Abstract
Asthma is a common chronic respiratory disease affecting more than 300 million people worldwide. Clinical features of asthma and its immunological and molecular etiology vary significantly among patients. An understanding of the complexities of asthma has evolved to the point where precision medicine approaches, including microbiome analysis, are being increasingly recognized as an important part of disease management. Lung and gut microbiota play several important roles in the development, regulation, and maintenance of healthy immune responses. Dysbiosis and subsequent dysregulation of microbiota-related immunological processes affect the onset of the disease, its clinical characteristics, and responses to treatment. Bacteria and viruses are the most extensively studied microorganisms relating to asthma pathogenesis, but other microbes, including fungi and even archaea, can potently influence airway inflammation. This review focuses on recently discovered connections between lung and gut microbiota, including bacteria, fungi, viruses, and archaea, and their influence on asthma.
Dysregulation of microbiota-related immunological processes affects the onset of asthma, its clinical characteristics, and responses to treatment. Finlay et al. review connections between gut and lungs microbiota and its influence on allergic airway inflammation. This review covers not only the most well-known populations of microbes in the context of asthma—bacteria—but also discusses viruses, fungi, and archaea.
Main Text
Asthma is a chronic disease, currently affecting more than 300 million people worldwide and anticipated to increase to 400 million by 2025. Approximately 250,000 asthma-related deaths are reported yearly, many of which are avoidable (Christiansen and Zuraw, 2019). The pathogenesis of asthma is still not well understood, but the disease has been linked to several genetic, environmental, infectious, and nutritional factors. In the early 1900s, asthma was described as a single disease, but our understanding of its complexity has evolved to the point where precision medicine approaches, including microbiome analysis, are starting to emerge for its diagnosis and treatment (Desai and Oppenheimer, 2016, Taylor et al., 2018).
Given the huge number of different microorganisms that colonize the human body, their intimate contact with human cells and their multiple influences on the host, the human microbiome is no longer considered inert. Microbial communities of the respiratory and gastrointestinal tracts are essential in maintaining human health. The healthy adult microbiota, which includes bacteria, viruses, fungi, and even archaea, is diverse, stable, resistant, and resilient, meaning it is rich in many different species and can normally return to the pre-perturbation state (Levy et al., 2017).
Microbial dysbiosis within the lungs and gut in asthma can be caused by many factors related to lifestyle and environmental exposures. Imbalance between the symbiotic and pathological bacterial strains in the gut and lung may lead to altered immune development and inappropriate inflammatory responses, but it is still unclear whether the imbalance is the cause or effect of disease. This review will address the question and focus on the role of the lung and gut microbiota in the development and maintenance of respiratory health and its disturbances in different subtypes of asthma.
Asthma Is Not a Single Disease
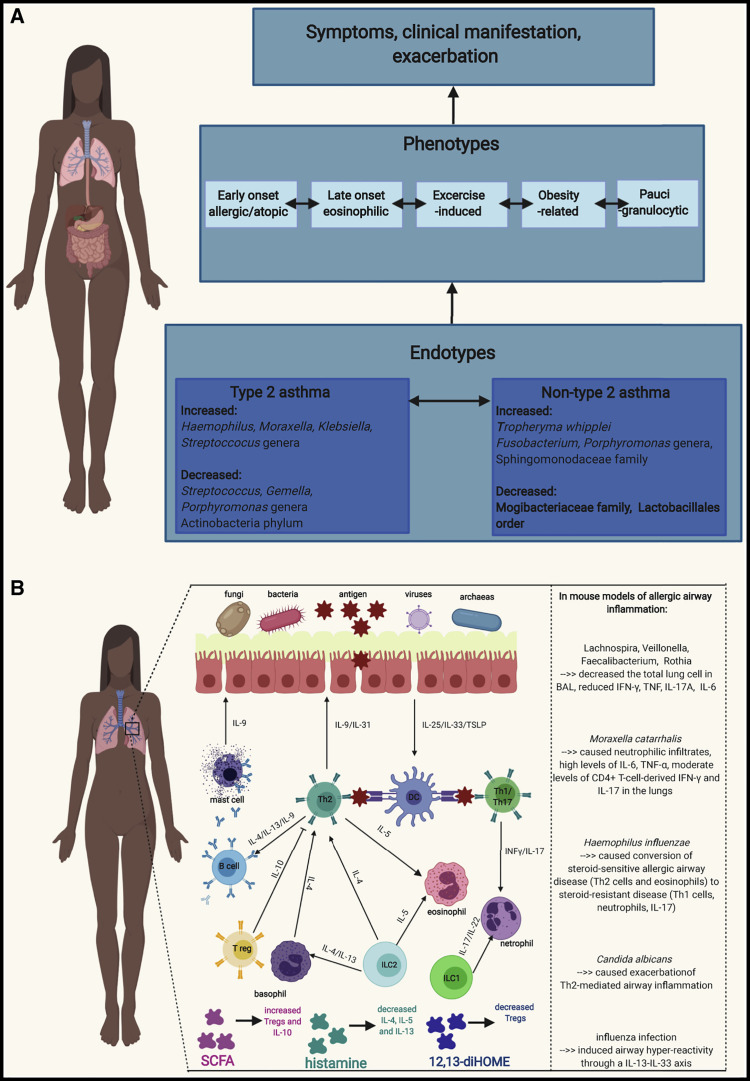
Asthma is characterized by a large spectrum of clinically observed phenotypes and an even larger range of underlying molecular and immunological mechanisms, called endotypes (Wenzel, 2012, Sugita et al., 2018, Boutin et al., 2017). Currently, the best understanding of these complex endotypes as mechanistic pathogenetic processes lays in their dualistic division as type 2 or non-type 2 asthma (Agache and Akdis, 2016) (Figure 1 A). However, often endotypes can co-exist and/or include other mixed processes, which cannot be easily categorized (Kuo et al., 2017, Agache and Akdis, 2016, Cosío et al., 2017). Less well-defined forms of asthma, situated between clinical phenotypes and molecular endotypes, include inflammatory phenotypes of eosinophilic, neutrophilic, mixed, and paucigranulocytic asthma (Wang et al., 2011, Tan et al., 2019).
Figure 1.
Schematic Representation of Asthma Phenotypes and Endotypes with Differences in Microbiota between Endotypes and Simplified Cellular Mechanisms Involved in Asthma and Its Correlation with Microbiota
(A) Schematic representation of asthma phenotypes and endotypes with differences in microbiota between endotypes. Endotypes classify asthma based on pathophysiological mechanisms, whereas phenotype refers to the clinical and morphological description of disease. The two best-described asthma endotypes are the type 2 endotype, which includes mostly Th2 cell responses, and the non-type 2 endotype. However, endotypes can co-exist and/or include other mixed processes. Type 2 asthma is usually observed in the clinic as early-onset allergic asthma, late-onset eosinophilic asthma, or exercise-induced asthma. Non-type 2 asthma mechanisms are observed usually in neutrophilic, obesity-related, and paucigranulocytic phenotypes.
(B) Simplified cellular mechanisms involved in asthma and its correlation with microbiota. Th2-cell-driven inflammation engages Th 2 cells, type 2 innate lymphoid cells, T follicular helper cells, type 2 B cells, eosinophils, and mast cells and results in increased amounts of IL-4, IL-5, IL-9, IL-13, prostaglandin D2, and CCR8 in sputum, BAL, serum, and bronchial biopsies. Non-type 2 asthma is characterized by infiltration of Th1 and Th17 cells, neutrophils, and the presence of type I interferons, NLRP3 inflammasome, and IL-1β and IL-17 cytokines.
Type 2 asthma is usually observed in the clinic as early-onset allergic asthma, late-onset eosinophilic asthma, or exercise-induced asthma (Fahy, 2015). These patients are usually allergic to common aeroallergens, have high blood eosinophilia, increased periostin, and high exhaled nitric oxide. They tend to respond well to treatment with glucocorticosteroids or novel biologicals, such as anti-immunoglobulin E (IgE), anti-interleukin-5 (IL-5), or anti-IL-4 and anti-IL-13 approaches (Castro et al., 2018, Corren et al., 2019). However, within high eosinophilic asthma, there are also those whose eosinophilia is resistant to steroid treatment and others with persistent severe asthma, who demonstrate a rapid decline in lung function, enhanced airway remodeling, and frequent exacerbations (Chung et al., 2014, Pavlidis et al., 2019). Mechanistically, type 2 inflammation engages T helper (Th) type 2 cells, type 2 innate lymphoid cells, T follicular helper cells, type 2 B cells, eosinophils, mast cells, and “signature” type 2 mediators including the cytokines IL-4, IL-5, and IL-13 as well as prostaglandin D2 (Ray and Kolls, 2017) (Figure 1B).
Non-type 2 asthma, observed more often in obesity-related asthma, neutrophilic asthma, and paucigranulocytic asthma (Kuo et al., 2017, Cosío et al., 2017, Ray and Kolls, 2017), is usually connected with poor responsiveness to steroids and more severe phenotypes, even in childhood (Ramratnam et al., 2017). Non-type 2 inflammation is characterized by infiltration of Th1 and Th17 cells and neutrophils, the presence of type I interferons, NLRP3 inflammasome activation, and an IL-1β and IL-17 signature (Rossios et al., 2018, Kim et al., 2017, Tan et al., 2019). The effect of microbiota dysbiosis on asthma heterogeneity seems intuitive, but, until recently (Taylor et al., 2018, Boutin et al., 2017), lung and gut microbiome composition have not been addressed in studies on clinical or inflammatory phenotypes and endotypes of asthma.
Healthy Lungs Are Not Sterile
Microbes had already inhabited every environmental niche on earth, including the most extreme, prior to humans. Therefore, it is not surprising that all surfaces of the human body, including the upper respiratory tract (URT) and lower respiratory tract (LRT), are populated with microbial communities. Yet, even upon initiation of the Human Microbiome Project in 2007 (Turnbaugh et al., 2007), lungs were not sampled, partially due to the existing dogma at the time that lungs are sterile. This dogma significantly delayed the initiation of studies on the lung microbiota relative to the gut microbiota (Dickson et al., 2016, Moffatt and Cookson, 2017) and likely persisted for several reasons: (1) healthy lungs were often not examined by invasive bronchoscopy techniques due to ethical concerns; (2) culture-based techniques were limited to known pathogens; and (3) bronchoscopy techniques were suspected to carry contamination from the oropharynx or nasal cavity. However, a pioneer publication using culture-independent 16S rRNA sequencing revealed differences in the microbial communities found in various locations of the healthy respiratory tract relative to those found in the respiratory tract of patients with asthma and chronic obstructive pulmonary disease (COPD) (Hilty et al., 2010). Numerous other studies confirming this observation have followed, elucidating the special role of the airway microbiota in health (Man et al., 2017) and disease (Wypych et al., 2019).
The respiratory system in humans consists of an URT, starting with the nasal cavity followed by the nasopharynx, oropharynx, and larynx above the vocal cords; and LRT, continuing in the larynx below the vocal cords and including the trachea, bronchi, bronchioles, and countless alveoli of the lungs. The 70 m2 (Weibel, 1979) surface area of the human respiratory tract is inhabited by the microbiota, differing in quantities and qualities along the tract in response to niche-specific conditions (Man et al., 2017, Huffnagle et al., 2017). The movement of air and particles, including microbiota, in the respiratory system is bidirectional. The composition of the lung microbiome in each site is thus a consequence of the balance reached between migration to the airways, elimination from the airway, site-specific immune processes (e.g., presence or absence of inflammation) and the growth rates of the microbes (Huffnagle et al., 2017). Highlighting the role of the URT as a gatekeeper to respiratory health (Man et al., 2017), microbiota enter the lung mainly via microaspiration from the nasopharynx and the more densely and diversely populated oropharynx. Microbes can also enter the lung via dispersion along the mucosal surface (Bassis et al., 2015, Dickson et al., 2017).
Bacteria of the healthy URT include the genera Staphylococcus, Propionibacterium, Dolosigranulum, Corynebacterium, Moraxella, Streptococcus, Haemophilus, Rothia, Veillonella, Prevotella, and Leptotrichia (Zhou et al., 2014, Frank et al., 2010, Wos-Oxley et al., 2016). In addition to bacteria, several viruses and fungi have been found in the URT of healthy people. Human rhinovirus (RV), bocavirus, adenovirus, coronavirus, polymavirus, and Anneloviridae family viruses have been identified in the URT of large proportions of healthy asymptomatic children using PCR-based methods and metagenomics, even in geographically isolated locations (van den Bergh et al., 2012, Bogaert et al., 2011, Wylie et al., 2012, Altan et al., 2019). Detection frequency of these viruses in many studies is not different in the URT of asymptomatic controls and children hospitalized due to the respiratory tract infections, in contrast to respiratory syncytial virus (RSV), human metapneumovirus (hMPV), parainfluenza viruses (PIVs), and influenza, being causally associated with hospitalizations, lower respiratory symptoms, and pneumonia (Singleton et al., 2010, Sarna et al., 2018, Bhuiyan et al., 2019). These data suggest that in the cases of these “commonly carried” URT viruses, not only their presence, but rather total viral load (Jansen et al., 2011) or complex interactions between viruses and bacteria (van den Bergh et al., 2012) or other environmental (e.g., allergen) and intrinsic host factors (e.g., sensitization) (Rubner et al., 2017) determine the occurrence of disease in the URT or LRT. Interestingly, in healthy adults these viruses are detected less frequently than in asymptomatic children, suggesting age-dependent and perhaps ecosystem-dependent changes in viral load of the URT (Sundell et al., 2019). Aspergillus, Penicillium, Candida, and Alternaria are fungal genera found in the URT of healthy people (Charlson et al., 2012).
