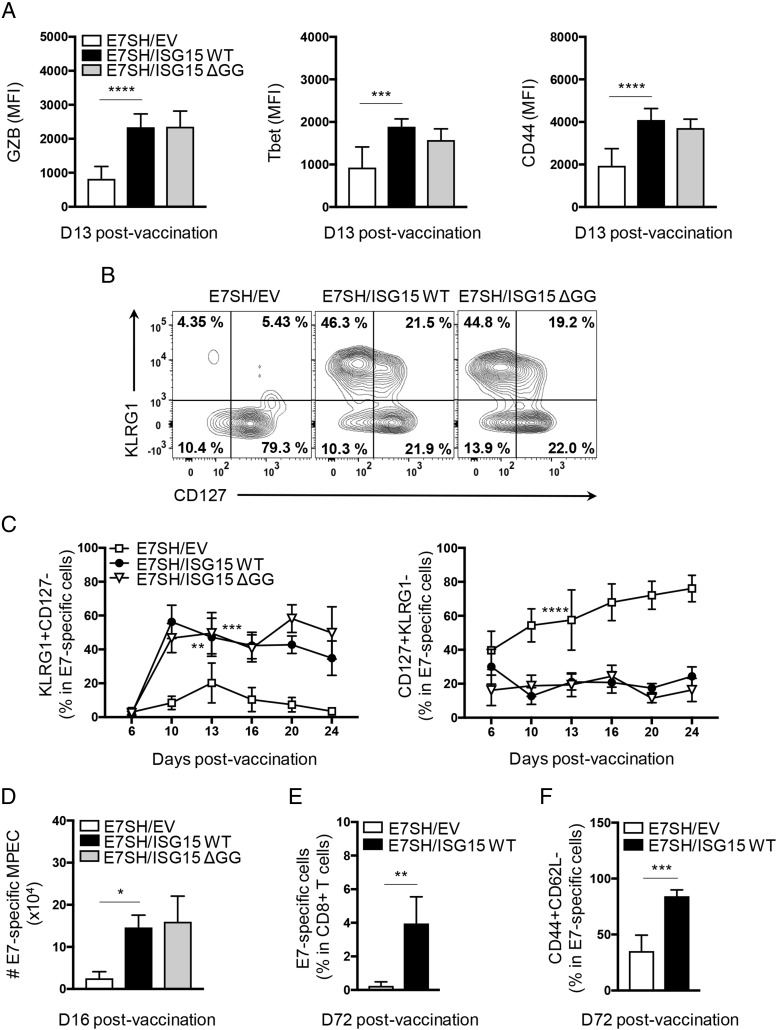
The CD8+ T cell response to vaccination with E7SH was increased in equal measure by concomitant vaccination with either ISG15 WT or ISG15 ΔGG throughout the entire kinetics, as monitored in blood (Fig. 3C). The CD8+ T cells raised after ISG15-adjuvanted vaccination had undergone CTL effector differentiation, as determined by expression of GZB and the transcription factor Tbet and the effector phenotype CD44+CD62L− (Fig. 3D). Both ISG15 WT and nonconjugatable ISG15 ΔGG promoted the CTL response in equal measure (Fig. 3C, 3D).
To validate that the increased magnitude of the CTL response observed in blood was a consequence of an increase in CD8+ T cell priming, we examined the response in dLN and spleen. Both ISG15 WT and ISG15 ΔGG significantly increased, in equal measure, the magnitude of the E7-specific CD8+ T cell response in dLN and spleen (Fig. 3E). Furthermore, the responder CD8+ T cells had differentiated into CTLs as determined on day 16 by expression of GZB and coexpression of IFN-γ and TNF-α (Fig. 3E). This analysis highlighted that vaccination with E7SH alone hardly generated functional CTLs, whereas ISG15 WT or ISG15 ΔGG-adjuvanted vaccination raised a sizeable CTL response. We conclude that ISG15 does not need to be conjugated to proteins to have an adjuvant effect on the CTL response to vaccination.
