Abstract
Rapid and reliable diagnosis of influenza is essential for identification of contagious patients and effective patient management. Near‐patient assays allow establishment of the diagnosis within minutes in young children, and this study aimed to evaluate near‐patient assays in relation to the patient’s age. A total of 194 patients with laboratory‐confirmed influenza A/H3N2 virus infection, diagnosed within a prospective cohort study, were included. Cryopreserved nasopharyngeal swabs collected from these patients were tested by four near‐patient assays (Binax Now Influenza A&B, Quick S‐Influ A/B, Influ‐A&B Respi‐Strip, and Actim Influenza A&B). The main outcome measure was sensitivity of the near‐patient assays in relation to the age of patients. The Binax Now, Quick S‐Influ, Influ‐A&B Respi‐Strip and Actim assays had overall sensitivities of 19%, 18%, 26%, and 40%, respectively. The estimated sensitivity for influenza A/H3N2 virus detection in nasopharyngeal swabs was 17–56% in children 1 year of age and decreased to 8–22% in patients 80 years of age (logistic regression). The sensitivity of the Influ‐A&B Respi‐Strip and Actim assays decreased significantly with increasing age (p 0.014 and p 0.033, respectively (logistic regression)), a trend for decrease was observed for the Binax Now assay (p 0.074 (logistic regression)), and the low sensitivity of the Quick S‐Influ assay was similar in children and adults. Less than one‐fourth of diagnosed influenza A/H3N2 virus infections can be identified in elderly patients using a near‐patient assay. Consequently, near‐patient assays are of limited value for confirming the diagnosis when influenza is clinically suspected in adults. Antiviral therapy and additional diagnostic procedures cannot be withheld on the basis of a negative near‐patient assay result, particularly in adult patients.
Keywords: Influenza virus infection, point‐of‐care diagnosis, respiratory tract disease, sensitivity, test evaluation study
Introduction
An estimated 200 000 hospitalizations and 36 000 deaths are attributable to influenza annually in the USA alone [1]. Elderly patients are most affected; that is, influenza‐related deaths, hospitalizations and use of health services are recorded mostly for patients >65 years of age [2, 3]. The virus can spread particularly rapidly in care homes, potentially affecting 60% of residents [4]. Accurate and rapid diagnosis of influenza is essential for identification of contagious patients and effective management of cases of influenza. Management decisions regarding infection control, referral to specialized units, use of additional diagnostic procedures and administration of antiviral therapy have to be made at the point of first contact with the patient. The antiviral drugs oseltamivir and zanamivir are highly effective against influenza A and B viruses, but must be administered as soon as possible, at least within the first 48 h after the onset of the acute respiratory disease, to be effective [5, 6].
Reliable identification of cases of influenza within this short time frame is challenging. The sensitivity and specificity of the clinical diagnosis of influenza are as low as 64–77% and 55–67%, respectively, even during the peak of the flu season [7, 8].
Laboratory‐based assays for the diagnosis of influenza, e.g. detection of virus or viral genome in respiratory secretions via virus culture or influenza virus‐specific PCR, are highly sensitive and specific [9]. Test results, however, usually become available more than 48 h after sample collection and several days after onset of the disease.
Near‐patient diagnostic assays provide results within 30 min, and almost 20 commercial assays are currently available for the diagnosis of influenza [10]. The specificity of these assays is well above 95% [10], but the values for test sensitivity stated in the manufacturers’ product information differ, sometimes considerably, from those described in investigator‐initiated studies [11, 12].
Previously, we observed that the sensitivity of a laboratory‐based diagnostic assay for detection of influenza virus or viral particles in respiratory specimen decreases significantly with increasing age of the patient [9]. Manufacturer‐initiated evaluations of near‐patient assays, to our knowledge, concern exclusively respiratory specimens collected from very young children (information provided by Binax Inc., Coris Bioconcept, Medix Biochemica, and Denka Seiken). However, studies that revealed a clearly lower sensitivity of these assays included samples from all age groups [11, 12]. Thus, it was suspected that the patient’s age may influence the detection rates achieved by near‐patient assays. It was therefore the aim of the present study to evaluate in parallel several different near‐patient assays by testing respiratory specimens from both children and adults with laboratory‐confirmed influenza A virus infection.