In the healthy lungs of the LRT, there are six dominant bacterial phyla: Firmicutes (including genera Streptococcus and Veillonella), Bacteroides (including genus Prevotella), Proteobacteria, Fusobacteria, Acidobacteria, and Actinobacteria (including Tropheryma whipplei) (Segal et al., 2016, Dickson et al., 2015, Dickson et al., 2017). The virome of the healthy lungs contains members of the Anelloviridae family and a high frequency of various bacteriophages (Abbas et al., 2017, Willner et al., 2009, Jankauskaitė et al., 2018), but their abundance is greater in the context of immunosuppression (Young et al., 2015) and/or chronic disease (Freer et al., 2018), suggesting that immunocompetence is a prerequisite to the low abundance and biodiversity of LRT virome. The lung mycobiome is composed of Eremothecium, Systenostrema, and Malassezia genera and the Davidiellaceae family (Charlson et al., 2012, van Woerden et al., 2013).
The majority of cited studies have used genetic techniques to identify microorganisms’ taxa, and the results were not usually confirmed by culture-dependent methods. However, sequencing allows identification of more diverse microbes and is not limited by the viability or ability to grow the microorganisms in vitro (Dickson et al., 2015). Most studies underscore that the healthy microbiome is rather transient in the lungs at steady state. In contrast, during disease, when inflammatory conditions enable the growth of certain bacteria, it can lead to dysbiosis and subsequent colonization with pathogens.
The healthy microbiome of the URT and LRT plays several important roles in the development and maintenance of homeostasis of the RT and whole organism. The composition of the microbiome at all mucosal sites changes dynamically from the first days of life, shaping the establishment of proper interactions between the microbiota and human body and constituting the critical “window of opportunity” for determining future health or disease (Pattaroni et al., 2018, Pammi et al., 2019, Stiemsma and Turvey, 2017). Directly after birth, the skin, gut, nasopharynx, and mouth microbial communities are undifferentiated and colonized by bacterial species reflective of delivery mode (de Steenhuijsen Piters et al., 2015). During birth and in the first days of life, maternal vaginal and skin microbiota only transiently transfer to the infant, whereas the mother’s gut microbiota becomes the primary source of microbial strains acquired by the baby by the age of four months (Ferretti et al., 2018, Yassour et al., 2018). The separation between the URT and LRT in the context of early life is important as these two anatomical sites represent different habitats. The URT experiences constant exposure to the external environment and its accompanying conditions from the first moment of life, which shapes the URT microbiota. However, it has been demonstrated that by 24 h after birth bacterial DNA can be detected in the LRT of neonates (Wypych et al., 2019). The transient nature of the LRT microbiota at the steady state is determined by the balance between microbiota that enter from the URT and microbiota that are eliminated by host immunity; therefore, URT samples cannot fully reflect on the interactions that occur in the LRT (Wypych et al., 2019). The healthy respiratory tract microbiome plays also important roles in conferring colonization resistance. The term “colonization resistance” refers to the ability of a healthy and timely microbiota to actively suppress or compete with pathogens to inhibit their outgrowth, as has been shown in vitro (Bomar et al., 2016) and in vivo (de Steenhuijsen Piters et al., 2019, Iwase et al., 2010).
The overall influence of the healthy airway microbiota on host immunity reflects cumulative effects of live microbes and components of their cells or metabolites on local and systemic innate and adaptive immune processes in the host (Segal et al., 2016, Wang et al., 2013). If the process of healthy and timely colonization is disrupted, early-life dysbiosis of the gut and lung microbiota becomes an important risk factor for the development of many respiratory diseases (Biesbroek et al., 2014, Teo et al., 2015). The mechanisms involved likely operate at different modes e.g., via inadequate training of host immunity (Gollwitzer et al., 2014), inefficient colonization resistance (de Steenhuijsen Piters et al., 2019), and/or improper lung morphogenesis (Yun et al., 2014). During in utero development and later in life, there are several direct and indirect factors that affect healthy microbiota colonization and subsequent host-microbiota interactions. These factors include mother’s health status (Stokholm et al., 2018), diet (Frei et al., 2012, Barcik et al., 2015), transfer of maternal antibodies and microbial molecules (Gomez de Agüero et al., 2016, Koch et al., 2016), mode of delivery (Bosch et al., 2016), and breastfeeding and early-life environment (day care, siblings [Wolsk et al., 2016], pets at home [Sitarik et al., 2018, Tun et al., 2017], traditional farm-like dust at home [Stein et al., 2016, Kirjavainen et al., 2019], and smoking and antibiotic and other drug usage in pregnancy and early childhood [Mitre et al., 2018, Loewen et al., 2018]).
Gut-Lung Axis Impacts on Respiratory and Gut Health
The human gut starts at the oral cavity, continues through the esophagus, stomach, small intestine, colon, and ends at the rectum. Its 150–200 m2 enormous surface area provides microbes opportunities for colonization or transient occupation. Indeed, the human gut is inhabited by between 100 thousand and 100 billion bacteria per mL of luminal content depending on the region and therefore is the most densely colonized organ (Sender et al., 2016). Bacteria within the gut confer several functions to the host, including vitamin production, absorption of ions, protection against pathogens, histological development, enhanced immune functions, and fermentation of food (Hillman et al., 2017). They are typically dominated by five phyla; Bacteroidetes, Firmicutes, Proteobacteria, Actinobacteria, and Tenericutes (Desai and Oppenheimer, 2016, Almeida et al., 2019), whereas Bacteroides, Faecalibacterium, and Bifidobacterium are the most prevalent genera in healthy adults (Marsland et al., 2015). The oral cavity is mainly colonized by bacteria from the Streptococcaceae, Pasteurellaceae, Veillonellaceae, Prevotellaeace, and Neisseriaceae families and Gemella genus; the stomach mostly contains bacteria from the Lactobacillaceae family; the small intestine is dominated by Lactobacillaceae, Enterobacteriaceae, and Streptococcaceae; and the most densely colonized large intestine contains bacteria from the families of Enterococcaceae, Clostridiaceae, Enterobacteriaceae, Bacteroidaceae, Bifidobacteriaceae, Fusobacteriaceae, Lactobacillaceae, Peptostreptococcaceae, Peptococcaceae, Prevotellaeace, Lachnospiraceae, Ruminococcaceae, Rikenelleace, and the phylum Verrucomimicrobia (Hillman et al., 2017, Pereira and Berry, 2017). Candida, Saccharomyces, Malassezia, and Cladosporium are among the most commonly and consistently identified fungal genera in human gut mycobiota studies (Hoffmann et al., 2013, Strati et al., 2016, Auchtung et al., 2018, Mahnic and Rupnik, 2018, Iliev and Leonardi, 2017). Besides bacteria and fungi, the human gut microbiota is additionally composed of viruses (primarily phages) and archaea (mainly Methanobrevibacter smithii) (Lozupone et al., 2012).
A connection between the lungs and gut has been repeatedly demonstrated in both human and mouse studies. Inducible bronchus-associated lymphoid tissue (iBALT) and gut-associated lymphoid tissue (GALT), which are part of the mucosal-associated lymphoid tissue, are related in morphology and function, which includes the regulation of local immune responses (Elmore, 2006). The main immunological functions of GALT and iBALT include production and secretion of IgA at mucosal surfaces, as well as Th cell and cytotoxic (Tc) responses (Cesta, 2006). Regulation of local immunity in GALT occurs in Peyer’s patches and mesenteric lymph nodes. Terminal transformation of B cells to plasma cells able to secrete antibodies is finalized in the lamina propria, where memory and effector T and B cells reside. Importantly, 80% of activated B cells in adults are found in gut mucosal tissue (Brandtzaeg, 2010).
The concept of the gut-lung connection was born out of the observation that different lung diseases can be influenced by intestinal microenvironment changes and vice versa. The microbiota is an important factor responsible for interactions between these two sites in asthma (Marsland et al., 2015). In addition, the gut microbiota of patients with severe bacterial pneumonia (Chung, 2017), cystic fibrosis (Marsland et al., 2015), and influenza differs from that of healthy controls (Qin et al., 2015). Many studies show that early life is the most important period during which microbiota dysbiosis in the gut may lead to the development of many respiratory diseases, as the gut microbiota has a significant influence on immune cell maturation and resistance to pathogens (Sokolowska et al., 2018).
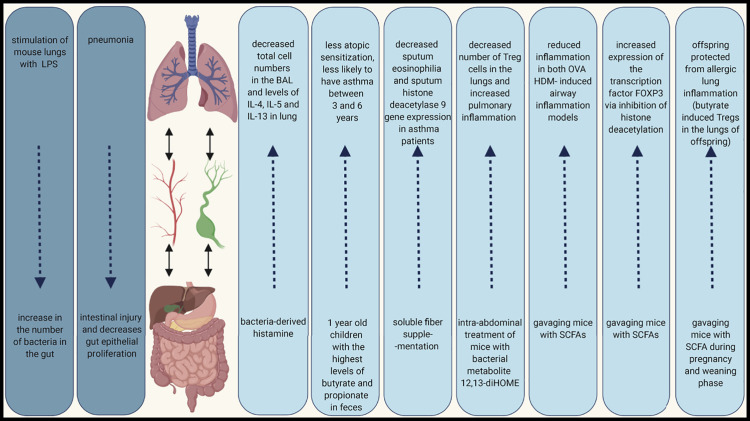
The mechanisms mediating communication between the gut and lungs are still unclear, but it has been suggested that epithelial cells, other structural cells, and immune cells absorb signals from the endothelium to form a local cytokine microenvironment, which leads to changes in immune responses at distal sites (Budden et al., 2017). Naive immune cells initially activated in the gut travel via lymph and blood vessels to the lung, where they have effector functions (He et al., 2017). Importantly, the local immune reaction in GALT and iBALT can influence systemic immune responses but the mucosal immune system can also act independently of the systemic site (Cesta, 2006). Interestingly, emerging evidence indicates that the gut-lung axis is bidirectional. For instance, stimulation of mouse lungs with lipopolysaccharide (LPS) causes a significant increase in the number of bacteria in the gut (Sze et al., 2014), and it has been shown that pneumonia induces intestinal injury (Perrone et al., 2012) and decreases gut epithelial proliferation (Coopersmith et al., 2003) (Figure 2 ).
Figure 2.
Gut-Lung Axis in Lung Inflammation Context
A connection between lungs and gut has been repeatedly demonstrated in both human and mouse studies. Many studies show that the microbiota is an important factor responsible for the interactions between these two sites. The interaction can be bidirectional; the gut microbiome can influence immune responses in lungs and lung stimulation can result in gut responses.
In addition, the gut-lung-liver axis has been described in COPD (Young et al., 2016). Short-chain fatty acids (SCFAs) derived from gut bacteria have inhibitory effects on proinflammatory responses in the lungs. One of the proposed mechanisms is that the liver can dampen the innate immune response by SCFAs binding to G-protein receptors or inhibiting the mevalonate pathway through HMGCoA reductase (Young et al., 2016). Furthermore, in a mouse model of pneumonia, it has been shown that cytokine expression in airspace macrophages was dependent on acute-phase protein (characteristic for hepatic acute-phase response) extravasation into the alveoli. These data suggest that the liver response enhances macrophage activation and pulmonary inflammation (Hilliard et al., 2015).
Bacteria Are the Largest Microbial Population in the Gut and Lungs
As the human gut is the most densely colonized site of the human body (Sender et al., 2016), its community of bacteria and their correlations with asthma both in early life and during established disease require special attention (Huang et al., 2017).
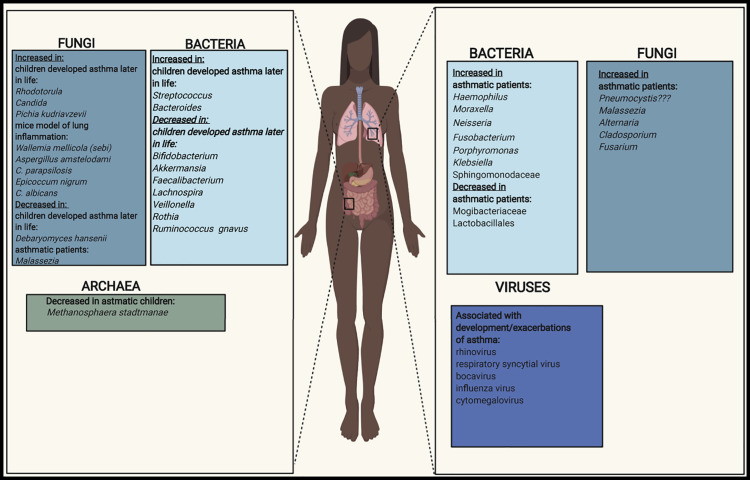
Lachnospira, Veillonella, Faecalibacterium, and Rothia genera are significantly decreased in relative abundance in fecal samples of three-month-old Canadian children at high risk of asthma. This bacterial signature is no longer evident at age 1 year and is accompanied by reduced amounts of fecal acetate and dysregulation of enterohepatic metabolites. Moreover, predicted bacterial community function analysis shows a reduction of LPS biosynthesis pathways in the microbiota of children at high risk of asthma (Arrieta et al., 2015). In an ovalbumin (OVA)-induced mouse model of airway inflammation, supplementation of a fecal slurry from an infant who developed asthma transplanted into germ-free mice with representative species from Lachnospira, Veillonella, Faecalibacterium, and Rothia genera decreased airway inflammation (Arrieta et al., 2015). In another human birth cohort study, Ecuadorian children with an increase in the relative abundance of Streptococcus and Bacteroides species and decrease in Bifidobacterium species and Ruminococcus gnavus in fecal samples at 3 months of age have a higher risk of developing atopy and wheeze at age 5 years (Arrieta et al., 2018). Finally, among neonates from the United States grouped into three clusters according to the composition of the gut microbiota, those with the lowest relative abundance of Bifidobacteria, Akkermansia, and Faecalibacterium genera and a higher relative abundance of Candida and Rhodotorula fungi have the highest risk of developing atopy and asthma (Fujimura et al., 2016) (Figure 3 ).
Figure 3.
