Abstract
Interferon-inducible transmembrane (IFITM) protein family is novel viral restriction factors with representative transmembrane structure. These proteins also exist in fish, however, their roles in the innate immune response remain unknown. Here, we report a characterization of teleost IFITM1 from flounder Paralichthys olivaceus (PoIFITM1), which exhibits conserved structure characteristic of the IFITM family but comprises a relatively longer N-terminal region. The expression and promoter activity of PoIFITM1 are markedly induced by aquatic animal viruses: Rana grylio virus (RGV) and Scophthalmus maximus rhabdovirus (SMRV). Overexpression and siRNA-mediated knockdown demonstrate that PoIFITM1 exhibits strong antiviral effects against both DNA virus (RGV) and RNA virus (SMRV), expanding the spectrum of viruses inhibited by IFITM proteins. Further analysis shows that PoIFITM1 suppresses viral entry into host cells, confirming that the IFITM-mediated restriction is conserved from lower vertebrates to mammals. Deletion mutagenesis reveals that PoIFITM1 exerts antiviral activity by targeting to Golgi complex and the N-terminal region is required for its subcellular localization, which is not observed in other known IFITM family members. Our current data provide the first evidence that IFITM1 functions as a key effector of the innate immune to restrict virus replication in lower vertebrates, through the action of impeding viral entry.
Keywords: Paralichthys olivaceus interferon-inducible transmembrane 1 (PoIFITM1), Antiviral effect, Rana grylio virus (RGV), Scophthalmus maximus rhabdovirus (SMRV), Golgi localization
Highlights
-
•
First cloning and functional characterization of IFITM1 in flounder Paralichthys olivaceus.
-
•
PoIFITM1 is induced by two aquatic animal viruses: RGV and SMRV.
-
•
PoIFITM1 exhibits strong antiviral effects against both DNA and RNA viruses.
-
•
PoIFITM1 suppresses viral entry into fish cells.
-
•
The N-terminus determines the antiviral activity through regulating the cellular localization.
1. Introduction
The interferon-inducible transmembrane (IFITM) proteins comprise a family of small interferon-stimulated proteins (~15 kD) that mediate the activities of interferons (IFNs) [1]. All members of IFITM family share a common topology, distinguished by the presence of two highly conserved transmembrane domains interspersed by a short cytoplasmic loop, with luminal amino- and carboxy-termini [2], [3]. Among this family, IFITM1, 2, and 3 are expressed basally in various tissues and cells [1], [4], which are involved in immune cell signaling, cell adhesion, oncogenesis, germ cell physiology, and regulation of endocytosis [5].
Recently, interest in IFITM proteins has increased with the discovery that IFITM1, 2 and 3 function as restriction factors against multiple pathogenic viruses, such as influenza A virus, dengue virus, West Nile virus, vesicular stomatitis virus, HIV-1, and SARS-coronavirus [6], [7], [8], [9], [10], [11]. IFITM proteins are the only known innate immune effector that inhibits viral entry. They suppressed infection of retroviruses pseudotyped with entry proteins of IFITM-restricted viruses [6], [7], [8], which firstly localized the IFITM-mediated restriction to the entry step in the virus replication cycle. Imaging studies provided direct evidence that IFITM proteins prevent viruses from fusing with late endosomal or lysosomal membranes [7], [12], most likely through reducing membrane fluidity and fusion potential [13].
The antiviral activity is shared by IFITM proteins, but the efficiency varies among them, likely owing to the sequence divergence lying at the termini [8], [11]. The N-terminal region is essential for IFITM proteins to restrict influenza A virus infection. IFITM3 allele that lacks the N-terminal 21 amino acids was significantly enriched in patients who were severely ill for H1N1 pandemic flu [14], [15]. Consistently, removing this region relocated IFITM3 from the endosomes to the cellular periphery and thereby abrogated its antiviral activity in vitro, highlighting the importance of the N-terminal region [16].