Free ISG15 enhances CTL differentiation, formation of short-lived effector and effector memory CTLs
To examine the impact of ISG15 on the intrinsic functionality of CTLs, we analyzed the protein expression levels of GZB, Tbet, and CD44 on a per-cell basis. In the ISG15-adjuvanted settings (WT or ΔGG), CTLs expressed higher levels of these molecules, indicating improved effector differentiation (Fig. 4A). Furthermore, we determined the formation of short-lived effector cells (SLECs) and memory precursor effector cells (MPECs) throughout the primary immune response. ISG15 WT and ΔGG increased in equal measure the frequency of SLECs (KLRG1+ CD127) within E7-specific CD8+ T cell pool raised upon vaccination, as measured in blood throughout the entire response kinetics. The frequency of MPECs (CD127+KLRG1−) within the E7-specific CD8+ T cell pool was correspondingly reduced in both ISG15-adjuvanted settings (Fig. 4B, 4C). Thus, CTL differentiation seemed to be geared more toward a short-lived effector- than an effector memory differentiation fate in the ISG15-adjuvanted setting. Nevertheless, the absolute numbers of E7-specific MPECs in spleen at day 16 postvaccination were significantly higher when ISG15 was included in the vaccination setting (Fig. 4D). This result correlated with an increased frequency of E7-specific CD8+ T cells in blood at day 72 postvaccination (steady-state memory) (Fig. 4E). Among these memory cells, effector memory phenotype cells (CD44+CD62L−) were drastically increased in the ISG15-adjuvanted setting (Fig. 4F). Thus, free ISG15 promotes the effector differentiation of CTLs, the formation of short-lived effector CTLs, as well as the formation of memory precursor effector CTLs and effector memory CTLs.
FIGURE 4.
The free form of ISG15 supports short-lived effector and effector memory differentiation of CTLs. (A–D) Mice (n = 7 per group) were vaccinated with plasmid encoding E7SH, either combined with EV, ISG15 WT, or ISG15 ΔGG as outlined in Fig. 2. (A) Flow cytometric analysis of protein expression as assessed by mean fluorescence intensity (MFI) of GZB, Tbet, and CD44 on E7-specific CD8+ T cells in blood at day 13 postvaccination. (B) Representative flow cytometric analysis of E7-specific CD8+ T cells stained with Abs to KLRG1 and CD127. Numbers indicate percentage of cells in each quadrant. (C) Quantification of percentage of SLECs (KLRG1+CD127−) (left) and MPECs (CD127+KLRG1−) (right) within E7-specific CD8+ T cells in blood over time. (D) On day 16, three mice per group were sacrificed, and total numbers of CD127+KLRG1− E7-specific cells per spleen were determined. (E) Quantification of the percentage of E7-specific cells within CD8+ T cells in blood on day 72. (F) Quantification of the percentage of CD44+CD62L− cells among E7-specific CD8+ T cells in blood on day 72. Results are representative of two experiments. Statistical analysis was performed using one-way ANOVA and Tukey posttest (A–D) or two-tailed Student t test (E). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Amino acid residues Y94 and Q100 of ISG15 are required for its adjuvant effect on the CTL response
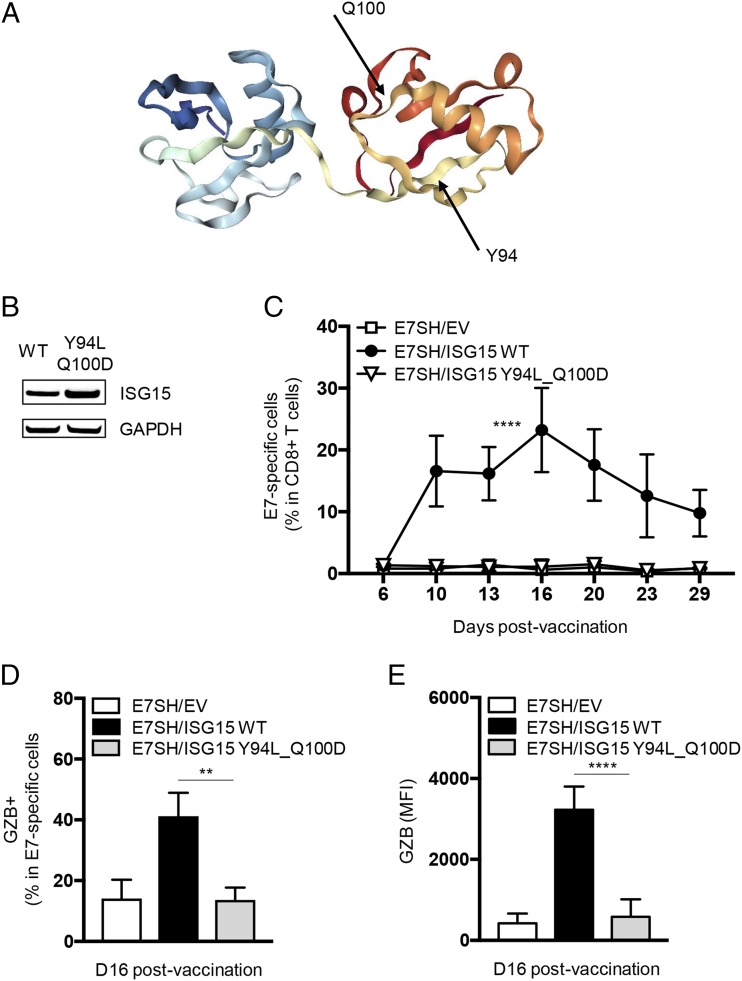
Recently, the integrin LFA-1 has been defined as a receptor for free ISG15 in a human cell culture system. Amino acid residues Y96 and Q102 in human ISG15 proved to be critical for LFA-1 binding (33) (Fig. 5A). Based on these findings, we hypothesized that the effect of free ISG15 on the CTL response that we observed in this study in vivo is a consequence of ISG15 binding to a receptor. We therefore tested the adjuvant activity of mouse ISG15 with the equivalent mutations, Y94L_Q100D (Fig. 5A), that should abrogate its ligand activity. The mutant, encoded by the pVAX expression vector, was expressed to at least equivalent levels as WT ISG15 at the protein level in transfected cells (Fig. 5B).