Bacteria, Fungi, Viruses, and Archaea in the Lungs and Gut that Dysbiosis Has the Influence on Asthma Development and Maintenance
The effects of the gut microbiota on asthma are at least partially mediated by bacterial metabolites, which may influence immune responses in distal parts of the body. The most known metabolites with demonstrated protective properties in human airway inflammation are SCFAs. Children with high amounts of butyrate and propionate in feces at 1 year of age have significantly less atopic sensitization and are less likely to have asthma between 3 and 6 years (Roduit et al., 2019). Moreover, soluble fiber supplementation has been found to decrease sputum eosinophilia and sputum histone deacetylase 9 gene expression in asthma patients (McLoughlin et al., 2019). In mice, SCFAs have been shown to increase the expression of the transcription factor FOXP3 via inhibition of histone deacetylation, thereby supporting the expansion of T regulatory cells (Tregs), and increasing the production of IL‐10 (Arpaia et al., 2013). SCFAs have also been shown to reduce inflammation in both OVA and house dust mite (HDM)-induced airway inflammation models (Trompette et al., 2014, Cait et al., 2018). Moreover, oral application of SCFAs to mice during pregnancy and weaning have protect offspring from allergic lung inflammation, with butyrate in particular potently inducing Tregs in the lungs of offspring (Roduit et al., 2019) (Figure 2).
Of recent interest are studies showing that gut bacteria in humans are able to produce other metabolites with pro- and anti-inflammatory potential, such as biogenic amines (including histamine) (Pugin et al., 2017) and oxylipins such as 12,13-diHOME (Levan et al., 2019). The number of histamine-secreting bacteria is significantly higher in fecal samples of asthma patients compared with non-asthmatic volunteers (Barcik et al., 2019). In addition, the number of histamine-secreting bacteria correlates with disease severity. However, in a model of OVA-induced allergic airway inflammation, bacteria-derived histamine decreased total cell number in the bronchoalveolar fluid (BAL) and amounts of IL-4, IL-5, and IL-13 in lung homogenate (Barcik et al., 2019), highlighting the complexity of bacteria-derived immune regulation. Conversely, intra-abdominal treatment of mice with 12,13-diHOME decreased the number of Treg cells in the lungs and increased pulmonary inflammation in a cockroach antigen mouse model of airway inflammation (Levan et al., 2019) (Figure 2). The metabolites secreted by lung and gut bacteria might prove useful for additional therapeutic approaches.
In the lungs, Proteobacteria appear repeatedly to be the most dominant phylum overrepresented in patients with asthma compared with non-asthmatic volunteers across several human studies (Huang et al., 2015, Marri et al., 2013, Zhang et al., 2016, Hilty et al., 2010). The Proteobacteria phylum is represented by potentially pathogenic bacteria, including those that belong to the genera Haemophilus, Moraxella, and Neisseria (Huang et al., 2015, Caverly et al., 2019).
Within asthmatic inflammatory phenotypes, patients with neutrophilic asthma, usually receiving high doses of inhaled corticosteroids (ICSs), demonstrate a less diverse bacterial load with relative enrichment in Haemophilus and Moraxella species, members of Proteobacteria phylum, and a reduction in the relative abundance of Streptococcus, Gemella, and Porphyromonas taxa compared with patients with eosinophilic asthma (Taylor et al., 2019, Simpson et al., 2016, Green et al., 2014). In patients with severe asthma, a Th17 cell epithelial gene signature representing the non-type 2 asthma has been associated with increased representation of the phylum Proteobacteria, which further correlates with worsening of asthma control (Huang et al., 2015) and neutrophilic exacerbations of asthma (Ghebre et al., 2018). Klebsiella is one genus of bacteria within the Proteobacteria phylum enriched specifically in patients with severe asthma, whereas the phylum Actinobacteria correlates with improvement or no change is asthma control. Indicating the complexities of identifying cause-and-effect relationships, changes to microbial communities in the lungs may also result from asthma therapies. Actinobacteria, for instance, was associated with molecular evidence of response to steroids (FKBP5 expression) (Huang et al., 2015). Certain airway microbiota may therefore have the potential to be used as indicators of asthma responsive to steroids, allowing clinicians to refine the therapies. Importantly, the substantial additive effects of obesity and asthma severity on host immunological responses and the airway and gut microbiota have been recently described. Both obese and non-obese asthma patients with severe disease have reduced fecal levels of Akkermansia muciniphila, which may have a causal relationship as shown in murine models of acute and chronic airway inflammation (Michalovich et al., 2019). Another genus present in the lungs of patients with severe asthma is Streptococcus (Zhang et al., 2016, Budden et al., 2019).
Subjects with atopic asthma, usually of the eosinophilic inflammatory phenotype, demonstrate enrichments in bacteria from the genera Fusobacterium and Porphyromonas and the Sphingomonodaceae family, and decreased relative abundance of members of Mogibacteriaceae family and Lactobacillales order. (Durack et al., 2017). In addition, eosinophils in the sputum of asthma patients have been connected with the presence of Tropheryma whipplei in human adults (Simpson et al., 2016). In contrast to surprisingly reproducible findings in neutrophilic asthma, the status of the microbiota in eosinophilic and type 2 asthma is less clear and more heterogeneous, potentially reflecting again the differences in underlying endotypes or mixed responses caused by ICS treatment.
It currently remains unclear to what extent the diversity and presence of specific bacteria in the airways of adult patients with asthma reflects the type of inflammation or the microbial response to corticosteroid treatment. It has been demonstrated that treatment with a combination of ICSs and oral glucocorticoids correlates positively with an increased abundance of Proteobacteria and Pseudomonas, and with a decreased abundance of Bacteroidetes, Fusobacteria, and Prevotella (Denner et al., 2016). On the other hand, a study in mild asthmatics not treated with ICSs has demonstrated a unique enrichment in members of the Haemophilus, Neisseria, Fusobacterium, and Porphyromonas species and the Sphingomonodaceae family with concurrent depletion in members of the Mogibacteriaceae family and Lactobacillales in their airways (Durack et al., 2017). Together these studies suggest that the lung microbiome composition in patients with asthma, who are often treated with corticosteroids, is probably a result of complex interactions between the inflammatory milieu and the drug effects.
Infection of mice with Moraxella catarrhalis (M. catarrhalis, Proteobacterium phyla) results in neutrophilic infiltrates, high concentrations of IL-6, IL-1β, and tumor necrosis factor α (TNF-α), and moderate concentration of CD4+ T cell-derived IFN-γ and IL-17 in the lungs 1 day after infection. Moreover, neutralization of IL-17 or TNF-α, but not IL-6, results in accelerated clearance of M. catarrhalis and effectively prevent infection-induced exacerbation of allergic airway inflammation (Alnahas et al., 2017). Furthermore, Haemophilus influenzae from Proteobacteroium phyla is able to convert steroid-sensitive allergic airway disease (AAD) associated with Th2 cells and eosinophils to steroid-resistant disease associated with Th1 cells, neutrophils, and dominant IL-17 responses (Essilfie et al., 2012).
An increasingly large body of evidence suggests a role for bacteria in asthma, but further studies are needed to more clearly define the most important species involved and to understand whether bacterial dysbiosis in the context of asthma is the cause or effect of disease. More detailed mechanistic studies are necessary to gain full understanding of the complex associations of lung and gut microbiota compositions and metabolism at different points in life with specific types of asthmatic inflammation. Even if some immunological mechanisms of early-life protection have been described, there are probably many others. In addition, the immunological and molecular mechanisms of already established dysbiosis in adult asthma need to be characterized in more detail, ideally to reverse these processes. Moreover, further attention should be paid to understanding the connection between a patient’s microbiome and response to asthma therapies, as these may influence one another. Microbial-derived mechanisms might be the reason of poor response to the treatment, as it has been elegantly demonstrated in vaccination (Hagan et al., 2019). Finally, future work should focus on continuing to characterize in detail the cellular and molecular mechanisms mediating communication between bacteria and the host in asthma.
Looking beyond Bacteria: Fungi: The Forgotten Microbes
Important groups of microbes, which together comprise the “rare biosphere” (Lynch and Neufeld, 2015, Jousset et al., 2017) of the microbiota, include fungi and other micro-eukaryotes. It remains unclear whether fungi colonize the healthy adult gut or are simply detected in sequencing studies as transient oral or food-derived fungi (Auchtung et al., 2018), but even transient colonizers of the gut have the potential to impact microbial community ecology and host immune responses (Hallen-Adams and Suhr, 2017). Although the mechanisms involved were not explored, two recent studies in human birth cohorts from the United States and Ecuador have identified fungal dysbiosis as a key feature of infant gut microbiota signatures associated with the development of high-risk asthma phenotypes in childhood (Fujimura et al., 2016, Arrieta et al., 2018). The specific signatures of fungal dysbiosis associated with asthma and related phenotypes differed in these studies, but it is notable that fungal dysbiosis was much more striking than bacterial dysbiosis in both studies. These results suggest that fungal dysbiosis may be a readily detectable biomarker of asthma risk. Moreover, both studies found that fungal dysbiosis co-associated with bacterial dysbiosis (Fujimura et al., 2016, Arrieta et al., 2018), calling for follow-up studies investigating both the microbial and immune mechanisms underlying these associations.
Microbes within the gut influence host immune responses to other microbes, interact physically and metabolically with one another, and compete for space and nutrients. Early studies in animal models aimed at establishing a causal link between fungal dysbiosis and asthma severity demonstrated that mice pre-treated with antibiotics are susceptible to fungal overgrowth following oral gavage with the common human gut commensal Candida albicans (C. albicans) (Noverr et al., 2004, Noverr et al., 2005). These mice further demonstrate exacerbated Th2 cell-mediated airway inflammation following airway challenge (Noverr et al., 2004, Noverr et al., 2005), an effect postulated to be mediated at least in part by the production of immunomodulatory prostaglandins by C. albicans (Noverr et al., 2001, Erb-Downward and Noverr, 2007). Congruent with this, overgrowth of another Candida species, C. parapsilosis, in the murine gut following antibiotic treatment has more recently been linked to prostaglandin PGE2-mediated polarization of lung macrophages to the M2 phenotype and exacerbated allergic inflammation following antigen sensitization and challenge. In this model, fungal overgrowth is correlated with inflammatory immune cell infiltration to the lung but also required specific antibiotic-induced changes to the bacterial microbiota. Only antibiotics causing reductions in the relative abundance of bacteria from the genus (among others) Lactobacillus are associated with fungal overgrowth and exacerbated airway inflammation following antigen challenge (Kim et al., 2014). Notably, Lactobacillus and other lactic acid-producing bacteria are of interest for their use as a probiotic to protect against asthma and allergy (Forsythe et al., 2007) and have known anti-fungal effects (Savage, 1969, Noverr and Huffnagle, 2004).
Further evidence that the historical and current microbial context within which fungal dysbiosis occurs influences the associated immunological consequences during asthma has emerged more recently. Mice with no pre-existing mycobiota demonstrate a mixed Th2 and Th17 cell lung infiltrate following inoculation with a dysbiotic fugal community, whereas the same treatment in conventional mice with a pre-existing mycobiome results only in increased Th2 cell inflammation during asthma (Li et al., 2018). Similarly, fungi-naive mice newly colonized with C. albicans demonstrate exacerbated AAD associated with systemic expansion of fungus-specific Th17 cells and IL-17-hyperresponsive neutrophils (Shao et al., 2019). Together these studies highlight the complex, dynamic nature of the gut microbiota and the importance of cross-kingdom microbial interactions in determining asthma outcomes.
In addition to antibiotic treatment, antifungal treatment induces changes to both the bacterial and fungal communities in the gut of specific pathogen-free (SPF) mice and has been shown to exacerbate type 2 allergic airway inflammation and eosinophilia in a HDM model of AAD (Wheeler et al., 2016, Li et al., 2018). Continuous supplementation with three fungal species identified in this model to be overrepresented in the gut following antifungal treatment (Aspergillus amstelodami, Wallemia mellicola [sebi], and Epicoccum nigrum) (Wheeler et al., 2016, Li et al., 2018) or with Wallemia mellicola alone following antibiotic treatment (Skalski et al., 2018) similarly increases asthma severity (Figure 3) (Wheeler et al., 2016, Skalski et al., 2018). Antifungal treatment and the introduction of dysbiotic fungal communities to SPF mice in these studies results in changes to gut bacterial communities, including depletions of bacteria from the Lactobacillaceae family (Li et al., 2018, Skalski et al., 2018). However, the AAD-exacerbating effects of supplementation with a dysbiotic fungal community can be recapitulated in altered Schaedler flora mice without pre-treatment with antibiotics and in the absence of substantial changes to the bacterial microbiota (Li et al., 2018, Skalski et al., 2018). Furthermore, Li et al. (2018) recently demonstrated that direct sensing of antifungal-induced fungal dysbiosis by gut-resident CX3CR1+ mononuclear phagocytes (Leonardi et al., 2018) increases numbers of Th2 cells in the lamina propria and is responsible for fungal dysbiosis-associated increased airway inflammation in a HDM model of AAD, independent of antifungal-associated changes to the bacterial communities (Li et al., 2018). Thus, in addition to having potential indirect effects on airway inflammation via restructuring of immunomodulatory gut bacterial populations or the production of systemic metabolites, asthma-associated fungal dysbiosis in the gut can have direct effects on host innate and adaptive immune cell populations.
Local fungal dysbiosis in the lung has also been linked to asthma in several studies in humans. It has been shown in a small sample of subjects that children with and without severe asthma exhibit differences in the relative abundance of specific fungi, but not overall fungal diversity, in BAL samples. These differences included increases in the relative abundance of Pneumocystis in the lungs of patients with severe asthma (Goldman et al., 2018), a microbe whose presence within the lungs has been linked to the induction of host type 2 immune responses in humans and animal models (Eddens et al., 2016). Subclinical colonization of the lungs with Pneumocystis at 2–5 months of age has been suggested to be one of the most common infections in infancy (Vargas et al., 2001, Vargas et al., 2005) and is associated with increased mucin gene expression (Pérez et al., 2014, Vargas et al., 2001, Vargas et al., 2005, Vargas et al., 2013, Rojas et al., 2019). Early lung colonization with Pneumocystis may therefore represent a risk factor for the induction of Th2 cell-skewed immune responses in the lung and predispose infants to asthma (Rojas et al., 2019).