Materials and Methods
Patient samples
Virological surveillance of respiratory virus activity in Austria is based on respiratory specimen collected by sentinel physicians (physicians of inpatient and outpatient hospital units, general practitioners and paediatricians throughout Austria) within two surveillance systems—the Diagnostic Influenza Network Austria (DINOE) and the Respiratory Network (RNW). The inclusion criterion for the DINOE is an influenza‐like illness (acute onset of fever with respiratory and constitutional symptoms) and that for the RNW is the clinical diagnosis of an acute respiratory tract infection including rhinitis, bronchitis and pneumonia. These two surveillance systems include patients of all age groups with acute respiratory tract infections. Nasopharyngeal swabs are taken by experienced physicians, with a swab being taken from both the nose and the throat of the patients. The swab content is tranferred into 1000 μL of transport medium consisting of minimal essential medium (MEM) (GIBCO; Life Technologies, Lofer, Austria) and discarded thereafter. All clinicians participating in the two surveillance systems were provided with transport medium by the Clinical Institute of Virology, Medical University of Vienna. Immediately after delivery, the clinical samples were vortexed vigorously, diluted in MEM (1 : 2), and split into aliquots before being screened for influenza A and B viruses by virus isolation or RT‐PCR, or both.
For the present investigation, influenza A/H3N2‐positive samples that were collected during the period of epidemic activity of influenza in the winter seasons of 2005–2006 and 2006–2007 were tested and cryopreserved for later analysis. Samples were frozen at −70°C and thawed only once for the present investigation. Only samples collected from patients younger than 10 years or older than 35 years were included. The separation of patients into these two age groups was done for practical reasons. In the two surveillance networks DINOE and RNW, the highest consultation rates were observed in patients younger than 10 years or older than 35 years during the influenza season. The aim was therefore to optimize the project for the comparative assessment of near‐patient assays in these age groups. For the purpose of standardization, only influenza A virus‐positive samples were included. The virological diagnosis was established in all patients by influenza A virus‐specific PCR and confirmed by virus isolation.
Virus isolation in tissue culture and typing of the isolates
Virus isolation in tissue culture and typing of the isolates were carried out on Madin–Darby canine kidney cells (American Type Culture Collection, Manassas, VA, USA) according to standard procedures [13]. The influenza A virus strains isolated during the study period were typed and subtyped as described previously [14].
Detection of influenza A virus RNA sequences
Viral genome RNA was detected in clinical samples by a nested PCR assay as described previously [9]. In brief, viral RNA was extracted from 140 μL of the clinical sample by using QIAamp Viral RNA mini kits (QIAGEN, Hilden, Germany). For reverse transcription and the two steps of the nested PCR, influenza A virus type‐specific primers that bind to the highly conserved region of the influenza A virus genome coding for non‐structural protein 1 were used. Each PCR experiment included two to five positive controls, several negative controls, and two to three respiratory virus‐positive specimens (respiratory syncytial virus, enteroviruses, rhinoviruses, coronavirus, parainfluenza viruses or adenoviruses) interposed between the samples tested. Quantification of influenza A virus‐specific RNA in clinical samples was done as described previously [15].
Detection of influenza virus antigen by ELISA
The in‐house ELISA used for detection of influenza virus antigen was carried out as described previously [9, 16].
Detection of influenza virus antigen using near‐patient assays
The commercially available rapid diagnostic tests are screening tests for influenza A and B virus infections that can provide results within 30 min. These tests are immunoassays that detect influenza viral antigen in respiratory specimens with the use of virus‐specific antibodies. They may also be referred to as rapid tests or point‐of‐care tests. All official Austrian representatives of the manufacturers of assays for the near‐patient diagnosis of influenza (n = 4) were invited to participate in the present study by donating kits sufficient in number for testing 30 clinical specimens from children and 50 clinical specimens from adults. Table 1 summarizes selected characteristics of the four rapid assays evaluated in the present study. According to the manufacturers’ product informations, nasopharyngeal swabs in transport medium may be used without significant loss of sensitivity and specificity of these assays. A total of 200 μL of clinical specimen was used for each test. This quantity equals the average volume of nasopharyngeal secretion collected by a standard cotton swab (Medix Biochemica and Denka Seiken, personal communication). Clinical specimens from 194 patients were included for the evaluation of the four assays. The volume of the 194 samples was not sufficient for testing by all four assays. Therefore, samples were assigned randomly within the two age groups to the four diagnostic assays to allow equal numbers of samples to be tested with the four assays.