FIGURE 5.
Residues Y94 and Q100 in ISG15 are critical for its adjuvant effect on the CTL response. (A) Three-dimensional structure of ISG15 and the position of amino acids Y94 and Q100 (http://www.rcsb.org/structure/5TLA) (61). (B) Validation of equal expression of WT and Y94L_Q100D mutant ISG15 constructs in HeLa cells, as determined by Western blotting on total cell lysates. GAPDH was used as loading control. Results are representative of two analyses with individual samples. (C–E) Mice (n = 4 per group) were vaccinated with plasmid encoding E7SH, either combined with EV, ISG15 WT, or ISG15 Y94L_Q100D as outlined in Fig. 2. (C) Quantification of the E7-specific CD8+ T cell response over time in blood. (D) Quantification of the percentage of GZB+ cells among H-2Db/E749–57 tetramer+ cells at day 16. (E) Protein quantification (mean fluorescence intensity [MFI]) of GZB in H-2Db/E749–57 tetramer+ cells at day 16. Results are representative of two experiments. Statistical analysis was performed using one-way ANOVA and Tukey posttest and is indicated for day 16. **p < 0.01, ****p < 0.0001.
Mice were vaccinated as described above with a vector encoding E7SH, in conjunction with either empty vector, vector encoding ISG15 WT, or ISG15 Y94L_Q100D. The E7-specific CD8+ T cell response was followed in blood over time. The two mutations in ISG15 completely abrogated its ability to improve the E7-specific CD8+ T cell response, as determined by response magnitude (Fig. 5C), generation of E7-specific GZB+ cells (Fig. 5D), and intrinsic CTL quality, as defined by GZB expression on a per cell basis (Fig. 5E).
ISG15 promotes the CTL response via NK cells
In the same study based on human cells culture as referred above (33), extracellular recombinant ISG15 was shown to promote IFN-γ production from IL-12–primed NK cells by binding to LFA-1 (Fig. 6A). We therefore examined whether ISG15 acted via NK cells to improve the CTL response in our vaccination setting. For this purpose, NK cells were depleted with antiserum to glycolipid asialo GM1 (39) before and during the vaccination regimen. This depletion did not affect CD8+ T cells (Fig. 6B) and was very effective in NK cell depletion as measured in blood throughout the T cell response kinetics (Fig. 6C). Interestingly, the response to vaccination with E7SH in combination with ISG15 WT was significantly reduced upon NK cell depletion (Fig. 6D). In the NK cell–depleted setting, the E7-specific CD8+ T cell response was negligible until day 10, which was the peak of the response in the control setting. At day 13 postvaccination, the CD8+ T cell response reached its peak in the NK cell–depleted setting, which was lower than in the control setting. Furthermore, NK cell depletion led to a significant reduction in the frequency of SLECs and a corresponding increase in MPECs at day 10 of the response, but no differences in SLEC and MPEC frequencies were evident at day 13 in the control and NK cell–depleted settings (Fig. 6E). This indicates that the CTLs were capable of acquiring an effector phenotype in the absence of NK cells. Thus, in absence of NK cells, the effects of ISG15 on the magnitude and effector quality of the CTL response were delayed. However, the response was not fully abrogated as in case of the ISG15 Y94L_Q100D mutant, suggesting that ISG15 additionally stimulates the CD8+ T cell response in an NK cell–independent manner.