Although the IFITM family has been widely studied in mammals, little is known about its function in lower vertebrates. Likemammals, fish possess IFN-mediated innate immune defense against viral infection [17], [18], [19]. Some studies suggested that the IFITM-mediated restriction might be a conserved characteristic in vertebrates [20]. Indeed, four members of IFITM family (IFITM1, 2, 3, and 5) have been annotated in fish genome, which display an orthologous relationship with mammalian counterparts [20]. The expression of IFITMs was induced by PolyI:C in rainbow trout (Oncorhynchus mykiss) and large yellow croaker (Pseudosciaena crocea) [21], [22], suggesting their roles in IFN-mediated antiviral immune. However, so far there are no direct evidences whether fish IFITMs possess the ability to restrict virus infection, and how they exert the antiviral activity.
In this study, we isolated fish IFITM1 gene from flounder Paralichthys olivaceus, and delineated its antiviral role against both DNA and RNA viruses. We provided significant evidence that PoIFITM1 blocked the entry stage of viral replication. Importantly, we observed that PoIFITM1 displayed a unique subcellular distribution in the Golgi apparatus, which determines its antiviral effect. These findings lead to a better understanding of the functional roles and action mechanisms of IFITM proteins in innate immune defense of lower vertebrates.
2. Materials and methods
2.1. Cells and viruses
Flounder embryonic cells (FEC) were cultured at 25 °C in Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS) [23], and Epithelioma papulosum cyprini (EPC) cells were maintained at 25 °C in medium 199 (Gibco) with 10% FBS. Rana grylio virus (RGV) and Scophthalmus maximus rhabdovirus (SMRV) were propagated and titered in EPC cells as described previously [24], [25].
2.2. Gene cloning and plasmids
PoIFITM1 was retrieved from a SMART cDNA library made with mRNAs derived from UV-inactivated grass carp reovirus (GCRV)-infected FEC cells [26]. RACE-PCR was used to clone the full-length cDNAs according to the previous report [27]. Multiple sequence alignments were generated by the Clustal X software, and phylogenetic tree was constructed by the Neighbor-Joining method using MEGA 5 program.
For overexpression assay, the full-length (aa 1–162) and ΔN (aa 77–162) PoIFITM1 were cloned into pcDNA3.1(+) vector (Invitrogen, USA), respectively. For promoter activity assay, the 5′-flanking region of PoIFITM1 was amplified by a GenomeWalker Universal Kit (Clontech, USA). The fragment (−1664/+1) was inserted into pGL3-Basic luciferase vector (Promega, USA). For subcellular localization study, the entire ORFs of PoIFITM1 and Caveolin-1 (CAV1) were cloned into pEGFP and pDsRed2 (Clontech, USA), respectively. The generated plasmids were confirmed by sequencing analyses. All primers used for constructions were list in Table 1 . pDsRed2-Mito, pDsRed2-ER, and pDsRed-Golgi were purchased from Clontech.
Table 1.
Primers used in this study.