An 18S pyrosequencing study in adult patients similarly found that, compared to non-atopic controls, patients with asthma demonstrate differences in the relative abundance of specific fungi in pooled sputum (van Woerden et al., 2013). In these adults, those with asthma demonstrate an increase in the relative abundance of species of Malassezia previously linked to the allergic condition of atopic dermatitis (van Woerden et al., 2013). Overall, a picture is emerging wherein patients with asthma exhibit an increase in the relative abundance of specific fungi associated with the induction of type 2 inflammatory immune pathways, some of which are either absent or present only in low abundance in the healthy lung.
Findings of altered fungal populations in the lungs of asthmatic patients are not surprising, as fungal components such as mannans, proteases (Porter et al., 2009), chitin, and β-glucans (Van Dyken et al., 2011) are often found in common human allergens and house dust. While the underlying mechanisms of sensitization remain unclear, many patients with severe and/or uncontrolled asthma are sensitized to fungi (Denning et al., 2014, Stern et al., 2008, Fraczek et al., 2018) and/or may show improvements in asthma symptoms following antifungal therapy (Chishimba et al., 2012, Denning et al., 2014). Lung fungi commonly associated with sensitization and asthma include those from the genera Aspergillus, Alternaria, and Cladosporium (Figure 3) (Denning et al., 2014, Fraczek et al., 2018). Interestingly, some evidence from animal models suggests that sensitization occurs secondary to the natural Th2 cell and eosinophilic immune response required for effective clearance of infection-causing fungi (Porter et al., 2009). Additionally, increased fungal load in BAL samples has been associated with corticosteroid therapy in patients with severe asthma with and without fungal sensitization (Fraczek et al., 2018). Thus, future work is needed to determine whether pathological fungal colonization and mycological dysbiosis of the lungs causes the onset of more severe asthma and occurs subsequent to severe asthma-associated corticosteroid treatment, and/or in response to asthma-associated changes in the lung microenvironment.
More recently, studies have begun to precisely characterize the airway mycobiota associated with certain asthma endotypes and clinical features. In a study examining the microbiota of endotracheal brushings and BAL fluid, Fusarium, Cladosporium, and Alternaria (Figure 3) species were among the fungal organisms identified to be enriched in the BAL of patients with asthma and type 2 high asthma relative to healthy controls and patients with type 2 low asthma, respectively (Sharma et al., 2019). Indicating their potential for use as biomarkers of disease, the relative abundance of Alternaria and Cladosporium in BAL fluid has also negatively associated with forced expiratory volume 1, a clinical measure of lung function (Sharma et al., 2019).
Over the past couple of decades, there has been an explosion of research into the composition and immunological effects of non-bacterial members of the gut microbiota, including fungi, on the host. Although much remains to be elucidated, a robust body of evidence now supports a substantial role for fungi in the dysbiosis-asthma paradigm. Fungal-bacterial and fungal-host interactions both play an important role in determining the effects of fungal dysbiosis on host health. Much of this work has been done in pediatric birth cohorts and mouse models, calling for more studies into the relevance of the gut mycobiota to asthma in human adults. Furthermore, if and how early-life fungal dysbiosis influences the onset and/or severity of asthma later in life when the dysbiosis may no longer be present remains to be mechanistically tested.
Virions Readily Colonize the Lungs
Although viral infections are known to be a primary cause of asthmatic exacerbations, the relationship between viruses and asthma is not completely understood, partly because of continuously evolving knowledge about the healthy virome of the respiratory tract. It has been observed that the composition and abundance of the URT and LRT virome differs among healthy subjects and asymptomatic and symptomatic patients with chronic respiratory diseases, but detailed longitudinal studies are needed for better comprehension of the causal relationships (Sundell et al., 2019, Freer et al., 2018). Complex interactions between viruses and additional environmental and intrinsic factors are probably responsible for the longer but transient presence of some viruses in the respiratory tract of asthmatic patients or the development of fully blown infections leading to severe exacerbations (Vandini et al., 2019, Sarna et al., 2018).
In the first years of human life, almost all wheezing episodes are correlated with viral infections (Mikhail and Grayson, 2019). The most common viruses known to induce wheeze or exacerbations of asthma are RV and RSV, but bocavirus, influenza virus, and cytomegalovirus (CMV) (Figure 3) have also been implicated (Meissner, 2016, Mikhail and Grayson, 2019). It has been reported that early-life RV-induced wheezing has additive effects on asthma risk later in life.
The most abundant cell type in the airways of infants with RV bronchiolitis is neutrophils, although infiltration of eosinophils also plays an important role (Heinonen et al., 2019). Moreover, it has been recently demonstrated that concentrations of mast cells IL-6, IL-8, TNF-α, and IFN-α was significantly higher after stimulation with RV (Liu et al., 2019). T cells participate in controlling RV infection through the recognition of viral antigens and subsequent initiation of both Tc and antibody-mediated immune responses (Heinonen et al., 2019). Of particular interest is a recent study showing in vitro that RV enters and forms viral replication centers in B lymphocytes and induces the proliferation of B cells (Aab et al., 2017).
Another virus linked with increased risk of asthma is RSV (Bacharier et al., 2012); however, its association with the disease is less clear (Kast et al., 2017). There are studies suggesting a connection between RSV-induced bronchiolitis and asthma (Bacharier et al., 2012), but others have not identified a correlation between RSV infection in infants and the development of asthma at school age (Wu et al., 2008). The long-term influence of RSV infection on asthma can be connected with fact that the virus typically affects neonates during lung development (Jartti and Gern, 2017). Neonatal regulatory B cells are highly permissive to RSV infection, and the frequency of this cell population can predict the severity of acute bronchiolitis (Zhivaki et al., 2017). Moreover, RSV rapidly adheres to human eosinophils and might be inactivated by these cells. The capacity of eosinophils to capture virus is reduced up to 75% with increasing severity of asthma (Sabogal Piñeros et al., 2019).
Influenza viruses are another viral infections connected with asthma exacerbations (Coverstone et al., 2019). Asthma in the setting of influenza infection is associated with worse severity, higher risk of hospitalization, need for intensive care, and mortality (Ritchie et al., 2015). IL-33 has been hypothesized to be necessary for driving influenza virus-induced asthma exacerbations. In a mouse model of asthma, IL-33 enhanced airway hyperresponsiveness and airway inflammation by suppressing innate and adaptive antiviral responses (Ravanetti et al., 2019). Another study has shown that influenza infection induced airway hyper-reactivity through a pathway that required the IL-13-IL-33 axis (Chang et al., 2011).
Besides the most well-studied viruses linked to asthma and described above, there are also a few less commonly encountered viruses that influence airway inflammation involved in asthma. Human bocavirus 1 (HBoV) and CMV are also connected with wheezing episodes in children. A study published by Del Rosal et al. (2016) reported that, in a small sample of children with HBoV aged ≥4 years who were previously hospitalized due to acute bronchiolitis, all of them developed recurrent wheezing, and half of them had asthma at age 5–7 years. It has been also demonstrated that subcutaneous injection of HBoV-VP1u or B19V-VP1u proteins in OVA-sensitized mice resulted in elevated airway inflammation (Chiang et al., 2019). Finally, CMV DNA was detected in 51.4% of patients with recurrent wheeze and was more prevalent among those aged 12–36 months with a positive modified asthma predictive index (mAPI) than in those of the same age group with a negative mAPI (Sun et al., 2018).
The connection between respiratory viral infections and asthma development has been recently investigated using more mechanistic approaches (Makris and Johnston, 2018, Toussaint et al., 2017). However, the precise mechanisms responsible for the correlation are still unclear and need further detailed investigations in animal models as well as in human studies using multi-omics approaches (Sokolowska et al., 2017). Analyses considering the global virome of patients should also be considered in the future. Finally, scientists should also ask the question: is it possible that gut viruses can influence lung inflammation?
Archaea-Emerging Players in Human Gut
Although understudied, archaea are consistently identified in human microbiota studies and their community composition differs according to body site (Koskinen et al., 2017). Methonoarchaea, including Methanobrevibacter smithii and Methanosphaera stadtmanae (M. stadtmanae), have been proposed as keystone members of this microbial community and possess immunogenic properties (Blais Lecours et al., 2011, Bang et al., 2014).
A role for intestinal archaea in the dysbiosis-asthma paradigm was recently suggested by a study in a subset of 472 children from the KOALA birth cohort in the Netherlands (Barnett et al., 2019). The presence of M. stadtmanae in stool samples from 6- to 10-year-old children has been associated in a dose-dependent manner with a reduced risk of being diagnosed with asthma (Barnett et al., 2019). This effect was independent of parental asthma status (Barnett et al., 2019). Only 8.3% of samples tested in this study were positive for M. stadtmanae, however, and certain archaeal proteins have actually been proposed to have pro-inflammatory (Blais Lecours et al., 2011) or allergenic properties (Bragin et al., 2018). Thus, more cohort studies and mechanistic studies into the immunomodulatory effects of gut archaea in the context of asthma are needed. As in the case of the other microbial communities discussed in this review, the microbial and immune context within which disrupted archaeal communities occur likely determine the ultimate immunological consequences for the host. Furthermore, as the human lung microbiota in childhood and adults continues to be characterized, the role of lung archaea in the dysbiosis-asthma paradigm will need to be determined.
Conclusions and Future Perspectives
It is now supported by a rich body of literature that the human microbiota influences the maturation and function of the host immune system. Both lung and gut microbiota have impacts on asthma development, phenotype, and severity, but the detailed cellular and molecular mechanisms of these correlations have not yet been fully characterized. One important tool that will be helpful to understand mechanisms underlying the connection between the human microbiota and the immune system is humanized microbiota mouse models for allergic airway inflammation. Special attention should also be given to improving in vivo animal models to establish studies that mimic human clinical conditions as closely as possible. Currently, there are only two common mouse models of allergic airway inflammation: OVA and HDM-induced acute models, which reflect mainly type 2 asthma. Other models, utilizing high allergen doses or LPS, Chlamydia muridarum, Haemophilus influenzae, paramyxovirus, or RV1, usually in addition to allergen, partially reflect non-type 2 inflammation and should be used to complement OVA and HDM models (Tan et al., 2019, Kim et al., 2017).
Thanks to advances in DNA and RNA sequencing technologies, it is now possible to characterize many unculturable microbial strains that were not known to be associated with asthma conditions just a few years ago. Recent analyses using currently available sequencing methods have allowed us to determine that microbial dysbiosis of the gut and lungs in airway inflammation is characterized by taxa-specific shifts in abundance at the family, genus, species, and even strain levels. These data clearly show that future studies should be more concentrated on specific changes of microbiota taxonomy, including strain differences, rather than at overall changes. On the other hand, global views of the microbiota and its correlations with host phenotypes should be taken into consideration. Studying isolated bacterial strains in the context of airway inflammation might have very different outcomes than if the same strains were investigated within a bacterial community. Moreover, more emphasis should be given to the detailed study of different parts of the human microbiota, including viruses, archaea, fungi, bacteriophages as well as parasites, and the possibility of global shifts or changes to inter-kingdom or inter-site microbial interactions within the microbiota in the context of asthma.
Moreover, it is well established that the distribution of microbiota differs according to physico-chemcial niches in the gut. The study of gut microbial compositions in mice and their correlations with airway inflammation should therefore take into consideration gut biogeography. The most common approach to analyzing the gut microbiota is analysis of the fecal microbiota while ignoring the microbes that remain along the gastrointestinal tract. This approach may lack important information and potential misinterpretation of the connection between the gut microbiota and host and is a limitation of most current studies.
Asthma is very complex disease characterized by many different phenotypes and endotypes. Human cohort studies should therefore include clearly defined clinical outcomes and collect longitudinal data from detailed and specified questions about symptoms, treatments, and clinical information. Complete information regarding patient diet and lifestyle should also be carefully prepared to ensure the best overview of patient conditions and potential influences on the microbiota. Future studies should be extended to include several time points during a patient’s life to determine whether changes to the earlier-life microbiota are connected to asthma development or preventions, or to an increase or improvement in airway inflammation. Importantly, longitudinal and prospective analyses of larger cohorts of patients are needed to understand the relationships between the course of the disease, its phenotype and endotype, susceptibility to disease progression, and response to treatment. Finally, considerable attention has and continues to be paid to early-life changes to the gut and lung microbiota and their influence on allergy symptoms later in life. While it is indisputable that early life is a very important period during which microbiome dysbiosis may lead to the development of immune-mediated diseases, we should also be able to answer the question: is it still possible to change, through modulation of the microbiota, allergic disease conditions later in life even if the early-life period was neglected?
To summarize, it is clear that the future diagnosis and treatment of patients with asthma should be assisted by analysis of the composition and metabolic activity of an individual’s microbiome. Clinical studies of new therapeutics should consider including microbiome and metabolite analyses to determine whether microbiome features correlate with responses to treatment. Further mechanistic and epidemiological studies are needed to uncover the functional, multidirectional associations between the specific microbiota strains, the host immune system and allergens.
Acknowledgments
We apologize to members of the community whose studies were not cited herein because of space constraints. The Finlay lab is supported by Canadian Institutes of Health Research Foundation grant no. FDN-159935; R.C.T.B. is supported by Vanier Canada Graduate Scholarship, a University of British Columbia Four Year Doctoral Fellowship, and a Vancouver Coastal Health-Canadian Institutes of Health Research (CIHR)-UBC MD/PhD Studentship Award; M.S. is supported by Swiss National Science Foundation grant no. 310030_189334, Switzerland; and B.B.F. is a University of British Columbia Peter Wall Distinguished Professor.
Declaration of Interests
The authors declare no competing interests.