Discussion
In the current study, we have identified a tissue-wide response to free ISG15. Gene expression profiling demonstrated that expression of ISG15 ΔGG in the epidermis stimulated the migration of innate immune cells and endothelial cells, as well as extracellular matrix degradation and remodeling. Endogenous ISG15 has been implicated in migration of different cancer cell lines (43). Furthermore, recombinant ISG15 was found to induce neutrophil chemotaxis in vitro (44) and influx of DCs to the site of infection in vivo (34). The gene expression signature induced by free ISG15 in the skin suggested myeloid cell recruitment and proinflammatory activity according to the upregulation of the cell surface receptors Trem1 and Trem2 (45). Free ISG15 promoted inflammation also as judged by the upregulation of chemokines CXCL1 and CXCL3, which are produced by neutrophils and promote vascular leakage (46), as well as the upregulation of IL-21R, which responds to T cell–derived IL-21 by induction of proinflammatory cytokines (47). However, the gene signature also suggested epithelial tissue repair, as indicated by collagen synthesis and increased expression of IL-24 (48) and ICOSL (49). Its collective properties as a molecule that is released from cells following pathogen challenge and/or cell death and is able to mobilize and activate various leukocytes suggest that free ISG15 acts as an alarmin (50), as has been proposed earlier (51). Another criterion to classify ISG15 as an alarmin is that the candidate molecule is able to activate innate and adaptive immune responses, which we indeed show to be the case for free ISG15. In our gene set obtained from the vaccinated skin, we did not observe upregulation of IL-10 or IL-6 that were previously shown to be produced by human blood–derived monocytes in response to extracellular ISG15 (51). We also did not find IFN-γ that can be produced by human blood–derived lymphoid cells in response to extracellular ISG15 (31–33). This may be explained by the fact that we determine a response in the skin rather than peripheral blood.
Vaccination using an MHC class I epitope only is known to be ineffective in eliciting a CTL response. That is why MHC class II–binding helper epitopes are included in therapeutic vaccines to cancer and infectious disease (52). An optimal CTL response to vaccination relies on cross-presentation and activation of the DCs presenting the vaccine Ag by CD4+ T cells (53). We and others have shown previously in the DNA vaccination model presented in this study how inclusion of helper epitopes supports the CTL response (37, 38, 41). In the current study, we find that free ISG15 can improve the CTL response to the vaccine as an alternative to CD4+ T cell help. Moreover, we find that this adjuvant activity depends on residues Y94 and Q100 of mouse ISG15, whose equivalents were implicated in LFA-1 binding in a human in vitro system (33). We now show that the in vivo function of free ISG15 depends on these amino acids. The fact that free ISG15 needs these residues to bind to LFA-1 in vitro and to optimize the CTL response in vivo supports the concept that free ISG15 acts extracellularly as an immunomodulatory molecule. Remarkably, we found that free ISG15 promoted the vaccine-specific CTL response, at least in part, via NK cells. This is in further agreement with the study of Swaim et al. (33), which found free ISG15 to promote human NK cell functin in vitro.
In our study, free ISG15 promoted the clonal expansion of CD8+ T cells that respond to the vaccine, as well as their differentiation into SLECs and effector memory cells. The improved effector quality of CTLs primed in presence of free ISG15 was testified by the increased expression of GZB on a per-cell basis. CD4+ T cell help also promotes the CTL response in this manner, but the “help” delivered by ISG15 was CD4+ T cell independent, because we have shown that CD4+ T cells do not respond to the vaccine that we have employed in the current study (38). Our findings are consistent with those of other authors who found that free ISG15 promoted the CTL response in a setting with i.m. DNA vaccination (34) that is less robust than intraepidermal vaccination (54). In our study, we show the underlying mechanisms. As discussed above, free ISG15 caused a tissue alert and promoted the CTL response. Free ISG15 may have acted directly on CD8+ T cells to promote their response, but, more likely, given the myeloid cell activity induced, DC function was affected. The limiting factor in the CTL response in this model is the appropriate activation of DCs. Migratory cDC1 deliver the vaccine Ag from the skin to the dLNs, and a deficiency in their activation limits the CTL response to vaccination (42). We hypothesize that free ISG15 acts as an alarmin in the skin, promoting an inflammatory phenotype and creating the required signals for adequate activation of migratory cDC1s presenting the vaccine Ag.