Table 1.
Selected characteristics of the near‐patient assays evaluated in the present study
| Assay | Manufacturer | Influenza type detected | Recommended specimen | Recommended incubation period | Sensitivity for influenza A viruses | Specificity for influenza A viruses | ||
|---|---|---|---|---|---|---|---|---|
| Manufacturer’s product information (%) | Investigator‐initiated studies (%) | Manufacturer’s product information (%) | Investigator‐initiated studies (%) | |||||
| Binax Now Influenza A&B | Binax Inc., Portland, ME, USA | A and B | Nasal wash, nasal aspirate, nasopharyngeal swabs | 15 min | 100 | 59–80 [12, 22, 23] | 92–93 | 98–99 [12, 22] |
| Quick S‐Influ A/B ‘Seiken’ | Denka Seiken Co., Ltd, Tokyo, Japan | A and B | Nasal swab, nasal aspirate | 15 min | 90–93 | 81 [11] | 98–99 | 96 [11] |
| Influ‐A&B Respi‐Strip | Coris BioConcept, Gembloux, Belgium | A and B | Nasopharyngeal aspirates, washings or swabs | 15 min | 99 | ND | 88 | ND |
| Actim Influenza A&B | Medix Biochemica Ab, Kauniainen, Finland | A and B | Nasal swabs and nasopharyngeal aspirates | 10 min | 88–92 | ND | 99–100 | ND |
ND, not done—only samples positive for influenza A viruses were included in these investigations.
Statistical analysis
Logistic regression was used to analyse the relationship between the test results obtained by the four near‐patient diagnostic assays and the patients’ age. Receiver operating characteristic (ROC) curves were used to assess the extent to which the concentration of influenza A virus‐specific RNA in clinical samples may be associated with a false‐negative result in a near‐patient assay.
Comparison of near‐patient assay results, with respect to concentration of influenza A virus, was carried out using the Mann–Whitney U‐test. In all analyses, a p‐value of <0.05 was considered to be statistically significant. All statistical analyses were performed using the commercial software SPSS 15.0 (SPSS Inc., Chicago, IL, USA).
Results
Detection of influenza A viruses in clinical specimens
All clinical specimens used for the present evaluation of near‐patient assays were positive for influenza A/H3N2 for the purpose of standardization, which corresponds to the seasonal predominance of influenza A/H3N2 during the study period [14] (http://www.eiss.org). These specimens were collected from patients 1–101 years of age. The mean age of the children was 4.6 years (standard deviation (SD), 2.9 years) and that of the adults was 57.6 years (SD, 15.9 years). Seven of these patients were younger than 1 year and 20 were older than 80 years. The Binax Now Influenza A&B, Quick S‐Influ A/B, Influ‐A&B Respi‐Strip, and Actim Influenza A&B had overall sensitivities of 19%, 18%, 26%, and 40%, respectively. As shown in Fig. 1, the sensitivity of the three more sensitive assays decreased with increasing age of the patients, whereas the low sensitivity of the Quick S‐Influ A/B assay was constant throughout all ages, from young children to elderly patients. The estimated probability of detecting influenza A virus in clinical specimens that tested positive according to RT‐PCR was 17–56% in children 1 year of age, and decreased to 8–22% in patients 80 years of age. The sensitivity of the near‐patient assays with comparably high overall sensitivity (Influ‐A&B Respi‐Strip and Actim Influenza A&B) decreased significantly with increasing age (Table 2). A trend for lower sensitivity in adults was observed also for the Binax Now Influenza A&B.
Figure 1.

Estimated sensitivity of near‐patient assays in relation to the patient’s age. Curves were obtained from logistic regression analysis and plotted with the use of the software SigmaPlot 10.0 (Systat Software GmbH, Erkratti, Germany).