| Primers | Sequence (5′–3′) | Usage |
|---|---|---|
| SMART-F | CAACGCAGAGTACGCGGG | 5′ RACE PCR |
| IFITM1-R1 | GCGGTGGAGCTGTGATGT | |
| SMART-R | TCAACGCAGAGTACT(16) | 3′ RACE PCR |
| IFITM1-F1 | TCTGCTGCCTGGGACTTG | |
| IFITM1-F2 | AGGACTACATCATCTGGTC | Real-time PCR |
| IFITM1-R2 | AATAATGACCAAGACGCCA | |
| IFITM1-F3 | CGGCTAGCATGGATCCTAAATGTCAGTC | IFITM1 plasmid for localization |
| IFITM1-R3 | CCAAGCTTGAAACTTGTTGACAAAAAATCT | |
| IFITM1-WT-F | CGGAATTCATGGATCCTAAATGTCAGT | Wild-type IFITM1 plasmid |
| IFITM1-WT-R | CCCTCGAGTTAAAACTTGTTGACGAAAAA | |
| IFITM1-ΔN-F | CGGAATTCATGATCATCTGGTCCA | IFITM1ΔN plasmid |
| IFITM1-WT-R | CCCTCGAGTTAAAACTTGTTGACGAAAAA | |
| IFITM1-P-F | CGGCTAGCTTGACACCGGCTTCT | Promoter activity assay |
| IFITM1-P-R | CCAAGCTTGGAAAGGGAAAGTCTCCTCCCTCCGACTGG | |
| β-actin-F | CACTGTGCCCATCTACGAG | Real-time PCR |
| β-actin-R | CCATCTCCTGCTCGAAGTC | |
| Mx-F | GCCGTCATAGGAGACCAAA | Real-time PCR |
| Mx-R | TTCCTCGTAGTCCCTGTAGC | |
| RGV-MCP-F | CAGTCAGGGACATGGTTGTG | Real-time PCR |
| RGV-MCP-R | GGGAGTGACGCAGGTGTAAT | |
| SMRV-N-F | CAAGGGTGGATATTGACCGATG | Real-time PCR |
| SMRV-N-R | GCACCAGTACAGCTCTGCTCTTC | |
| CAV1-F | CCAAGCTTATGTTCTTCCTTCTGCCTCC | CAV1 plasmid for localization |
| CAV1-R | CGGGATCCCGCACCTCCTTGGACATGC |
2.3. Induction of PoIFITM1 by viruses
For virus induction, FEC cells cultured in 25 cm2 culture plates were incubated with RGV at a multiplicity of infection (MOI) of 0.5, or with SMRV at an MOI of 0.3. The cells treated with FBS-free DMEM were used as control in parallel. After 6 h, the transfection mixture was replaced with fresh medium. The cells were harvested at various times (3, 6, 12, 24, 48, 72 and 96 h) post-infection. Total RNA isolation, cDNA synthesis, and real-time PCR assay of gene expression were performed as described previously [28]. The relative expression levels of target genes were determined using β-actin as an internal control with the comparative Ct (2−ΔΔCt) method [29].
2.4. Promoter activity assay
FEC cells seeded in 24-well plates overnight were transfected with 1 μg promoter/pGL3 constructs and 0.1 μg Renilla Luciferase vector pRL-TK (Promega, USA) using Lipofectamine 2000. At 24 h post-transfection, the cells were treated with RGV (0.2 MOI) and SMRV (0.1 MOI) or left untreated. After 48 h, the cells were lysed with the Dual-Luciferase Reporter Assay System (Promega, USA). Luciferase activities were measured by Junior LB9509 Luminometer (Berthold, Germany). All samples were tested in triplicate and the results were presented as relative light units (RLU) normalized to the amounts of Renilla Luciferase activities.
2.5. Antiviral activity evaluation
For overexpression assay, FEC or EPC cells seeded in 6-well plates were transfected with 4 μg pcDNA3.1-PoIFITM1 or empty vector (pcDNA3.1) control. After 24 h, the cells were infected with RGV (0.5 MOI), or with SMRV (0.3 MOI), and incubated for 24 h. The cell monolayers were subjected to total RNA extraction for real-time PCR assay of viral replication. The supernatant aliquots were subjected to 50% tissue culture infective dose (TCID50) assay of viral titers as described previously [28].
For knockdown assay, small interfering RNA (siRNA) oligonucleotides targeting PoIFITM1 (Sense: 5′-GACCAUACCACUGUGGAAATT-3′) and non-targeting control siRNA (Sense: 5′-UUCUCCGAACGUGUCACGUTT-3′) were synthesized by GenePharma (Shanghai, China). FEC cells seeded in 6-well plates were transfected with PoIFITM1-specific siRNA or control siRNA at 100 nM final concentration using Lipofectamine 2000. After 48 h, the cells were infected with RGV (0.5 MOI), or with SMRV(0.3 MOI) for 48 h. The cells were then collected to total RNA extraction for real-time PCR assay of viral replication. The supernatant aliquots were harvested for measurement of virus yields by TCID50 assay. A rescue experiment was performed by the introduction of pcDNA3.1-PoIFITM1 plasmid (4 μg/well) into siRNA-transfected FEC cells at 24 h post-transfection. The viral replication was determined as described above.