Free ISG15 proved to act, at least in part, via NK cells to optimize the CTL response. NK cells and DCs are known to communicate in a bidirectional fashion: DCs can help activate NK cells and thereby promote innate immunity, and reciprocally, NK cells can help activate DCs and thereby promote adaptive immunity (55). Therefore, we hypothesize that ISG15 impacts NK cell–DC cross-talk and thereby creates an optimal CTL response. This cross-talk may take place in the skin, because NK cells have been found in both healthy and inflamed skin (56, 57), and migratory DCs are known to play a role in our vaccination model (42). Therefore, in our model, ISG15 may have promoted IFN-γ production by NK cells, which is known to enhance expression of costimulatory molecules and IL-12 by DCs (58). The (migratory) DCs would thereby be optimized for CTL priming. Because NK cell depletion did not fully abrogate the ISG15 effect on the CTL response in our model in contrast to mutation of Y94 and Q100, ISG15 likely acts as an immunostimulatory ligand on other cells as well. NKT cells are a good candidate, because this cell type was not depleted by our strategy, and reciprocal NKT cell–DC activation has been described (59). We conclude that ISG15 is part of an innate route to promote the CTL response as an alternative or supplement to CD4+ T cell help (53). An earlier study reports that DC activation promoted by NK cell–derived IFN-γ could replace CD4+ T cell help in inducing a protective antitumor CD8+ T cell response (60) that supports this concept.
Our data tie together various independent observations on free ISG15 function that were primarily made in vitro and, even in human cells, into one concept that explains how free ISG15 can act as an alarmin and adjuvant to bridge innate and adaptive immunity. Our study relies on deliberate expression of ISG15, which has likely widened the window to observe this immune stimulatory function of ISG15 and its underlying mechanisms. By binding to USP18 and stabilizing it, intracellular free ISG15 promotes negative feedback on the type I IFN response in human, but not in mouse (7, 26). The question is whether this process would limit the application of free ISG15 as immune adjuvant in human. We show, in this study, that local and transient production of free ISG15 in the mouse skin is beneficial for the T cell response to therapeutic vaccination. In human, such a setting might be created as well, formulating free ISG15 and Ag either in pDNA or in protein form. In such vaccine-adjuvant settings, free ISG15 in proteinaceous form can only act extracellularly, and free ISG15 encoded by pDNA is expressed locally and transiently. For these reasons, the vaccine-adjuvant effect of free ISG15 may be reproducible in human.
Supplementary Material
Acknowledgments
We thank Astrid Bovens and the personnel of the experimental animal, flow cytometry, and genomics core facilities of the Netherlands Cancer Institute for expert assistance.
This work was supported by The Netherlands Organisation for Health Research and Development Gravitation Subgrant 00002 from the Institute for Chemical Immunology.
Data have been deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE139469.
The online version of this article contains supplemental material.
- BM
- bone marrow
- DC
- dendritic cell
- dLN
- draining lymph node
- GZB
- granzyme B
- HPV
- human papilloma virus
- IPA
- Ingenuity Pathway Analysis
- ISG
- IFN-stimulated gene
- MPEC
- memory precursor effector cell
- pDNA
- plasmid DNA
- SLEC
- short-lived effector cell
- WT
- wild-type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Schneider W. M., Chevillotte M. D., Rice C. M. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32: 513–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Narasimhan J., Wang M., Fu Z., Klein J. M., Haas A. L., Kim J. J. 2005. Crystal structure of the interferon-induced ubiquitin-like protein ISG15. J. Biol. Chem. 280: 27356–27365. [DOI] [PubMed] [Google Scholar]