Table 2.
Sensitivity of the near‐patient diagnostic assays with respect to the age of the patients
| Diagnostic assay/manufacturer | No. of specimens | Sensitivity (%) | Mean age of the patients (SD) (years) | pa | ||
|---|---|---|---|---|---|---|
| 0–10 years | >35 years | Negative | Positive | |||
| Binax Now Influenza A&B/Binax Inc. | 80 | 30 | 12 | 39 (27) | 25 (28) | 0.074 |
| Quick S‐Influ A/B/Seiken | 80 | 17 | 18 | 37 (27) | 37 (28) | 0.997 |
| Influ‐A&B Respi‐Strip/Coris Bioconcept | 80 | 43 | 16 | 40 (26) | 23 (24) | 0.014 |
| Actim Influenza A&B/Medix Biochemica | 80 | 53 | 32 | 41 (28) | 28 (25) | 0.033 |
SD, standard deviation.
aLogistic regression.
In vitro sensitivity of near‐patient diagnostic assays
In order to evaluate the sensitivity of the near‐patient assays under standardized conditions, cell‐adapted virus stock preparations of influenza A/Wisconsin/67/05(H3N2), A/New Caledonia/20/99(H1N1) and B/Jiangsu/10/03 were tested with three of the four near‐patient assays and, for comparison, with an in‐house antigen‐ELISA and an in‐house RT‐PCR assay (in vitro evaluation of the Quick S‐Influ A/B assay was not possible because the manufacturer provided kits for only 80 tests). These preparations were diluted in ten‐fold steps in MEM, vortexed vigorously, separated into five aliquots, and tested with the five assays. In these experiments, the detection limits of the three near‐patient assays and the in‐house antigen‐ELISA ranged between 102 and 106 tissue culture infective doses (TCID50)/mL, whereas RT‐PCR had an in vitro sensitivity of 0.01–0.001 TCID50/mL (Table 3). The in vitro sensitivity of the Actim Influenza A&B assay was one log titre higher for the influenza A/H3N2 strain used than the sensitivity of the other near‐patient assay. The sensitivity of all assays used was lower for the influenza A/H1N1 than for the influenza B virus strain used.
Table 3.
In vitro sensitivity of the near‐patient assays for influenza A and B viruses in comparison to an in‐house ELISA and influenza A virus‐specific RT‐PCR
| Diagnostic assay/manufacturer | Lowest concentration of stock virus yielding a positive result (TCID50/mL) | ||
|---|---|---|---|
| A/Wisconsin/67/05 (H3N2) | A/New Caledonia/20/99 (H1N1) | B/Jiangsu/ 10/03 | |
| Binax Now Influenza A&B/Binax Inc. | 104 | 105 | 103 |
| Influ‐A&B Respi‐Strip/Coris Bioconcept | 104 | 106 | 103 |
| Actim Influenza A&B/Medix Biochemica | 103 | 105 | 102 |
| Antigen‐ELISA/in‐house | 103 | 105 | 103 |
| RT‐PCR/in‐house | 10−3 | 10−2 | ND |
ND, not done because RT‐PCR is only specific for influenza A viruses; TCID50, 50% tissue culture infective dose.
Quantification of influenza A virus‐specific RNA in clinical specimens
Clinical samples with a sufficient volume available (n = 53) were additionally tested using a quantitative influenza A virus‐specific RT‐PCR, for comparison with the qualitative results obtained by the near‐patient assays. Positive results of the near‐patient assays were associated with significantly higher concentrations of influenza A virus RNA in the clinical samples than negative results (Table 4). The ROC cut‐off points for a predicted sensitivity of 100% for near‐patient assays in detecting influenza A virus in clinical samples were similar to the in vitro sensitivities observed when testing virus stock preparations of influenza A virus (compare 3, 4). The area under the ROC curve for a positive near‐patient assay result was similar for all four assays and ranged between 0.81 and 0.88 (Fig. 2).
Table 4.