2.6. Virus binding and entry assay
To analyze virus binding, FEC cells transfected with pcDNA3.1-PoIFITM1 or PoIFITM1-specific siRNA were seeded in 6-well plates. After 24 h, the cells were infected with RGV at an MOI of 5 for 1 h at 4 °C to permit viral attachment but prevent entry, and then washed with cold PBS. Total cellular RNA was extracted for real-time PCR assay to quantify the cell-associated virus. To analyze virus entry, the cells were incubated with RGV, as described above, for 1 h at 4 °C. The virus inocula were removed and the cells were washed with cold PBS. Then prewarmed medium was added and the cells were cultured for another 4 h at 25 °C. Noninternalized viruses were removed by washing the cells with citrate buffer (40 mM sodium citrate, 135 mM NaCl, 10 mM KCl [pH 3.0]) for 5 min. Total cellular RNA was extracted to measure the amount of virus that entered cells.
2.7. Subcellular localization
To determine the localization of PoIFITM1 and its mutant, FEC cells were grown on microscopic coverslips in 6-well plates, and cotransfected with 2 μg pEGFP-IFITM1 or pEGFP-IFITM1 ΔN, together with 2 μg plasmids pDsRed-Golgi, pDsRed2-Mito, pDsRed2-ER, or pDsRed2-CAV1. After 48 h, the cells were washed with PBS, fixed in 4% paraformaldehyde (PFA) for 30 min, and stained with Hoechst33342 (Sigma, USA) for 5 min. The cells were then visualized under a Leica DM IRB fluorescence microscope (objective 100×). To analyze the association of IFITM1 with endocytosis, FEC cells were transfected with 2 μg pEGFP-IFITM1 or pEGFP-IFITM1 ΔN. After 24 h, the cells were incubated with 4 μg/ml Alexa Fluor 555-conjugated cholera toxin subunit B (CTxB-AF555) (Invitrogen, USA) at 25 °C for 40 min. After washing with PBS to remove noninternalized CTxB-AF555, the cells were fixed for viewing as described above.
3. Results
3.1. Identification of PoIFITM1
The full-length cDNA of PoIFITM1 consists of a 489 bp open reading frame (ORF) encoding 162 amino acids, flanked by 84 bp of 5′UTR and 165 bp of 3′UTR (GenBank accession no: KC777348). Multiple sequence alignment reveals that it contains two putative transmembrane domains located at amino acid residues 77–97 and 127–147 (Fig. 1 A). The transmembrane domains and intervening intracellular loop are highly conserved, whereas the N- and C-terminal regions are variable. Compared to mammalian IFITM members, PoIFITM1 has a longer N-terminal region. Two highly conserved cysteine residues of S-palmitoylation [30], [31] were also found in the first transmembrane domain (Cys 90, Cys 91) (Fig. 1B). Phylogenetic analysis shows that PoIFITM1 forms a cluster with IFITM1 of sea bass and large yellow croaker, apart from mammalian and amphibian IFITMs (Fig. 1C). Consistently, it exhibits 40–51% identity to other known fish IFITM1 and 35–42% identity to mammalian counterparts (Table 2 ), indicating that PoIFITM1 is closely related to fish IFITMs whose sequences correspond to orthologs of mammalian IFITMs.
Fig. 1.

Identification of flounder IFITM1. (A) Schematic diagram of flounder IFITM1 domain structure. IFITM1 comprises five domains: N- and C-termius, two transmembrane domains (TM1, TM2), and cytoplasmic domain. The numbers refer to the amino acid residues. (B) Multiple alignment of flounder IFITM1 amino acid sequence with several typical IFITMs derived from flounder (Po), human (Hs), mouse (Mm), large yellow croaker (Lyc), European seabass (Dl), and frog (Xt). Characteristic domains are marked above the alignment and transmembrane helixes are indicated by box. Cysteine residues of S-palmitoylation are shown with asterisk. Gaps used to maximize the alignment are shown by dashes. (C) Phylogenetic analysis of IFITMs in vertebrates. The bootstrap confidence values shown at the nodes of the tree derived from 1000 replicates. GenBank accession numbers of sequences are listed in Table 2.