References
- Aab A., Wirz O., van de Veen W., Söllner S., Stanic B., Rückert B., Aniscenko J., Edwards M.R., Johnston S.L., Papadopoulos N.G. Human rhinoviruses enter and induce proliferation of B lymphocytes. Allergy. 2017;72:232–243. doi: 10.1111/all.12931. [DOI] [PubMed] [Google Scholar]
- Abbas A.A., Diamond J.M., Chehoud C., Chang B., Kotzin J.J., Young J.C., Imai I., Haas A.R., Cantu E., Lederer D.J. The Perioperative Lung Transplant Virome: Torque Teno Viruses Are Elevated in Donor Lungs and Show Divergent Dynamics in Primary Graft Dysfunction. Am. J. Transplant. 2017;17:1313–1324. doi: 10.1111/ajt.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agache I., Akdis C.A. Endotypes of allergic diseases and asthma: An important step in building blocks for the future of precision medicine. Allergol. Int. 2016;65:243–252. doi: 10.1016/j.alit.2016.04.011. [DOI] [PubMed] [Google Scholar]
- Almeida A., Mitchell A.L., Boland M., Forster S.C., Gloor G.B., Tarkowska A., Lawley T.D., Finn R.D. A new genomic blueprint of the human gut microbiota. Nature. 2019;568:499–504. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnahas S., Hagner S., Raifer H., Kilic A., Gasteiger G., Mutters R., Hellhund A., Prinz I., Pinkenburg O., Visekruna A. IL-17 and TNF-α Are Key Mediators of Moraxella catarrhalis Triggered Exacerbation of Allergic Airway Inflammation. Front. Immunol. 2017;8:1562. doi: 10.3389/fimmu.2017.01562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altan E., Dib J.C., Gulloso A.R., Escribano Juandigua D., Deng X., Bruhn R., Hildebrand K., Freiden P., Yamamoto J., Schultz-Cherry S., Delwart E. Effect of Geographic Isolation on the Nasal Virome of Indigenous Children. J. Virol. 2019;93 doi: 10.1128/JVI.00681-19. e00681–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N., Campbell C., Fan X., Dikiy S., van der Veeken J., deRoos P., Liu H., Cross J.R., Pfeffer K., Coffer P.J., Rudensky A.Y. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta M.C., Arévalo A., Stiemsma L., Dimitriu P., Chico M.E., Loor S., Vaca M., Boutin R.C.T., Morien E., Jin M. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J. Allergy Clin. Immunol. 2018;142:424–434. doi: 10.1016/j.jaci.2017.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta M.C., Stiemsma L.T., Dimitriu P.A., Thorson L., Russell S., Yurist-Doutsch S., Kuzeljevic B., Gold M.J., Britton H.M., Lefebvre D.L., CHILD Study Investigators Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- Auchtung T.A., Fofanova T.Y., Stewart C.J., Nash A.K., Wong M.C., Gesell J.R., Auchtung J.M., Ajami N.J., Petrosino J.F. Investigating Colonization of the Healthy Adult Gastrointestinal Tract by Fungi. MSphere. 2018;3:1–16. doi: 10.1128/mSphere.00092-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacharier L.B., Cohen R., Schweiger T., Yin-Declue H., Christie C., Zheng J., Schechtman K.B., Strunk R.C., Castro M. Determinants of asthma after severe respiratory syncytial virus bronchiolitis. J. Allergy Clin. Immunol. 2012;130:91–100. doi: 10.1016/j.jaci.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang C., Weidenbach K., Gutsmann T., Heine H., Schmitz R.A. The intestinal archaea Methanosphaera stadtmanae and Methanobrevibacter smithii activate human dendritic cells. PLoS ONE. 2014;9:e99411. doi: 10.1371/journal.pone.0099411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcik W., Untersmayr E., Pali-Schöll I., O’Mahony L., Frei R. Influence of microbiome and diet on immune responses in food allergy models. Drug Discov. Today Dis. Models. 2015;17-18:71–80. doi: 10.1016/j.ddmod.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcik W., Pugin B., Brescó M.S., Westermann P., Rinaldi A., Groeger D., Van Elst D., Sokolowska M., Krawczyk K., Frei R. Bacterial secretion of histamine within the gut influences immune responses within the lung. Allergy. 2019;74:899–909. doi: 10.1111/all.13709. [DOI] [PubMed] [Google Scholar]
- Barnett D.J.M., Mommers M., Penders J., Arts I.C.W., Thijs C. Intestinal archaea inversely associated with childhood asthma. J. Allergy Clin. Immunol. 2019;143:2305–2307. doi: 10.1016/j.jaci.2019.02.009. [DOI] [PubMed] [Google Scholar]
- Bassis C.M., Erb-Downward J.R., Dickson R.P., Freeman C.M., Schmidt T.M., Young V.B., Beck J.M., Curtis J.L., Huffnagle G.B. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio. 2015;6:e00037. doi: 10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiyan M.U., Snelling T.L., West R., Lang J., Rahman T., Granland C., de Gier C., Borland M.L., Thornton R.B., Kirkham L.S. The contribution of viruses and bacteria to community-acquired pneumonia in vaccinated children: a case-control study. Thorax. 2019;74:261–269. doi: 10.1136/thoraxjnl-2018-212096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesbroek G., Tsivtsivadze E., Sanders E.A., Montijn R., Veenhoven R.H., Keijser B.J., Bogaert D. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am. J. Respir. Crit. Care Med. 2014;190:1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- Bogaert D., Keijser B., Huse S., Rossen J., Veenhoven R., van Gils E., Bruin J., Montijn R., Bonten M., Sanders E. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS ONE. 2011;6:e17035. doi: 10.1371/journal.pone.0017035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomar L., Brugger S.D., Yost B.H., Davies S.S., Lemon K.P. Corynebacterium accolens Releases Antipneumococcal Free Fatty Acids from Human Nostril and Skin Surface Triacylglycerols. MBio. 2016;7 doi: 10.1128/mBio.01725-15. e01725–e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch A.A.T.M., Levin E., van Houten M.A., Hasrat R., Kalkman G., Biesbroek G., de Steenhuijsen Piters W.A.A., de Groot P.C.M., Pernet P., Keijser B.J.F. Development of Upper Respiratory Tract Microbiota in Infancy is Affected by Mode of Delivery. EBioMedicine. 2016;9:336–345. doi: 10.1016/j.ebiom.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin R.C.T., Petersen C., Finlay B.B. Microbial Insights into Asthmatic Immunopathology. A Forward-Looking Synthesis and Commentary. Ann. Am. Thorac. Soc. 2017;14(Supplement_5):S316–S325. doi: 10.1513/AnnalsATS.201707-534AW. [DOI] [PubMed] [Google Scholar]
- Bragin A.O., Sokolov V.S., Demenkov P.S., Ivanisenko T.V., Bragina E.Y., Matushkin Y.G., Ivanisenko V.A. [Prediction of Bacterial and Archaeal Allergenicity with AllPred Program] Mol. Biol. (Mosk.) 2018;52:326–332. doi: 10.7868/S0026898418020179. [DOI] [PubMed] [Google Scholar]
- Brandtzaeg P. The mucosal immune system and its integration with the mammary glands. J. Pediatr. 2010;156(2, Suppl):S8–S15. doi: 10.1016/j.jpeds.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Budden K.F., Gellatly S.L., Wood D.L., Cooper M.A., Morrison M., Hugenholtz P., Hansbro P.M. Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 2017;15:55–63. doi: 10.1038/nrmicro.2016.142. [DOI] [PubMed] [Google Scholar]
- Budden K.F., Shukla S.D., Rehman S.F., Bowerman K.L., Keely S., Hugenholtz P., Armstrong-James D.P.H., Adcock I.M., Chotirmall S.H., Chung K.F., Hansbro P.M. Functional effects of the microbiota in chronic respiratory disease. Lancet Respir. Med. 2019;7:907–920. doi: 10.1016/S2213-2600(18)30510-1. [DOI] [PubMed] [Google Scholar]
- Cait A., Hughes M.R., Antignano F., Cait J., Dimitriu P.A., Maas K.R., Reynolds L.A., Hacker L., Mohr J., Finlay B.B. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. 2018;11:785–795. doi: 10.1038/mi.2017.75. [DOI] [PubMed] [Google Scholar]
- Castro M., Corren J., Pavord I.D., Maspero J., Wenzel S., Rabe K.F., Busse W.W., Ford L., Sher L., FitzGerald J.M. Dupilumab Efficacy and Safety in Moderate-to-Severe Uncontrolled Asthma. N. Engl. J. Med. 2018;378:2486–2496. doi: 10.1056/NEJMoa1804092. [DOI] [PubMed] [Google Scholar]
- Caverly L.J., Huang Y.J., Sze M.A. Past, Present, and Future Research on the Lung Microbiome in Inflammatory Airway Disease. Chest. 2019;156:376–382. doi: 10.1016/j.chest.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesta M.F. Normal structure, function, and histology of mucosa-associated lymphoid tissue. Toxicol. Pathol. 2006;34:599–608. doi: 10.1080/01926230600865531. [DOI] [PubMed] [Google Scholar]
- Chang Y.J., Kim H.Y., Albacker L.A., Baumgarth N., McKenzie A.N., Smith D.E., Dekruyff R.H., Umetsu D.T. Innate lymphoid cells mediate influenza-induced airway hyper-reactivity independently of adaptive immunity. Nat. Immunol. 2011;12:631–638. doi: 10.1038/ni.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlson E.S., Diamond J.M., Bittinger K., Fitzgerald A.S., Yadav A., Haas A.R., Bushman F.D., Collman R.G. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am. J. Respir. Crit. Care Med. 2012;186:536–545. doi: 10.1164/rccm.201204-0693OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang S.R., Lin C.Y., Chen D.Y., Tsai H.F., Lin X.C., Hsu T.C., Tzang B.S. The effects of human parvovirus VP1 unique region in a mouse model of allergic asthma. PLoS ONE. 2019;14:e0216799. doi: 10.1371/journal.pone.0216799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chishimba L., Niven R.M., Cooley J., Denning D.W. Voriconazole and posaconazole improve asthma severity in allergic bronchopulmonary aspergillosis and severe asthma with fungal sensitization. J. Asthma. 2012;49:423–433. doi: 10.3109/02770903.2012.662568. [DOI] [PubMed] [Google Scholar]
- Christiansen S.C., Zuraw B.L. Treatment of Hypertension in Patients with Asthma. N. Engl. J. Med. 2019;381:1046–1057. doi: 10.1056/NEJMra1800345. [DOI] [PubMed] [Google Scholar]
- Chung K.F. Airway microbial dysbiosis in asthmatic patients: A target for prevention and treatment? J. Allergy Clin. Immunol. 2017;139:1071–1081. doi: 10.1016/j.jaci.2017.02.004. [DOI] [PubMed] [Google Scholar]
- Chung K.F., Wenzel S.E., Brozek J.L., Bush A., Castro M., Sterk P.J., Adcock I.M., Bateman E.D., Bel E.H., Bleecker E.R. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur. Respir. J. 2014;43:343–373. doi: 10.1183/09031936.00202013. [DOI] [PubMed] [Google Scholar]
- Coopersmith C.M., Stromberg P.E., Davis C.G., Dunne W.M., Amiot D.M., 2nd, Karl I.E., Hotchkiss R.S., Buchman T.G. Sepsis from Pseudomonas aeruginosa pneumonia decreases intestinal proliferation and induces gut epithelial cell cycle arrest. Crit. Care Med. 2003;31:1630–1637. doi: 10.1097/01.CCM.0000055385.29232.11. [DOI] [PubMed] [Google Scholar]
- Corren J., Castro M., O’Riordan T., Hanania N.A., Pavord I.D., Quirce S., Chipps B.E., Wenzel S.E., Thangavelu K., Rice M.S. Dupilumab Efficacy in Patients with Uncontrolled, Moderate-to-Severe Allergic Asthma. J. Allergy Clin. Immunol. Pract. 2019 doi: 10.1016/j.jaip.2019.08.050. [DOI] [PubMed] [Google Scholar]
- Cosío B.G., Pérez de Llano L., Lopez Viña A., Torrego A., Lopez-Campos J.L., Soriano J.B., Martinez Moragon E., Izquierdo J.L., Bobolea I., Callejas J., on behalf of the CHACOS study group Th-2 signature in chronic airway diseases: towards the extinction of asthma-COPD overlap syndrome? Eur. Respir. J. 2017;49:49. doi: 10.1183/13993003.02397-2016. [DOI] [PubMed] [Google Scholar]
- Coverstone A.M., Wang L., Sumino K. Beyond Respiratory Syncytial Virus and Rhinovirus in the Pathogenesis and Exacerbation of Asthma: The Role of Metapneumovirus, Bocavirus and Influenza Virus. Immunol. Allergy Clin. North Am. 2019;39:391–401. doi: 10.1016/j.iac.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Steenhuijsen Piters W.A., Sanders E.A., Bogaert D. The role of the local microbial ecosystem in respiratory health and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370 doi: 10.1098/rstb.2014.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Steenhuijsen Piters W.A.A., Jochems S.P., Mitsi E., Rylance J., Pojar S., Nikolaou E., German E.L., Holloway M., Carniel B.F., Chu M.L.J.N. Interaction between the nasal microbiota and S. pneumoniae in the context of live-attenuated influenza vaccine. Nat. Commun. 2019;10:2981. doi: 10.1038/s41467-019-10814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rosal T., García-García M.L., Calvo C., Gozalo F., Pozo F., Casas I. Recurrent wheezing and asthma after bocavirus bronchiolitis. Allergol. Immunopathol. (Madr.) 2016;44:410–414. doi: 10.1016/j.aller.2015.07.004. [DOI] [PubMed] [Google Scholar]
- Denner D.R., Sangwan N., Becker J.B., Hogarth D.K., Oldham J., Castillo J., Sperling A.I., Solway J., Naureckas E.T., Gilbert J.A., White S.R. Corticosteroid therapy and airflow obstruction influence the bronchial microbiome, which is distinct from that of bronchoalveolar lavage in asthmatic airways. J. Allergy Clin. Immunol. 2016;137:1398–1405. doi: 10.1016/j.jaci.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D.W., Pashley C., Hartl D., Wardlaw A., Godet C., Del Giacco S., Delhaes L., Sergejeva S. Fungal allergy in asthma-state of the art and research needs. Clin. Transl. Allergy. 2014;4:14. doi: 10.1186/2045-7022-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M., Oppenheimer J. Elucidating asthma phenotypes and endotypes: progress towards personalized medicine. Ann. Allergy Asthma Immunol. 2016;116:394–401. doi: 10.1016/j.anai.2015.12.024. [DOI] [PubMed] [Google Scholar]