Results of near‐patient assays for the diagnosis of influenza virus infection in relation to influenza A virus RNA concentration in clinical samples
| Near‐patient assay/manufacturer | No. of samples available | Median concentration of influenza A virus RNA (copies/mL) | p‐valuea | ROC curve cut‐off for 100% sensitivity (copies/mL) | |
|---|---|---|---|---|---|
| Negative | Positive | ||||
| Binax Now Influenza A&B/Binax Inc. | 28 | 1.7 × 103 | 5.1 × 104 | 0.002 | 2.7 × 103 |
| Quick S‐Influ A/B/Seiken | 24 | 1.9 × 103 | 8.0 × 104 | 0.048 | 2.6 × 103 |
| Influ‐A&B Respi‐Strip/Coris Bioconcept | 22 | 2.1 × 103 | 7.2 × 104 | 0.014 | 9.3 × 102 |
| Actim Influenza A&B/Medix Biochemica | 32 | 3.4 × 103 | 3.1 × 104 | 0.004 | 6.8 × 102 |
ROC, receiver operating characteristic.
aMann–Whitney U‐test.
Figure 2.
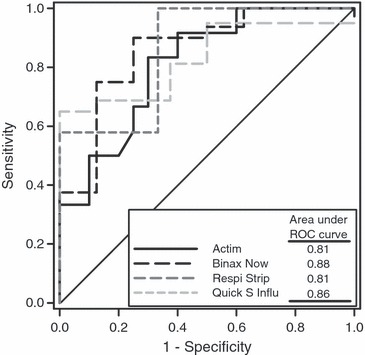
Receiver operating characteristic (ROC) curves for prediction of a positive result of near‐patient assays for detection of influenza A virus in clinical samples from patients with laboratory‐confirmed influenza A virus infection.
Discussion
A low sensitivity of near‐patient assays may have been suspected on the basis of previous studies, but the present study demonstrated unambiguously, for the first time, that the effectiveness of near‐patient assays for the diagnosis of influenza A/H3N2 virus infection decreases significantly with increasing age of the patient. The low sensitivity of near‐patient assays for the diagnosis of influenza, in comparison to virus isolation or RT‐PCR, does not allow a reliable exclusion of the diagnosis of influenza. The present study supports the belief that antiviral therapy and additional diagnostic procedures should not be withheld in cases where influenza is clinically suspected on the basis of a negative near‐patient assay, particularly in adult patients. On the other hand, the high specificity of near‐patient assays, together with the advantage of results being available within 30 min, may provide important information for the clinical evaluation of patients with an acute respiratory illness, provided that the result of the near‐patient assay is positive.
Reliable and rapid identification of influenza cases is of particular importance in elderly patients. The age group >65 years is at high risk of a severe course of influenza; 90% of all influenza‐related deaths occur in this age group [1], and the estimated risk of influenza‐related hospitalizations is 19–56 per 10 000 individuals during the influenza season [17]. Highly effective antiviral drugs are available for the treatment of influenza but must be administered within 48 h after onset of the disease and are effective only against influenza viruses. Respiratory syncytial virus co‐circulates frequently with influenza viruses, causing a significant disease burden also in elderly patients, and the clinical features are difficult to distinguish from those of influenza [18]. The usefulness of near‐patient assays is particularly low in the elderly. As a consequence, a considerable number of patients are treated unnecessarily and ineffectively with antiviral or antibacterial drugs, or are exposed to radiation due to additional diagnostic procedures. Therefore, the importance of annual vaccination against influenza, for elderly patients and their care‐givers, cannot be overemphasized [19].
In our previous study, the detection rate for influenza A viruses was clearly lower when testing respiratory specimens from adults than when testing specimens from young children, even with the use of highly sensitive assays such as RT‐PCR [9]. The results of previous evaluations of near‐patient assay sensitivity, with regard to patient age, were discordant. Fader [12] noted a clear decrease in assay sensitivity with increasing age, whereas Weinberg and Walker [20] did not, although both studies evaluated the same assay (Binax Now; Binax Inc., Portland, ME, USA). The respiratory specimens used were heterogeneous (nasal washes, nasal aspirates and bronchoalveolar lavages) and differed between the two studies. In adults, nasal washes are more convenient to collect than nasopharyngeal aspirates, but a diluting effect is inherent in this technique of sample collection and results in lower detection rates [20]. The strength of the present study is the use of comparable samples; all clinical specimens were collected with an identical technique by experienced and well‐trained physicians, with the use of a standardized sampling kit. Furthermore, in vitro evaluation of near‐patient assay sensitivity with the use of virus stock preparations could be confirmed by determination of the influenza A/H3N2 virus RNA concentrations in clinical samples. These features reduce the potential for bias and support the notion that the findings can be generalized.