Table 2.
Amino acid identity comparison of PoIFITM1 with other known IFITM proteins.
| Gene | Accession number | Identity (%) |
|---|---|---|
| European seabass IFITM1 | CBJ56265.1 | 51 |
| Rainbow trout IFITM1 | AJ291989 | 40 |
| Rainbow trout IFITM2 | CAC85160 | 38 |
| Large yellow croaker IFITM1 | EU200363 | 42 |
| Elephant shark IFITM | AFM89728 | 45 |
| Frog IFITM1 | NP_001123403 | 36 |
| Frog IFITM3 | NP_001015758 | 34 |
| Mouse IFITM1 | NP_081096 | 35 |
| Mouse IFITM2 | NP_109619 | 33 |
| Mouse IFITM3 | NP_079654 | 36 |
| Rat IFITM1 | NP_001099784 | 42 |
| Rat IFITM2 | NP_110460 | 43 |
| Rat IFITM3 | NP_001129596 | 38 |
| Pig IFITM1 | XP_003124278 | 40 |
| Pig IFITM2 | NP_00123314 | 35 |
| Pig IFITM3 | NP_001188311 | 37 |
| Cow IFITM1 | NP_776976 | 37 |
| Cow IFITM2 | NP_001071522 | 37 |
| Cow IFITM3 | NP_001071609 | 39 |
| Human IFITM1 | CAA59337 | 37 |
| Human IFITM2 | CAG46672 | 32 |
| Human IFITM3 | NP_066362 | 36 |
3.2. PoIFITM1 expression is induced by viruses
Real-time PCR assay showed that the kinetics was similar in the cells infected with two types of fish viruses: RGV (DNA virus) and SMRV (RNA virus). There was a detectable basal level of PoIFITM1 in the control cells. After RGV and SMRV infection, its transcripts increased as early as 3 h, and reached a peak at 96 h, with about 14- and 8.5-fold upregulation, respectively (Fig. 2 A; p < 0.05). Under the same condition, we also detected the expression status of Mx, a known hallmark of the IFN response both in mammals and fish [32]. Similar but more pronounced Mx upregulation was observed (40- and 1718-fold induced by RGV and SMRV, respectively; Fig. 2B; p < 0.05). This demonstrates the effectiveness of virus induction, suggesting that PoIFITM1 can be induced by IFN derived from virus infection.
Fig. 2.

Inducible expression of flounder IFITM1 by fish viruses. (A) Real-time PCR analysis of IFITM1 transcriptional level in FEC cells infected with RGV and SMRV for the indicated times. Values were normalized against that of the control cells (Con), and expressed as the common logarithm of the relative quantity (Log ΔRQ). (B) Real-time PCR detection of Mx expression under the same conditions. Values were normalized against that of the sample in which Mx level was the lowest. Data represent averages of three independent experiments, with the range indicated (±SD). ND, not detected. Statistical analysis was performed using Student’ t-test. *, P < 0.05; **, P < 0.005.
3.3. PoIFITM1 promoter is induced by viruses
5′-flanking regulatory sequence analysis revealed that PoIFITM1 promoter possesses characteristics of genes responsive to type I and type II IFNs, with an IFN-stimulated response element (ISRE), a gamma IFN activated sequence (GAS) and 21 GAAA/TTTC motifs. A TATA box is found in IFITM1 promoter, which is absent in mammalian IFITMs [4]. Other putative transcription factor binding sites that can regulate immune gene are also identified, such as AP-1, IRF-2, NF-κB, Oct-1, GATA-1, GATA-2, GATA-3, SP-1 (Fig. 3 A).