- 3.Perng Y. C., Lenschow D. J. 2018. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 16: 423–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sampson D. L., Fox B. A., Yager T. D., Bhide S., Cermelli S., McHugh L. C., Seldon T. A., Brandon R. A., Sullivan E., Zimmerman J. J., et al. 2017. A four-biomarker blood signature discriminates systemic inflammation due to viral infection versus other etiologies. Sci. Rep. 7: 2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogunovic D., Byun M., Durfee L. A., Abhyankar A., Sanal O., Mansouri D., Salem S., Radovanovic I., Grant A. V., Adimi P., et al. 2012. Mycobacterial disease and impaired IFN-γ immunity in humans with inherited ISG15 deficiency. Science 337: 1684–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radoshevich L., Impens F., Ribet D., Quereda J. J., Nam Tham T., Nahori M. A., Bierne H., Dussurget O., Pizarro-Cerdá J., Knobeloch K. P., Cossart P. 2015. ISG15 counteracts Listeria monocytogenes infection. eLife 4: e06848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dzimianski J. V., Scholte F. E. M., Bergeron É., Pegan S. D. 2019. ISG15: it’s complicated. J. Mol. Biol. 431: 4203–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hermann M., Bogunovic D. 2017. ISG15: in sickness and in health. Trends Immunol. 38: 79–93. [DOI] [PubMed] [Google Scholar]
- 9.Loeb K. R., Haas A. L. 1992. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J. Biol. Chem. 267: 7806–7813. [PubMed] [Google Scholar]
- 10.Yuan W., Krug R. M. 2001. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 20: 362–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim K. I., Giannakopoulos N. V., Virgin H. W., Zhang D. E. 2004. Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol. Cell. Biol. 24: 9592–9600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao C., Beaudenon S. L., Kelley M. L., Waddell M. B., Yuan W., Schulman B. A., Huibregtse J. M., Krug R. M. 2004. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proc. Natl. Acad. Sci. USA 101: 7578–7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dastur A., Beaudenon S., Kelley M., Krug R. M., Huibregtse J. M. 2006. Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J. Biol. Chem. 281: 4334–4338. [DOI] [PubMed] [Google Scholar]
- 14.Ketscher L., Basters A., Prinz M., Knobeloch K. P. 2012. mHERC6 is the essential ISG15 E3 ligase in the murine system. Biochem. Biophys. Res. Commun. 417: 135–140. [DOI] [PubMed] [Google Scholar]
- 15.Malakhov M. P., Malakhova O. A., Kim K. I., Ritchie K. J., Zhang D. E. 2002. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 277: 9976–9981. [DOI] [PubMed] [Google Scholar]
- 16.Durfee L. A., Lyon N., Seo K., Huibregtse J. M. 2010. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol. Cell 38: 722–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu M., Li X. L., Hassel B. A. 2003. Proteasomes modulate conjugation to the ubiquitin-like protein, ISG15. J. Biol. Chem. 278: 1594–1602. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X., Bogunovic D., Payelle-Brogard B., Francois-Newton V., Speer S. D., Yuan C., Volpi S., Li Z., Sanal O., Mansouri D., et al. 2015. Human intracellular ISG15 prevents interferon-α/β over-amplification and auto-inflammation. Nature 517: 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan J. B., Arimoto K., Motamedchaboki K., Yan M., Wolf D. A., Zhang D. E. 2015. Identification and characterization of a novel ISG15-ubiquitin mixed chain and its role in regulating protein homeostasis. Sci. Rep. 5: 12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basters A., Knobeloch K. P., Fritz G. 2018. USP18 - a multifunctional component in the interferon response. Biosci. Rep. 38: BSR20180250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ketscher L., Hannß R., Morales D. J., Basters A., Guerra S., Goldmann T., Hausmann A., Prinz M., Naumann R., Pekosz A., et al. 2015. Selective inactivation of USP18 isopeptidase activity in vivo enhances ISG15 conjugation and viral resistance. Proc. Natl. Acad. Sci. USA 112: 1577–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez M. R., Monte K., Thackray L. B., Lenschow D. J. 2014. ISG15 functions as an interferon-mediated antiviral effector early in the murine norovirus life cycle. J. Virol. 88: 9277–9286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Werneke S. W., Schilte C., Rohatgi A., Monte K. J., Michault A., Arenzana-Seisdedos F., Vanlandingham D. L., Higgs S., Fontanet A., Albert M. L., Lenschow D. J. 2011. ISG15 is critical in the control of Chikungunya virus infection independent of UbE1L mediated conjugation. PLoS Pathog. 7: e1002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meuwissen M. E., Schot R., Buta S., Oudesluijs G., Tinschert S., Speer S. D., Li Z., van Unen L., Heijsman D., Goldmann T., et al. 2016. Human USP18 deficiency underlies type 1 interferonopathy leading to severe pseudo-TORCH syndrome. J. Exp. Med. 213: 1163–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malakhova O. A., Kim K. I., Luo J. K., Zou W., Kumar K. G., Fuchs S. Y., Shuai K., Zhang D. E. 2006. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 25: 2358–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Speer S. D., Li Z., Buta S., Payelle-Brogard B., Qian L., Vigant F., Rubino E., Gardner T. J., Wedeking T., Hermann M., et al. 2016. ISG15 deficiency and increased viral resistance in humans but not mice. Nat. Commun. 7: 11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D’Cunha J., Ramanujam S., Wagner R. J., Witt P. L., Knight E., Jr., Borden E. C. 1996. In vitro and in vivo secretion of human ISG15, an IFN-induced immunomodulatory cytokine. J. Immunol. 157: 4100–4108. [PubMed] [Google Scholar]
- 28.Knight E., Jr., Cordova B. 1991. IFN-induced 15-kDa protein is released from human lymphocytes and monocytes. J. Immunol. 146: 2280–2284. [PubMed] [Google Scholar]
- 29.Padovan E., Terracciano L., Certa U., Jacobs B., Reschner A., Bolli M., Spagnoli G. C., Borden E. C., Heberer M. 2002. Interferon stimulated gene 15 constitutively produced by melanoma cells induces e-cadherin expression on human dendritic cells. Cancer Res. 62: 3453–3458. [PubMed] [Google Scholar]
- 30.Taylor J. L., D’Cunha J., Tom P., O’Brien W. J., Borden E. C. 1996. Production of ISG-15, an interferon-inducible protein, in human corneal cells. J. Interferon Cytokine Res. 16: 937–940. [DOI] [PubMed] [Google Scholar]
- 31.Recht M., Borden E. C., Knight E., Jr 1991. A human 15-kDa IFN-induced protein induces the secretion of IFN-gamma. J. Immunol. 147: 2617–2623. [PubMed] [Google Scholar]
- 32.D’Cunha J., Knight E., Jr., Haas A. L., Truitt R. L., Borden E. C. 1996. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc. Natl. Acad. Sci. USA 93: 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swaim C. D., Scott A. F., Canadeo L. A., Huibregtse J. M. 2017. Extracellular ISG15 signals cytokine secretion through the LFA-1 integrin receptor. Mol. Cell 68: 581–590.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villarreal D. O., Wise M. C., Siefert R. J., Yan J., Wood L. M., Weiner D. B. 2015. Ubiquitin-like molecule ISG15 acts as an immune adjuvant to enhance antigen-specific CD8 T-cell tumor immunity. Mol. Ther. 23: 1653–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Napolitano A., van der Veen A. G., Bunyan M., Borg A., Frith D., Howell S., Kjaer S., Beling A., Snijders A. P., Knobeloch K. P., Frickel E. M. 2018. Cysteine-reactive free ISG15 generates IL-1β-producing CD8α+ dendritic cells at the site of infection. J. Immunol. 201: 604–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osiak A., Utermöhlen O., Niendorf S., Horak I., Knobeloch K. P. 2005. ISG15, an interferon-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis virus. Mol. Cell. Biol. 25: 6338–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oosterhuis K., Aleyd E., Vrijland K., Schumacher T. N., Haanen J. B. 2012. Rational design of DNA vaccines for the induction of human papillomavirus type 16 E6- and E7-specific cytotoxic T-cell responses. Hum. Gene Ther. 23: 1301–1312. [DOI] [PubMed] [Google Scholar]
- 38.Ahrends T., Bąbała N., Xiao Y., Yagita H., van Eenennaam H., Borst J. 2016. CD27 agonism plus PD-1 blockade recapitulates CD4+ T-cell help in therapeutic anticancer vaccination. Cancer Res. 76: 2921–2931. [DOI] [PubMed] [Google Scholar]
- 39.Kasai M., Iwamori M., Nagai Y., Okumura K., Tada T. 1980. A glycolipid on the surface of mouse natural killer cells. Eur. J. Immunol. 10: 175–180. [DOI] [PubMed] [Google Scholar]
- 40.Love M. I., Huber W., Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahrends T., Spanjaard A., Pilzecker B., Bąbała N., Bovens A., Xiao Y., Jacobs H., Borst J. 2017. CD4+ T cell help confers a cytotoxic T cell effector program including coinhibitory receptor downregulation and increased tissue invasiveness. Immunity 47: 848–861.e5. [DOI] [PubMed] [Google Scholar]
- 42.Bąbała N., Bovens A., de Vries E., Iglesias-Guimarais V., Ahrends T., Krummel M. F., Borst J., Bins A. D. 2018. Subcellular localization of antigen in keratinocytes dictates delivery of CD4+ T-cell help for the CTL response upon therapeutic DNA vaccination into the skin. Cancer Immunol. Res. 6: 835–847. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y. L., Wu W. L., Jang C. W., Yen Y. C., Wang S. H., Tsai F. Y., Shen Y. Y., Chen Y. W. 2019. Interferon-stimulated gene 15 modulates cell migration by interacting with Rac1 and contributes to lymph node metastasis of oral squamous cell carcinoma cells. Oncogene 38: 4480–4495. [DOI] [PubMed] [Google Scholar]