The present study also has limitations. Only samples positive for influenza A/H3N2 viruses were tested, for the purpose of standardization, and conclusions cannot be drawn concerning the sensitivity of near‐patient assays for influenza A/H1N1 and B viruses. In the present study, the in vitro sensitivity of near‐patient assays differed for influenza A/H3N2, A/H1N1 and B viruses. Direct comparison of the in vitro sensitivities of near‐patient assays with that of RT‐PCR has to be made with caution, because of the use of cell culture‐grown influenza strains. Cell culture preparations contain a considerable amount of defective virus particles. Different structures of these particles may be detected by use of near‐patient assays and RT‐PCR, and degradation of virus particles follows different kinetics in cell culture than in the human host. Furthermore, the assay was evaluated retrospectively in a virological laboratory, as in most previous studies [20, 21]. The laboratory setting allows a more standardized inter‐assay comparison, but transfer of respiratory secretions into the medium used for preservation of virus and solubilization of mucus adds a dilution step that may reduce the detection rates achievable with near‐patient assays. In the present investigation, preservation of nasopharyngeal swabs in transport medium resulted in a final dilution of one log titre. This dilution effect cannot explain the differences in sensitivity observed between age groups or the differences among the four near‐patient assays. Nevertheless, the effectiveness of near‐patient assays for the diagnosis of influenza A/H3N2 virus infections may be expected to be somewhat higher when bedside testing is performed.
In conclusion, the sensitivity of near‐patient assays is not only significantly lower than that of influenza virus‐specific RT‐PCR, but is also significantly affected by the patient’s age. Antiviral therapy and additional diagnostic procedures should not be withheld on the basis of a near‐patient assay with a negative result, particularly in adult patients in whom influenza is clinically suspected. Because of the high specificity of the near‐patient assay, however, a positive test result may be helpful in making management decisions, including those concerning administration of antiviral therapy.
Transparency Declaration
Roche Pharmaceuticals funded, in part, the surveillance system DINOE. The manufacturers of the four near‐patient assays (Binax Inc., Coris Bioconcept, Medix Biochemica, and Denka Seiken) donated the test kits. The funding sources had no role in designing or conducting the study, in collection and evaluation of the data, in writing the manuscript, or in submission of the manuscript for publication. The corresponding author had full access to all of the data in the study and had final responsibility for the decision to submit for publication. There are no dual or conflicting interests to declare.
Acknowledgements
We thank D. Klinger for her excellent technical assistance and are particularly grateful for the dedication and hard work of the staff of the participating general and paediatric practices.