Fig. 3.
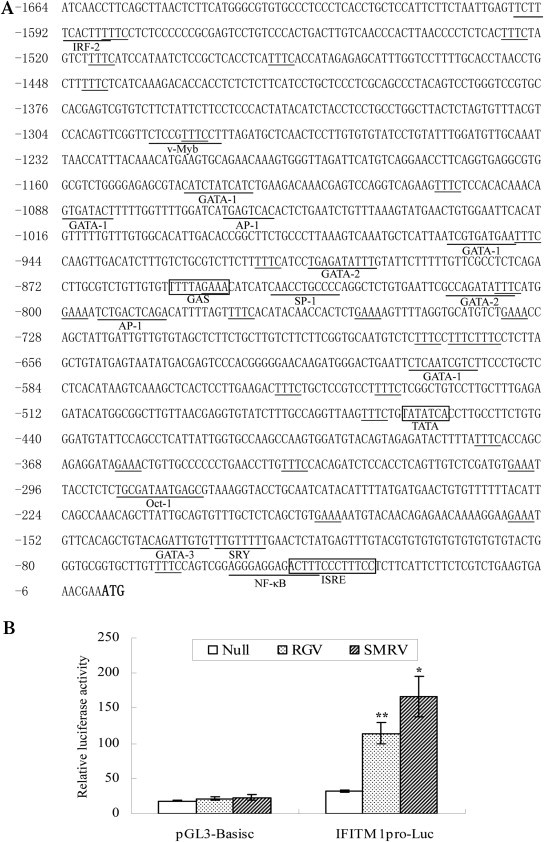
Induction of promoter activity of flounder IFITM1 by fish viruses. (A) Sequence of the 5′-flanking region of IFITM1. Characteristic motifs for IFN-stimulate genes including ISRE and GAS, as well as TATA box are indicated with boxes. GAAA/TTTC motifs and potential transcription factor binding sites are underlined. Nucleotide positions are counted from the initiation codon ATG (in bold) that is designated as +1. (B) Luciferase activity assay in FEC cells transfected with IFITM1 promoter construct (IFITM1pro-Luc) or empty vector (pGL3-Basic). The data were normalized by pRL-TK and represent averages of three independent experiments (*, P < 0.05; **, P < 0.005).
We further generated PoIFITM1 promoter-driven luciferase construct to analyze the promoter activity. In transient-transfected FEC cells, pGL3-Basic vector showed a low level of luciferase activity and no inducibility further by virus infection (Fig. 3B; p > 0.05). In contrast, the PoIFITM1pro-Luc displayed significant increase of activities after stimulation. Compared to the control cells, the relative luciferase activities of PoIFITM1 promoter were induced 3.5- and 5.2-fold in the presence of RGV and SMRV (p < 0.005 and p < 0.05, respectively) (Fig. 3B), indicating that the ISRE and GAS motifs are essential for IFITM1 induction.
3.4. PoIFITM1 exhibits antiviral effect against DNA and RNA viruses
FEC cells were transfected with PoIFITM1 construct and empty vector followed by infection with viruses. Compared with vector-transfected control cells, the titers of RGV and SMRV in PoIFITM1-transfected cells decreased about 40-fold (107.5TCID50/ml versus 105.9TCID50/ml; p < 0.05) and 69-fold (108.5TCID50/ml versus 106.6TCID50/ml; p < 0.05) at 24 h after infection (Fig. 4 A). Simultaneously, the mRNA expression of RGV major capsid protein (MCP) and SMRV nucleoprotein (N) reduced 6-fold and 3-fold, respectively (Fig. 4B; p < 0.005). The antiviral effect of PoIFITM1 was also examined in a heterologous system, the EPC cell line, which is more susceptible to virus infection. In agreement with the results in FEC cells, overexpression of IFITM1 in EPC cells resulted in 2- and 6-fold reduction of RGV MCP and SMRV N expression (Fig. 4C; p < 0.005). These data show that the expression of PoIFITM1 in fish cells elicits antiviral effects against both DNA and RNA viruses.