- 44.Owhashi M., Taoka Y., Ishii K., Nakazawa S., Uemura H., Kambara H. 2003. Identification of a ubiquitin family protein as a novel neutrophil chemotactic factor. Biochem. Biophys. Res. Commun. 309: 533–539. [DOI] [PubMed] [Google Scholar]
- 45.Ford J. W., McVicar D. W. 2009. TREM and TREM-like receptors in inflammation and disease. Curr. Opin. Immunol. 21: 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DiStasi M. R., Ley K. 2009. Opening the flood-gates: how neutrophil-endothelial interactions regulate permeability. Trends Immunol. 30: 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monteleone G., Pallone F., Macdonald T. T. 2009. Interleukin-21 (IL-21)-mediated pathways in T cell-mediated disease. Cytokine Growth Factor Rev. 20: 185–191. [DOI] [PubMed] [Google Scholar]
- 48.Sa S. M., Valdez P. A., Wu J., Jung K., Zhong F., Hall L., Kasman I., Winer J., Modrusan Z., Danilenko D. M., Ouyang W. 2007. The effects of IL-20 subfamily cytokines on reconstituted human epidermis suggest potential roles in cutaneous innate defense and pathogenic adaptive immunity in psoriasis. J. Immunol. 178: 2229–2240. [DOI] [PubMed] [Google Scholar]
- 49.Maeda S., Fujimoto M., Matsushita T., Hamaguchi Y., Takehara K., Hasegawa M. 2011. Inducible costimulator (ICOS) and ICOS ligand signaling has pivotal roles in skin wound healing via cytokine production. Am. J. Pathol. 179: 2360–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang D., Wei F., Tewary P., Howard O. M., Oppenheim J. J. 2013. Alarmin-induced cell migration. Eur. J. Immunol. 43: 1412–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dos Santos P. F., Van Weyenbergh J., Delgobo M., Oliveira Patricio D., Ferguson B. J., Guabiraba R., Dierckx T., Menezes S. M., Báfica A., Mansur D. S. 2018. ISG15-induced IL-10 is a novel anti-inflammatory myeloid axis disrupted during active tuberculosis. J. Immunol. 200: 1434–1442. [DOI] [PubMed] [Google Scholar]
- 52.Melief C. J., van Hall T., Arens R., Ossendorp F., van der Burg S. H. 2015. Therapeutic cancer vaccines. J. Clin. Invest. 125: 3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borst J., Ahrends T., Bąbała N., Melief C. J. M., Kastenmüller W. 2018. CD4+ T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 18: 635–647. [DOI] [PubMed] [Google Scholar]
- 54.Verstrepen B. E., Bins A. D., Rollier C. S., Mooij P., Koopman G., Sheppard N. C., Sattentau Q., Wagner R., Wolf H., Schumacher T. N., et al. 2008. Improved HIV-1 specific T-cell responses by short-interval DNA tattooing as compared to intramuscular immunization in non-human primates. Vaccine 26: 3346–3351. [DOI] [PubMed] [Google Scholar]
- 55.Pampena M. B., Levy E. M. 2015. Natural killer cells as helper cells in dendritic cell cancer vaccines. Front. Immunol. 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buentke E., Heffler L. C., Wilson J. L., Wallin R. P., Löfman C., Chambers B. J., Ljunggren H. G., Scheynius A. 2002. Natural killer and dendritic cell contact in lesional atopic dermatitis skin--Malassezia-influenced cell interaction. J. Invest. Dermatol. 119: 850–857. [DOI] [PubMed] [Google Scholar]
- 57.Ebert L. M., Meuter S., Moser B. 2006. Homing and function of human skin gammadelta T cells and NK cells: relevance for tumor surveillance. J. Immunol. 176: 4331–4336. [DOI] [PubMed] [Google Scholar]
- 58.Ferlazzo G., Morandi B. 2014. Cross-talks between natural killer cells and distinct subsets of dendritic cells. Front. Immunol. 5: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fujii S., Shimizu K., Smith C., Bonifaz L., Steinman R. M. 2003. Activation of natural killer T cells by alpha-galactosylceramide rapidly induces the full maturation of dendritic cells in vivo and thereby acts as an adjuvant for combined CD4 and CD8 T cell immunity to a coadministered protein. J. Exp. Med. 198: 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adam C., King S., Allgeier T., Braumüller H., Lüking C., Mysliwietz J., Kriegeskorte A., Busch D. H., Röcken M., Mocikat R. 2005. DC-NK cell cross talk as a novel CD4+ T-cell-independent pathway for antitumor CTL induction. Blood 106: 338–344. [DOI] [PubMed] [Google Scholar]
- 61.Daczkowski C. M., Dzimianski J. V., Clasman J. R., Goodwin O., Mesecar A. D., Pegan S. D. 2017. Structural insights into the interaction of coronavirus papain-like proteases and interferon-stimulated gene product 15 from different species. J. Mol. Biol. 429: 1661–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.