References
- 1. Thompson WW, Shay DK, Weintraub E et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 2003; 289: 179–186. [DOI] [PubMed] [Google Scholar]
- 2. McBean AM, Hebert PL. New estimates of influenza‐related pneumonia and influenza hospitalizations among the elderly. Int J Infect Dis 2004; 8: 227–235. [DOI] [PubMed] [Google Scholar]
- 3. Hayward AC, Harling R, Wetten S et al. Effectiveness of an influenza vaccine programme for care home staff to prevent death, morbidity, and health service use among residents: cluster randomised controlled trial. BMJ 2006; 333: 1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Impact of influenza on a nursing home population—New York. MMWR 1983; 32: 32–34. [PubMed] [Google Scholar]
- 5. Nicholson KG, Aoki FY, Osterhaus AD et al. Efficacy and safety of oseltamivir in treatment of acute influenza: a randomised controlled trial. Neuraminidase inhibitor flu treatment investigator group. Lancet 2000; 355: 1845–1850. [DOI] [PubMed] [Google Scholar]
- 6. Monto AS, Fleming DM, Henry D et al. Efficacy and safety of the neuraminidase inhibitor zanamivir in the treatment of influenza A and B virus infections. J Infect Dis 1999; 180: 254–261. [DOI] [PubMed] [Google Scholar]
- 7. Boivin G, Hardy I, Tellier G, Maziade J. Predicting influenza infections during epidemics with use of a clinical case definition. Clin Infect Dis 2000; 31: 1166–1169. [DOI] [PubMed] [Google Scholar]
- 8. Monto AS, Gravenstein S, Elliott M, Colopy M, Schweinle J. Clinical signs and symptoms predicting influenza infection. Arch Intern Med 2000; 160: 3243–3247. [DOI] [PubMed] [Google Scholar]
- 9. Steininger C, Kundi M, Aberle SW, Aberle JH, Popow‐Kraupp T. Effectiveness of reverse transcription‐PCR, virus isolation, and enzyme‐linked immunosorbent assay for diagnosis of influenza A virus infection in different age groups. J Clin Microbiol 2002; 40: 2051–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. World Health Organization . WHO recommendations on the use of rapid testing for influenza diagnosis. Geneva: WHO, 2005; 1–16. [Google Scholar]
- 11. Dunn JJ, Gordon C, Kelley C, Carroll KC. Comparison of the Denka‐Seiken INFLU A.B‐Quick and BD directigen flu A+B kits with direct fluorescent‐antibody staining and shell vial culture methods for rapid detection of influenza viruses. J Clin Microbiol 2003; 41: 2180–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fader RC. Comparison of the Binax NOW flu A enzyme immunochromatographic assay and R‐mix shell vial culture for the 2003–2004 influenza season. J Clin Microbiol 2005; 43: 6133–6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. American Public Health Association . Diagnostic procedures for viral, rickettsial and chlamydial infections, 5th edn Washington, DC: APHA, 1979. [Google Scholar]
- 14. Redlberger M, Aberle SW, Heinz FX, Popow‐Kraupp T. Dynamics of antigenic and genetic changes in the hemagglutinins of influenza A/H3N2 viruses of three consecutive seasons (2002/2003 to 2004/2005) in Austria. Vaccine 2007; 25: 6061–6069. [DOI] [PubMed] [Google Scholar]
- 15. Ward CL, Dempsey MH, Ring CJ et al. Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J Clin Virol 2004; 29: 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sarkkinen HK, Halonen PE, Salmi AA. Detection of influenza A virus by radioimmunoassay and enzyme‐immunoassay from nasopharyngeal specimens. J Med Virol 1981; 7: 213–220. [DOI] [PubMed] [Google Scholar]
- 17. Mullooly JP, Bridges CB, Thompson WW et al. Influenza‐ and RSV‐associated hospitalizations among adults. Vaccine 2007; 25: 846–855. [DOI] [PubMed] [Google Scholar]
- 18. Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high‐risk adults. N Engl J Med 2005; 352: 1749–1759. [DOI] [PubMed] [Google Scholar]
- 19. Poland GA, Tosh P, Jacobson RM. Requiring influenza vaccination for health care workers: seven truths we must accept. Vaccine 2005; 23: 2251–2255. [DOI] [PubMed] [Google Scholar]
- 20. Weinberg A, Walker ML. Evaluation of three immunoassay kits for rapid detection of influenza virus A and B. Clin Diagn Lab Immunol 2005; 12: 367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thomas Y, Kaiser L, Wunderli W. The use of near patient tests in influenza surveillance: Swiss experience and EISS recommendations. Euro Surveill 2003; 8: 240–246. [PubMed] [Google Scholar]
- 22. Booth S, Baleriola C, Rawlinson WD. Comparison of two rapid influenza A/B test kits with reference methods showing high specificity and sensitivity for influenza A infection. J Med Virol 2006; 78: 619–622. [DOI] [PubMed] [Google Scholar]
- 23. Smit M, Beynon KA, Murdoch DR, Jennings LC. Comparison of the NOW influenza A & B, NOW flu A, NOW flu B, and directigen flu A+B assays, and immunofluorescence with viral culture for the detection of influenza A and B viruses. Diagn Microbiol Infect Dis 2007; 57: 67–70. [DOI] [PubMed] [Google Scholar]