- Dickson R.P., Erb-Downward J.R., Freeman C.M., McCloskey L., Beck J.M., Huffnagle G.B., Curtis J.L. Spatial Variation in the Healthy Human Lung Microbiome and the Adapted Island Model of Lung Biogeography. Ann. Am. Thorac. Soc. 2015;12:821–830. doi: 10.1513/AnnalsATS.201501-029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R.P., Erb-Downward J.R., Freeman C.M., McCloskey L., Falkowski N.R., Huffnagle G.B., Curtis J.L. Bacterial Topography of the Healthy Human Lower Respiratory Tract. MBio. 2017;8 doi: 10.1128/mBio.02287-16. e02287–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R.P., Erb-Downward J.R., Martinez F.J., Huffnagle G.B. The Microbiome and the Respiratory Tract. Annu. Rev. Physiol. 2016;78:481–504. doi: 10.1146/annurev-physiol-021115-105238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durack J., Lynch S.V., Nariya S., Bhakta N.R., Beigelman A., Castro M., Dyer A.M., Israel E., Kraft M., Martin R.J., National Heart, Lung and Blood Institute’s “AsthmaNet” Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J. Allergy Clin. Immunol. 2017;140:63–75. doi: 10.1016/j.jaci.2016.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddens T., Campfield B.T., Serody K., Manni M.L., Horne W., Elsegeiny W., McHugh K.J., Pociask D., Chen K., Zheng M. A Novel CD4+ T Cell-Dependent Murine Model of Pneumocystis-driven Asthma-like Pathology. Am. J. Respir. Crit. Care Med. 2016;194:807–820. doi: 10.1164/rccm.201511-2205OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore S.A. Enhanced histopathology of mucosa-associated lymphoid tissue. Toxicol. Pathol. 2006;34:687–696. doi: 10.1080/01926230600939989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb-Downward J.R., Noverr M.C. Characterization of prostaglandin E2 production by Candida albicans. Infect. Immun. 2007;75:3498–3505. doi: 10.1128/IAI.00232-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essilfie A.T., Simpson J.L., Dunkley M.L., Morgan L.C., Oliver B.G., Gibson P.G., Foster P.S., Hansbro P.M. Combined Haemophilus influenzae respiratory infection and allergic airways disease drives chronic infection and features of neutrophilic asthma. Thorax. 2012;67:588–599. doi: 10.1136/thoraxjnl-2011-200160. [DOI] [PubMed] [Google Scholar]
- Fahy J.V. Type 2 inflammation in asthma--present in most, absent in many. Nat. Rev. Immunol. 2015;15:57–65. doi: 10.1038/nri3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti P., Pasolli E., Tett A., Asnicar F., Gorfer V., Fedi S., Armanini F., Truong D.T., Manara S., Zolfo M. Mother-to-Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome. Cell Host Microbe. 2018;24:133–145. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsythe P., Inman M.D., Bienenstock J. Oral treatment with live Lactobacillus reuteri inhibits the allergic airway response in mice. Am. J. Respir. Crit. Care Med. 2007;175:561–569. doi: 10.1164/rccm.200606-821OC. [DOI] [PubMed] [Google Scholar]
- Fraczek M.G., Chishimba L., Niven R.M., Bromley M., Simpson A., Smyth L., Denning D.W., Bowyer P. Corticosteroid treatment is associated with increased filamentous fungal burden in allergic fungal disease. J. Allergy Clin. Immunol. 2018;142:407–414. doi: 10.1016/j.jaci.2017.09.039. [DOI] [PubMed] [Google Scholar]
- Frank D.N., Feazel L.M., Bessesen M.T., Price C.S., Janoff E.N., Pace N.R. The human nasal microbiota and Staphylococcus aureus carriage. PLoS ONE. 2010;5:e10598. doi: 10.1371/journal.pone.0010598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freer G., Maggi F., Pifferi M., Di Cicco M.E., Peroni D.G., Pistello M. The Virome and Its Major Component, Anellovirus, a Convoluted System Molding Human Immune Defenses and Possibly Affecting the Development of Asthma and Respiratory Diseases in Childhood. Front. Microbiol. 2018;9:686. doi: 10.3389/fmicb.2018.00686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei R., Lauener R.P., Crameri R., O’Mahony L. Microbiota and dietary interactions: an update to the hygiene hypothesis? Allergy. 2012;67:451–461. doi: 10.1111/j.1398-9995.2011.02783.x. [DOI] [PubMed] [Google Scholar]
- Fujimura K.E., Sitarik A.R., Havstad S., Lin D.L., Levan S., Fadrosh D., Panzer A.R., LaMere B., Rackaityte E., Lukacs N.W. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016;22:1187–1191. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghebre M.A., Pang P.H., Diver S., Desai D., Bafadhel M., Haldar K., Kebadze T., Cohen S., Newbold P., Rapley L. Biological exacerbation clusters demonstrate asthma and chronic obstructive pulmonary disease overlap with distinct mediator and microbiome profiles. J. Allergy Clin. Immunol. 2018;141:2027–2036. doi: 10.1016/j.jaci.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D.L., Chen Z., Shankar V., Tyberg M., Vicencio A., Burk R. Lower airway microbiota and mycobiota in children with severe asthma. J. Allergy Clin. Immunol. 2018;141:808–811. doi: 10.1016/j.jaci.2017.09.018. [DOI] [PubMed] [Google Scholar]
- Gollwitzer E.S., Saglani S., Trompette A., Yadava K., Sherburn R., McCoy K.D., Nicod L.P., Lloyd C.M., Marsland B.J. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat. Med. 2014;20:642–647. doi: 10.1038/nm.3568. [DOI] [PubMed] [Google Scholar]
- Gomez de Agüero M., Ganal-Vonarburg S.C., Fuhrer T., Rupp S., Uchimura Y., Li H., Steinert A., Heikenwalder M., Hapfelmeier S., Sauer U. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351:1296–1302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- Green B.J., Wiriyachaiporn S., Grainge C., Rogers G.B., Kehagia V., Lau L., Carroll M.P., Bruce K.D., Howarth P.H. Potentially pathogenic airway bacteria and neutrophilic inflammation in treatment resistant severe asthma. PLoS ONE. 2014;9:e100645. doi: 10.1371/journal.pone.0100645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan T., Cortese M., Rouphael N., Boudreau C., Linde C., Maddur M.S., Das J., Wang H., Guthmiller J., Zheng N.Y. Antibiotics-Driven Gut Microbiome Perturbation Alters Immunity to Vaccines in Humans. Cell. 2019;178:1313–1328. doi: 10.1016/j.cell.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallen-Adams H.E., Suhr M.J. Fungi in the healthy human gastrointestinal tract. Virulence. 2017;8:352–358. doi: 10.1080/21505594.2016.1247140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Wen Q., Yao F., Xu D., Huang Y., Wang J. Gut-lung axis: The microbial contributions and clinical implications. Crit. Rev. Microbiol. 2017;43:81–95. doi: 10.1080/1040841X.2016.1176988. [DOI] [PubMed] [Google Scholar]
- Heinonen S., Rodriguez-Fernandez R., Diaz A., Rodriguez-Pastor S.O., Ramilo O., Mejias A. Infant Immune Response to Respiratory Viral Infections. Immunol. Allergy Clin. North Am. 2019;39:361–376. doi: 10.1016/j.iac.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard K.L., Allen E., Traber K.E., Yamamoto K., Stauffer N.M., Wasserman G.A., Jones M.R., Mizgerd J.P., Quinton L.J. The Lung-Liver Axis: A Requirement for Maximal Innate Immunity and Hepatoprotection during Pneumonia. Am. J. Respir. Cell Mol. Biol. 2015;53:378–390. doi: 10.1165/rcmb.2014-0195OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman E.T., Lu H., Yao T., Nakatsu C.H. Microbial Ecology along the Gastrointestinal Tract. Microbes Environ. 2017;32:300–313. doi: 10.1264/jsme2.ME17017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilty M., Burke C., Pedro H., Cardenas P., Bush A., Bossley C., Davies J., Ervine A., Poulter L., Pachter L. Disordered microbial communities in asthmatic airways. PLoS ONE. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C., Dollive S., Grunberg S., Chen J., Li H., Wu G.D., Lewis J.D., Bushman F.D. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS ONE. 2013;8:e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.J., Marsland B.J., Bunyavanich S., O’Mahony L., Leung D.Y., Muraro A., Fleisher T.A. The microbiome in allergic disease: Current understanding and future opportunities-2017 PRACTALL document of the American Academy of Allergy, Asthma & Immunology and the European Academy of Allergy and Clinical Immunology. J. Allergy Clin. Immunol. 2017;139:1099–1110. doi: 10.1016/j.jaci.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.J., Nariya S., Harris J.M., Lynch S.V., Choy D.F., Arron J.R., Boushey H. The airway microbiome in patients with severe asthma: Associations with disease features and severity. J. Allergy Clin. Immunol. 2015;136:874–884. doi: 10.1016/j.jaci.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagle G.B., Dickson R.P., Lukacs N.W. The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol. 2017;10:299–306. doi: 10.1038/mi.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev I.D., Leonardi I. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat. Rev. Immunol. 2017;17:635–646. doi: 10.1038/nri.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase T., Uehara Y., Shinji H., Tajima A., Seo H., Takada K., Agata T., Mizunoe Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465:346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- Jankauskaitė L., Misevičienė V., Vaidelienė L., Kėvalas R. Lower Airway Virology in Health and Disease-From Invaders to Symbionts. Medicina (Kaunas) 2018;54:72. doi: 10.3390/medicina54050072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R.R., Wieringa J., Koekkoek S.M., Visser C.E., Pajkrt D., Molenkamp R., de Jong M.D., Schinkel J. Frequent detection of respiratory viruses without symptoms: toward defining clinically relevant cutoff values. J. Clin. Microbiol. 2011;49:2631–2636. doi: 10.1128/JCM.02094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jartti T., Gern J.E. Role of viral infections in the development and exacerbation of asthma in children. J. Allergy Clin. Immunol. 2017;140:895–906. doi: 10.1016/j.jaci.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousset A., Bienhold C., Chatzinotas A., Gallien L., Gobet A., Kurm V., Küsel K., Rillig M.C., Rivett D.W., Salles J.F. Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J. 2017;11:853–862. doi: 10.1038/ismej.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast J.I., McFarlane A.J., Głobińska A., Sokolowska M., Wawrzyniak P., Sanak M., Schwarze J., Akdis C.A., Wanke K. Respiratory syncytial virus infection influences tight junction integrity. Clin. Exp. Immunol. 2017;190:351–359. doi: 10.1111/cei.13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R.Y., Pinkerton J.W., Essilfie A.T., Robertson A.A.B., Baines K.J., Brown A.C., Mayall J.R., Ali M.K., Starkey M.R., Hansbro N.G. Role for NLRP3 Inflammasome-mediated, IL-1β-Dependent Responses in Severe, Steroid-Resistant Asthma. Am. J. Respir. Crit. Care Med. 2017;196:283–297. doi: 10.1164/rccm.201609-1830OC. [DOI] [PubMed] [Google Scholar]
- Kim Y.-G., Udayanga K.G.S., Totsuka N., Weinberg J.B., Nunez G., Shibuya A. Gut Dysbiosis Promotes M2 Macrophage Polarization and Allergic Airway Inflammation via Fungi-Induced PGE2. Cell Host & Microbe. 2014 doi: 10.1016/j.chom.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirjavainen P.V., Karvonen A.M., Adams R.I., Täubel M., Roponen M., Tuoresmäki P., Loss G., Jayaprakash B., Depner M., Ege M.J. Farm-like indoor microbiota in non-farm homes protects children from asthma development. Nat. Med. 2019;25:1089–1095. doi: 10.1038/s41591-019-0469-4. [DOI] [PubMed] [Google Scholar]
- Koch M.A., Reiner G.L., Lugo K.A., Kreuk L.S., Stanbery A.G., Ansaldo E., Seher T.D., Ludington W.B., Barton G.M. Maternal IgG and IgA Antibodies Dampen Mucosal T Helper Cell Responses in Early Life. Cell. 2016;165:827–841. doi: 10.1016/j.cell.2016.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinen K., Pausan M.R., Perras A.K., Beck M., Bang C., Mora M., Schilhabel A., Schmitz R., Moissl-Eichinger C. First insights into the diverse human archaeome: Specific detection of Archaea in the gastrointestinal tract, lung, and nose and on skin. MBio. 2017;8:1–17. doi: 10.1128/mBio.00824-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.S., Pavlidis S., Loza M., Baribaud F., Rowe A., Pandis I., Sousa A., Corfield J., Djukanovic R., Lutter R., U-BIOPRED Study Group T-helper cell type 2 (Th2) and non-Th2 molecular phenotypes of asthma using sputum transcriptomics in U-BIOPRED. Eur. Respir. J. 2017;49:1602135. doi: 10.1183/13993003.02135-2016. [DOI] [PubMed] [Google Scholar]