Fig. 4.

Inhibition of virus replication by overexpression of flounder IFITM1. (A) TCID50 measurement of virus yields in FEC cells transfected with IFITM1 or empty vector control. (B) Real-time PCR analysis of the expression of RGV MCP and SMRV N, which were shown as fold increase values relative to the control (set as 1). (C) The experiments are similar to those in (B), except EPC cells were used. The results are representative of three independent experiments performed in triplicate (means ± SD) (*, P < 0.05; **, P < 0.005).
Subsequently, the physiological function of PoIFITM1 in controlling virus infection was further investigated by using siRNA technology. Compared with the cells transfected with nontargeting siRNA, transfection of PoIFITM1-specific siRNA enhanced the yields of RGV and SMRV by 68-fold (107.0TCID50/ml versus 108.8TCID50/ml; p < 0.005) and 100-fold (107.5TCID50/ml versus 109.5TCID50/ml; p < 0.05), respectively (Fig. 5 A). Consistently, knockdown of PoIFITM1 led to 1.7- and 2.1-fold increase of RGV MCP and SMRV N expression (Fig. 5B; p < 0.05). Moreover, overexpression of PoIFITM1 in the knockdown cells could rescue the interference of siRNA (Fig. 5C; p < 0.05). Similar viral replication was observed in untreated or control siRNA-transfected cells (Fig. 5D; p > 0.05), suggesting that the control siRNA does not have any inhibitory effect. These data mean that the loss of PoIFITM1 function increases susceptibility and infection of viruses, further confirming that PoIFITM1 serves as an antiviral effector.
Fig. 5.
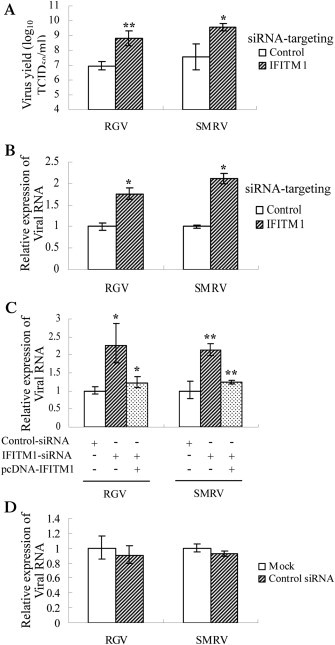
Enhancement of virus replication by knockdown of flounder IFITM1. (A) TCID50 measurement of virus yields in FEC cells transfected with IFITM1 siRNA or non-targeting control siRNA. (B) Real-time PCR analysis of the expression of RGV MCP and SMRV N, which were shown as fold increase values relative to the control (set as 1). (C) Experiment similar to (B), except the introduction of IFITM1 after transfection of siRNAs. (D) Real-time PCR analysis of viral replication in FEC cells transfected with control siRNA or mock treated. The results are representative of three independent experiments performed in triplicate (means ± SD) (*, P < 0.05; **, P < 0.005).
3.5. PoIFITM1 impedes the entry step of RGV replication
A classical virus binding and entry assay [33] was performed to investigate the mechanism underlying PoIFITM1-mediated restriction. For virus binding assay, the cells were incubated with RGV at 4 °C to permit virus-host binding, but not internalization. As shown in Fig. 6 , PoIFITM1 overexpression or knockdown cells showed comparable levels of virus binding with the control cells (p > 0.05), indicating that virus-host binding interaction is not affected by the expression of PoIFITM1. For virus entry assay, the cells were infected with RGV at 4 °C as above, and then incubated at 25 °C. The results of real-time PCR revealed that the amount of endocytosed RGV particles decreased 1.5-fold in PoIFITM1-overexpressing cell (Fig. 6A; p < 0.005), whereas it increased 3-fold in PoIFITM1 knockdown cells (Fig. 6B; p < 0.005). These data indicate that PoIFITM1 impedes viral entry into host cells.