- Leonardi I., Li X., Semon A., Li D., Doron I., Putzel G., Bar A., Prieto D., Rescigno M., McGovern D.P.B. CX3CR1+ mononuclear phagocytes control immunity to intestinal fungi. Science. 2018;359:232–236. doi: 10.1126/science.aao1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levan S.R., Stamnes K.A., Lin D.L., Panzer A.R., Fukui E., McCauley K., Fujimura K.E., McKean M., Ownby D.R., Zoratti E.M. Elevated faecal 12,13-diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat. Microbiol. 2019;4:1851–1861. doi: 10.1038/s41564-019-0498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais Lecours P., Duchaine C., Taillefer M., Tremblay C., Veillette M., Cormier Y., Marsolais D. Immunogenic properties of archaeal species found in bioaerosols. PLoS ONE. 2011;6:e23326. doi: 10.1371/journal.pone.0023326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M., Kolodziejczyk A.A., Thaiss C.A., Elinav E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017;17:219–232. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- Li X., Leonardi I., Semon A., Doron I., Gao I.H., Putzel G.G., Kim Y., Kabata H., Artis D., Fiers W.D. Response to Fungal Dysbiosis by Gut-Resident CX3CR1+ Mononuclear Phagocytes Aggravates Allergic Airway Disease. Cell Host Microbe. 2018;24:847–856. doi: 10.1016/j.chom.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Tan J., Liu J., Feng H., Pan D. Altered mast cell activity in response to rhinovirus infection provides novel insight into asthma. J. Asthma. 2019;18:1–9. doi: 10.1080/02770903.2019.1585870. [DOI] [PubMed] [Google Scholar]
- Loewen K., Monchka B., Mahmud S.M., ’t Jong G., Azad M.B. Prenatal antibiotic exposure and childhood asthma: a population-based study. Eur. Respir. J. 2018;52:1702070. doi: 10.1183/13993003.02070-2017. [DOI] [PubMed] [Google Scholar]
- Lozupone C.A., Stombaugh J.I., Gordon J.I., Jansson J.K., Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M.D.J., Neufeld J.D. Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 2015;13:217–229. doi: 10.1038/nrmicro3400. [DOI] [PubMed] [Google Scholar]
- Mahnic A., Rupnik M. Different host factors are associated with patterns in bacterial and fungal gut microbiota in Slovenian healthy cohort. PLoS ONE. 2018;13:e0209209. doi: 10.1371/journal.pone.0209209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makris S., Johnston S. Recent advances in understanding rhinovirus immunity. F1000Res. 2018;7 doi: 10.12688/f1000research.15337.1. F1000 Faculty Rev-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man W.H., de Steenhuijsen Piters W.A., Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017;15:259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marri P.R., Stern D.A., Wright A.L., Billheimer D., Martinez F.D. Asthma-associated differences in microbial composition of induced sputum. J. Allergy Clin. Immunol. 2013;131:346–352. doi: 10.1016/j.jaci.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsland B.J., Trompette A., Gollwitzer E.S. The Gut-Lung Axis in Respiratory Disease. Ann. Am. Thorac. Soc. 2015;12(Suppl 2):S150–S156. doi: 10.1513/AnnalsATS.201503-133AW. [DOI] [PubMed] [Google Scholar]
- McLoughlin R., Berthon B.S., Rogers G.B., Baines K.J., Leong L.E.X., Gibson P.G., Williams E.J., Wood L.G. Soluble fibre supplementation with and without a probiotic in adults with asthma: A 7-day randomised, double blind, three way cross-over trial. EBioMedicine. 2019;46:473–485. doi: 10.1016/j.ebiom.2019.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner H.C. Viral Bronchiolitis in Children. N. Engl. J. Med. 2016;374:1793–1794. doi: 10.1056/NEJMc1601509. [DOI] [PubMed] [Google Scholar]
- Michalovich D., Rodriguez-Perez N., Smolinska S., Pirozynski M., Mayhew D., Uddin S., Van Horn S., Sokolowska M., Altunbulakli C., Eljaszewicz A. Obesity and disease severity magnify disturbed microbiome-immune interactions in asthma patients. Nature Communication. 2019 doi: 10.1038/s41467-019-13751-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhail I., Grayson M.H. Asthma and viral infections: An intricate relationship. Ann. Allergy Asthma Immunol. 2019;123:352–358. doi: 10.1016/j.anai.2019.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitre E., Susi A., Kropp L.E., Schwartz D.J., Gorman G.H., Nylund C.M. Association Between Use of Acid-Suppressive Medications and Antibiotics During Infancy and Allergic Diseases in Early Childhood. JAMA Pediatr. 2018;172:e180315. doi: 10.1001/jamapediatrics.2018.0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt M.F., Cookson W.O. The lung microbiome in health and disease. Clin. Med. (Lond.) 2017;17:525–529. doi: 10.7861/clinmedicine.17-6-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noverr M.C., Falkowski N.R., McDonald R.A., McKenzie A.N., Huffnagle G.B. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect. Immun. 2005;73:30–38. doi: 10.1128/IAI.73.1.30-38.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noverr M.C., Huffnagle G.B. Regulation of Candida albicans morphogenesis by fatty acid metabolites. Infect. Immun. 2004;72:6206–6210. doi: 10.1128/IAI.72.11.6206-6210.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noverr M.C., Noggle R.M., Toews G.B., Huffnagle G.B. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect. Immun. 2004;72:4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noverr M.C., Phare S.M., Toews G.B., Coffey M.J., Huffnagle G.B. Pathogenic yeasts Cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infect. Immun. 2001;69:2957–2963. doi: 10.1128/IAI.69.5.2957-2963.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pammi M., Lal C.V., Wagner B.D., Mourani P.M., Lohmann P., Luna R.A., Sisson A., Shivanna B., Hollister E.B., Abman S.H. Airway Microbiome and Development of Bronchopulmonary Dysplasia in Preterm Infants: A Systematic Review. J. Pediatr. 2019;204:126–133. doi: 10.1016/j.jpeds.2018.08.042. [DOI] [PubMed] [Google Scholar]
- Pattaroni C., Watzenboeck M.L., Schneidegger S., Kieser S., Wong N.C., Bernasconi E., Pernot J., Mercier L., Knapp S., Nicod L.P. Early-Life Formation of the Microbial and Immunological Environment of the Human Airways. Cell Host Microbe. 2018;24:857–865. doi: 10.1016/j.chom.2018.10.019. [DOI] [PubMed] [Google Scholar]
- Pavlidis S., Takahashi K., Ng Kee Kwong F., Xie J., Hoda U., Sun K., Elyasigomari V., Agapow P., Loza M., Baribaud F., on behalf of the U-BIOPRED Study Group “T2-high” in severe asthma related to blood eosinophil, exhaled nitric oxide and serum periostin. Eur. Respir. J. 2019;53:1800938. doi: 10.1183/13993003.00938-2018. [DOI] [PubMed] [Google Scholar]
- Pereira F.C., Berry D. Microbial nutrient niches in the gut. Environ. Microbiol. 2017;19:1366–1378. doi: 10.1111/1462-2920.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez F.J., Ponce C.A., Rojas D.A., Iturra P.A., Bustamante R.I., Gallo M., Hananias K., Vargas S.L. Fungal colonization with Pneumocystis correlates to increasing chloride channel accessory 1 (hCLCA1) suggesting a pathway for up-regulation of airway mucus responses, in infant lungs. Results Immunol. 2014;4:58–61. doi: 10.1016/j.rinim.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone E.E., Jung E., Breed E., Dominguez J.A., Liang Z., Clark A.T., Dunne W.M., Burd E.M., Coopersmith C.M. Mechanisms of methicillin-resistant Staphylococcus aureus pneumonia-induced intestinal epithelial apoptosis. Shock. 2012;38:68–75. doi: 10.1097/SHK.0b013e318259abdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter P., Susarla S.C., Polikepahad S., Qian Y., Hampton J., Kiss A., Vaidya S., Sur S., Ongeri V., Yang T. Link between allergic asthma and airway mucosal infection suggested by proteinase-secreting household fungi. Mucosal Immunol. 2009;2:504–517. doi: 10.1038/mi.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugin B., Barcik W., Westermann P., Heider A., Wawrzyniak M., Hellings P., Akdis C.A., O’Mahony L. A wide diversity of bacteria from the human gut produces and degrades biogenic amines. Microb. Ecol. Health Dis. 2017;28:1353881. doi: 10.1080/16512235.2017.1353881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin N., Zheng B., Yao J., Guo L., Zuo J., Wu L., Zhou J., Liu L., Guo J., Ni S. Influence of H7N9 virus infection and associated treatment on human gut microbiota. Sci. Rep. 2015;5:14771. doi: 10.1038/srep14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramratnam S.K., Bacharier L.B., Guilbert T.W. Severe Asthma in Children. J. Allergy Clin. Immunol. Pract. 2017;5:889–898. doi: 10.1016/j.jaip.2017.04.031. [DOI] [PubMed] [Google Scholar]
- Ravanetti L., Dijkhuis A., Dekker T., Sabogal Pineros Y.S., Ravi A., Dierdorp B.S., Erjefält J.S., Mori M., Pavlidis S., Adcock I.M. IL-33 drives influenza-induced asthma exacerbations by halting innate and adaptive antiviral immunity. J. Allergy Clin. Immunol. 2019;143:1355–1370. doi: 10.1016/j.jaci.2018.08.051. [DOI] [PubMed] [Google Scholar]
- Ray A., Kolls J.K. Neutrophilic Inflammation in Asthma and Association with Disease Severity. Trends Immunol. 2017;38:942–954. doi: 10.1016/j.it.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie A.I., Farne H.A., Singanayagam A., Jackson D.J., Mallia P., Johnston S.L. Pathogenesis of Viral Infection in Exacerbations of Airway Disease. Ann. Am. Thorac. Soc. 2015;12(Suppl 2):S115–S132. doi: 10.1513/AnnalsATS.201503-151AW. [DOI] [PubMed] [Google Scholar]
- Roduit C., Frei R., Ferstl R., Loeliger S., Westermann P., Rhyner C., Schiavi E., Barcik W., Rodriguez-Perez N., Wawrzyniak M., PASTURE/EFRAIM study group High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy. 2019;74:799–809. doi: 10.1111/all.13660. [DOI] [PubMed] [Google Scholar]
- Rojas D.A., Iturra P.A., Méndez A., Ponce C.A., Bustamante R., Gallo M., Bórquez P., Vargas S.L. Increase in secreted airway mucins and partial Muc5b STAT6/FoxA2 regulation during Pneumocystis primary infection. Sci. Rep. 2019;9:2078. doi: 10.1038/s41598-019-39079-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossios C., Pavlidis S., Hoda U., Kuo C.H., Wiegman C., Russell K., Sun K., Loza M.J., Baribaud F., Durham A.L., Unbiased Biomarkers for the Prediction of Respiratory Diseases Outcomes (U-BIOPRED) Consortia Project Team Sputum transcriptomics reveal upregulation of IL-1 receptor family members in patients with severe asthma. J. Allergy Clin. Immunol. 2018;141:560–570. doi: 10.1016/j.jaci.2017.02.045. [DOI] [PubMed] [Google Scholar]
- Rubner F.J., Jackson D.J., Evans M.D., Gangnon R.E., Tisler C.J., Pappas T.E., Gern J.E., Lemanske R.F., Jr. Early life rhinovirus wheezing, allergic sensitization, and asthma risk at adolescence. J. Allergy Clin. Immunol. 2017;139:501–507. doi: 10.1016/j.jaci.2016.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabogal Piñeros Y.S., Bal S.M., Dijkhuis A., Majoor C.J., Dierdorp B.S., Dekker T., Hoefsmit E.P., Bonta P.I., Picavet D., van der Wel N.N. Eosinophils capture viruses, a capacity that is defective in asthma. Allergy. 2019;74:1898–1909. doi: 10.1111/all.13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarna M., Lambert S.B., Sloots T.P., Whiley D.M., Alsaleh A., Mhango L., Bialasiewicz S., Wang D., Nissen M.D., Grimwood K., Ware R.S. Viruses causing lower respiratory symptoms in young children: findings from the ORChID birth cohort. Thorax. 2018;73:969–979. doi: 10.1136/thoraxjnl-2017-210233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage D.C. Microbial interference between indigenous yeast and lactobacilli in the rodent stomach. J. Bacteriol. 1969;98:1278–1283. doi: 10.1128/jb.98.3.1278-1283.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal L.N., Clemente J.C., Tsay J.C., Koralov S.B., Keller B.C., Wu B.G., Li Y., Shen N., Ghedin E., Morris A. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat. Microbiol. 2016;1:16031. doi: 10.1038/nmicrobiol.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R., Fuchs S., Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell. 2016;164:337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- Shao T.Y., Ang W.X.G., Jiang T.T., Huang F.S., Andersen H., Kinder J.M., Pham G., Burg A.R., Ruff B., Gonzalez T. Commensal Candida albicans Positively Calibrates Systemic Th17 Immunological Responses. Cell Host Microbe. 2019;25:404–417. doi: 10.1016/j.chom.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Laxman B., Naureckas E.T., Hogarth D.K., Sperling A.I., Solway J., Ober C., Gilbert J.A., White S.R. Associations between fungal and bacterial microbiota of airways and asthma endotypes. Journal of Allergy and Clinical immunology. 2019 doi: 10.1016/j.jaci.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J.L., Daly J., Baines K.J., Yang I.A., Upham J.W., Reynolds P.N., Hodge S., James A.L., Hugenholtz P., Willner D., Gibson P.G. Airway dysbiosis: Haemophilus influenzae and Tropheryma in poorly controlled asthma. Eur. Respir. J. 2016;47:792–800. doi: 10.1183/13993003.00405-2015. [DOI] [PubMed] [Google Scholar]
- Singleton R.J., Bulkow L.R., Miernyk K., DeByle C., Pruitt L., Hummel K.B., Bruden D., Englund J.A., Anderson L.J., Lucher L. Viral respiratory infections in hospitalized and community control children in Alaska. J. Med. Virol. 2010;82:1282–1290. doi: 10.1002/jmv.21790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitarik A.R., Havstad S., Levin A.M., Lynch S.V., Fujimura K.E., Ownby D.R., Johnson C.C., Wegienka G. Dog introduction alters the home dust microbiota. Indoor Air. 2018;28:539–547. doi: 10.1111/ina.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalski J.H., Limon J.J., Sharma P., Gargus M.D., Nguyen C., Tang J., Coelho A.L., Hogaboam C.M., Crother T.R., Underhill D.M. Expansion of commensal fungus Wallemia mellicola in the gastrointestinal mycobiota enhances the severity of allergic airway disease in mice. PLoS Pathog. 2018;14:e1007260. doi: 10.1371/journal.ppat.1007260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowska M., Chen L.Y., Liu Y., Martinez-Anton A., Logun C., Alsaaty S., Cuento R.A., Cai R., Sun J., Quehenberger O. Dysregulation of lipidomic profile and antiviral immunity in response to hyaluronan in patients with severe asthma. J. Allergy Clin. Immunol. 2017;139:1379–1383. doi: 10.1016/j.jaci.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowska M., Frei R., Lunjani N., Akdis C.A., O’Mahony L. Microbiome and asthma. Asthma Res. Pract. 2018;4:1. doi: 10.1186/s40733-017-0037-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M.M., Hrusch C.L., Gozdz J., Igartua C., Pivniouk V., Murray S.E., Ledford J.G., Marques Dos Santos M., Anderson R.L., Metwali N. Innate Immunity and Asthma Risk in Amish and Hutterite Farm Children. N. Engl. J. Med. 2016;375:411–421. doi: 10.1056/NEJMoa1508749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.A., Morgan W.J., Halonen M., Wright A.L., Martinez F.D. Wheezing and bronchial hyper-responsiveness in early childhood as predictors of newly diagnosed asthma in early adulthood: a longitudinal birth-cohort study. Lancet. 2008;372:1058–1064. doi: 10.1016/S0140-6736(08)61447-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiemsma L.T., Turvey S.E. Asthma and the microbiome: defining the critical window in early life. Allergy Asthma Clin. Immunol. 2017;13:3. doi: 10.1186/s13223-016-0173-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokholm J., Blaser M.J., Thorsen J., Rasmussen M.A., Waage J., Vinding R.K., Schoos A.M., Kunøe A., Fink N.R., Chawes B.L. Maturation of the gut microbiome and risk of asthma in childhood. Nat. Commun. 2018;9:141. doi: 10.1038/s41467-017-02573-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strati F., Di Paola M., Stefanini I., Albanese D., Rizzetto L., Lionetti P., Calabrò A., Jousson O., Donati C., Cavalieri D., De Filippo C. Age and gender affect the composition of fungal population of the human gastrointestinal tract. Front. Microbiol. 2016;7:1227. doi: 10.3389/fmicb.2016.01227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita K., Sokolowska M., Akdis C.A. Key points for moving the endotypes field forward. In: Agache I.X., Hellings P., editors. Implementing Precision Medicine in Best Practices of Chronic Airway Diseases. Academic Press; 2018. pp. 107–114. [Google Scholar]
- Sun H., Li S., Yan Y., Chen Z., Wang Y., Hao C., Ji W. Associations between patient clinical characteristics and the presence of cytomegalovirus DNA in the bronchoalveolar lavage fluid of children with recurrent wheezing. BMC Infect. Dis. 2018;18:458. doi: 10.1186/s12879-018-3345-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundell N., Andersson L.M., Brittain-Long R., Sundvall P.D., Alsiö Å., Lindh M., Gustavsson L., Westin J. PCR Detection of Respiratory Pathogens in Asymptomatic and Symptomatic Adults. J. Clin. Microbiol. 2019;57:e00716–e00718. doi: 10.1128/JCM.00716-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze M.A., Tsuruta M., Yang S.W., Oh Y., Man S.F., Hogg J.C., Sin D.D. Changes in the bacterial microbiota in gut, blood, and lungs following acute LPS instillation into mice lungs. PLoS ONE. 2014;9:e111228. doi: 10.1371/journal.pone.0111228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H.T., Hagner S., Ruchti F., Radzikowska U., Tan G., Altunbulakli C., Eljaszewicz A., Moniuszko M., Akdis M., Akdis C.A. Tight junction, mucin, and inflammasome-related molecules are differentially expressed in eosinophilic, mixed, and neutrophilic experimental asthma in mice. Allergy. 2019;74:294–307. doi: 10.1111/all.13619. [DOI] [PubMed] [Google Scholar]
- Taylor S.L., Leong L.E.X., Choo J.M., Wesselingh S., Yang I.A., Upham J.W., Reynolds P.N., Hodge S., James A.L., Jenkins C. Inflammatory phenotypes in patients with severe asthma are associated with distinct airway microbiology. J. Allergy Clin. Immunol. 2018;141:94–103. doi: 10.1016/j.jaci.2017.03.044. [DOI] [PubMed] [Google Scholar]
- Taylor S.L., Leong L.E.X., Mobegi F.M., Choo J.M., Wesselingh S., Yang I.A., Upham J.W., Reynolds P.N., Hodge S., James A.L. Long-Term Azithromycin Reduces Haemophilus influenzae and Increases Antibiotic Resistance in Severe Asthma. Am. J. Respir. Crit. Care Med. 2019;200:309–317. doi: 10.1164/rccm.201809-1739OC. [DOI] [PubMed] [Google Scholar]
- Teo S.M., Mok D., Pham K., Kusel M., Serralha M., Troy N., Holt B.J., Hales B.J., Walker M.L., Hollams E. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint M., Jackson D.J., Swieboda D., Guedán A., Tsourouktsoglou T.D., Ching Y.M., Radermecker C., Makrinioti H., Aniscenko J., Bartlett N.W. Host DNA released by NETosis promotes rhinovirus-induced type-2 allergic asthma exacerbation. Nat. Med. 2017;23:681–691. doi: 10.1038/nm.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette A., Gollwitzer E.S., Yadava K., Sichelstiel A.K., Sprenger N., Ngom-Bru C., Blanchard C., Junt T., Nicod L.P., Harris N.L., Marsland B.J. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- Tun H.M., Konya T., Takaro T.K., Brook J.R., Chari R., Field C.J., Guttman D.S., Becker A.B., Mandhane P.J., Turvey S.E., CHILD Study Investigators Exposure to household furry pets influences the gut microbiota of infant at 3-4 months following various birth scenarios. Microbiome. 2017;5:40. doi: 10.1186/s40168-017-0254-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., Gordon J.I. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bergh M.R., Biesbroek G., Rossen J.W., de Steenhuijsen Piters W.A., Bosch A.A., van Gils E.J., Wang X., Boonacker C.W., Veenhoven R.H., Bruin J.P. Associations between pathogens in the upper respiratory tract of young children: interplay between viruses and bacteria. PLoS ONE. 2012;7:e47711. doi: 10.1371/journal.pone.0047711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken S.J., Garcia D., Porter P., Huang X., Quinlan P.J., Blanc P.D., Corry D.B., Locksley R.M. Fungal chitin from asthma-associated home environments induces eosinophilic lung infiltration. J. Immunol. 2011;187:2261–2267. doi: 10.4049/jimmunol.1100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Woerden H.C., Gregory C., Brown R., Marchesi J.R., Hoogendoorn B., Matthews I.P. Differences in fungi present in induced sputum samples from asthma patients and non-atopic controls: a community based case control study. BMC Infect. Dis. 2013;13:69. doi: 10.1186/1471-2334-13-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandini S., Biagi C., Fischer M., Lanari M. Impact of Rhinovirus Infections in Children. Viruses. 2019;11:E521. doi: 10.3390/v11060521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas S.L., Hughes W.T., Santolaya M.E., Ulloa A.V., Ponce C.A., Cabrera C.E., Cumsille F., Gigliotti F. Search for primary infection by Pneumocystis carinii in a cohort of normal, healthy infants. Clin. Infect. Dis. 2001;32:855–861. doi: 10.1086/319340. [DOI] [PubMed] [Google Scholar]
- Vargas S.L., Ponce C.A., Gallo M., Pérez F., Astorga J.F., Bustamante R., Chabé M., Durand-Joly I., Iturra P., Miller R.F. Near-universal prevalence of Pneumocystis and associated increase in mucus in the lungs of infants with sudden unexpected death. Clin. Infect. Dis. 2013;56:171–179. doi: 10.1093/cid/cis870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas S.L., Ponce C.A., Luchsinger V., Silva C., Gallo M., López R., Belletti J., Velozo L., Avila R., Palomino M.A. Detection of Pneumocystis carinii f. sp. hominis and viruses in presumably immunocompetent infants who died in the hospital or in the community. J. Infect. Dis. 2005;191:122–126. doi: 10.1086/426451. [DOI] [PubMed] [Google Scholar]
- Wang F., He X.Y., Baines K.J., Gunawardhana L.P., Simpson J.L., Li F., Gibson P.G. Different inflammatory phenotypes in adults and children with acute asthma. Eur. Respir. J. 2011;38:567–574. doi: 10.1183/09031936.00170110. [DOI] [PubMed] [Google Scholar]
- Wang J., Li F., Sun R., Gao X., Wei H., Li L.J., Tian Z. Bacterial colonization dampens influenza-mediated acute lung injury via induction of M2 alveolar macrophages. Nat. Commun. 2013;4:2106. doi: 10.1038/ncomms3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibel E.R. Morphometry of the human lung: the state of the art after two decades. Bull. Eur. Physiopathol. Respir. 1979;15:999–1013. [PubMed] [Google Scholar]
- Wenzel S.E. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat. Med. 2012;18:716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- Wheeler M.L., Limon J.J., Bar A.S., Leal C.A., Gargus M., Tang J., Brown J., Funari V.A., Wang H.L., Crother T.R. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe. 2016;19:865–873. doi: 10.1016/j.chom.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner D., Furlan M., Haynes M., Schmieder R., Angly F.E., Silva J., Tammadoni S., Nosrat B., Conrad D., Rohwer F. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS ONE. 2009;4:e7370. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolsk H.M., Chawes B.L., Følsgaard N.V., Rasmussen M.A., Brix S., Bisgaard H. Siblings Promote a Type 1/Type 17-oriented immune response in the airways of asymptomatic neonates. Allergy. 2016;71:820–828. doi: 10.1111/all.12847. [DOI] [PubMed] [Google Scholar]
- Wos-Oxley M.L., Chaves-Moreno D., Jáuregui R., Oxley A.P., Kaspar U., Plumeier I., Kahl S., Rudack C., Becker K., Pieper D.H. Exploring the bacterial assemblages along the human nasal passage. Environ. Microbiol. 2016;18:2259–2271. doi: 10.1111/1462-2920.13378. [DOI] [PubMed] [Google Scholar]
- Wu P., Dupont W.D., Griffin M.R., Carroll K.N., Mitchel E.F., Gebretsadik T., Hartert T.V. Evidence of a causal role of winter virus infection during infancy in early childhood asthma. Am. J. Respir. Crit. Care Med. 2008;178:1123–1129. doi: 10.1164/rccm.200804-579OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie K.M., Mihindukulasuriya K.A., Sodergren E., Weinstock G.M., Storch G.A. Sequence analysis of the human virome in febrile and afebrile children. PLoS ONE. 2012;7:e27735. doi: 10.1371/journal.pone.0027735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wypych T.P., Wickramasinghe L.C., Marsland B.J. The influence of the microbiome on respiratory health. Nat. Immunol. 2019;20:1279–1290. doi: 10.1038/s41590-019-0451-9. [DOI] [PubMed] [Google Scholar]
- Yassour M., Jason E., Hogstrom L.J., Arthur T.D., Tripathi S., Siljander H., Selvenius J., Oikarinen S., Hyoty H., Virtanen S.M. Strain-Level Analysis of Mother-to-Child Bacterial Transmission during the First Few Months of Life. Cell Host Microbe. 2018;24:146–154. doi: 10.1016/j.chom.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J.C., Chehoud C., Bittinger K., Bailey A., Diamond J.M., Cantu E., Haas A.R., Abbas A., Frye L., Christie J.D. Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients. Am. J. Transplant. 2015;15:200–209. doi: 10.1111/ajt.13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.P., Hopkins R.J., Marsland B. The Gut-Liver-Lung Axis. Modulation of the Innate Immune Response and Its Possible Role in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Cell Mol. Biol. 2016;54:161–169. doi: 10.1165/rcmb.2015-0250PS. [DOI] [PubMed] [Google Scholar]
- Yun Y., Srinivas G., Kuenzel S., Linnenbrink M., Alnahas S., Bruce K.D., Steinhoff U., Baines J.F., Schaible U.E. Environmentally determined differences in the murine lung microbiota and their relation to alveolar architecture. PLoS ONE. 2014;9:e113466. doi: 10.1371/journal.pone.0113466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Cox M., Liang Z., Brinkmann F., Cardenas P.A., Duff R., Bhavsar P., Cookson W., Moffatt M., Chung K.F. Airway Microbiota in Severe Asthma and Relationship to Asthma Severity and Phenotypes. PLoS ONE. 2016;11:e0152724. doi: 10.1371/journal.pone.0152724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivaki D., Lemoine S., Lim A., Morva A., Vidalain P.O., Schandene L., Casartelli N., Rameix-Welti M.A., Hervé P.L., Dériaud E. Respiratory Syncytial Virus Infects Regulatory B Cells in Human Neonates via Chemokine Receptor CX3CR1 and Promotes Lung Disease Severity. Immunity. 2017;46:301–314. doi: 10.1016/j.immuni.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Mihindukulasuriya K.A., Gao H., La Rosa P.S., Wylie K.M., Martin J.C., Kota K., Shannon W.D., Mitreva M., Sodergren E., Weinstock G.M. Exploration of bacterial community classes in major human habitats. Genome Biol. 2014;15:R66. doi: 10.1186/gb-2014-15-5-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]