Fig. 6.

Flounder IFITM1 restricts RGV entry. (A) Real-time PCR assessment of viral binding and entry in FEC cell transfected with IFITM1 or empty vector. In IFITM1-transfected cells, the amount of cell-bound virus was similar, but that of virus entry into cells reduced as relative to the control. (B) Experiment similar to (A), except FEC cells transfected with IFITM1-siRNA were used. All samples were tested in triplicate and the data represent the results from three independent experiments (*, P < 0.05; **, P < 0.005).
3.6. The N-terminal region of PoIFITM1 is required for its antiviral activity
A truncated mutant (ΔN) was generated by deleting the first 1–76 amino acids to assess the role of N-terminal region in PoIFITM1 antiviral function. Real-time PCR assay showed that transfection of wide-type IFITM1 led to a significant reduction of MCP expression (2.6-fold against empty vector; p < 0.05), whereas deletion of N-terminus yielded a slight reduction (1.2-fold against empty vector, p > 0.05; 0.5-fold against wide-type IFITM1, p < 0.05) (Fig. 7 A). Consistently, measurement of the virus production revealed that overexpression of IFITM1 ΔN resulted in a moderate decrease in viral titer as relative to control cells (from 107.2 TCID50/ml to 106.9 TCID50/ml; p > 0.05) (Fig. 7B). It therefore appears that removing of N-terminus induces a loss in the ability of IFITM1 to inhibit viral replication, suggesting this region is essential for its antiviral function.
Fig. 7.
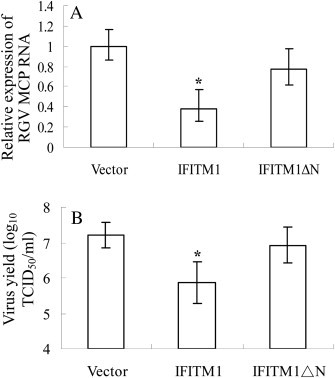
Role of the N-terminal region in restricting virus replication. (A) Real-time PCR analysis of MCP expression in FEC cells transfected with IFITM1, N-terminal deletion mutant (IFITM1 ΔN), or empty vector. (B) TCID50 measurement of virus titers in the cells described in (A). Deletion of the N-terminal region impaired the inhibitory activity of IFITM1 toward virus infection. All samples were tested in triplicate and the data represent the results from three independent experiments. *, P < 0.05; **, P < 0.005.
3.7. The N-terminal region determines the cellular localization
Fluorescence analysis showed that PoIFITM1 colocalizaed with the endocytic marker proteins CTxB and CAV1, whereas the N-terminal truncation barely colocalizaed with these proteins (compare row 1 and 3 or row 2 and 4; Fig. 8 A). This indicates that the N-terminal region contributes to associate with the endocytosis compartments.
Fig. 8.
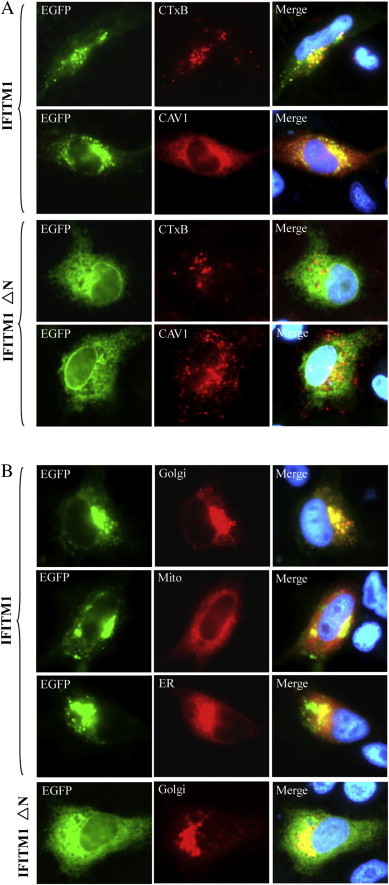
Subcellular localization of full-length and truncated (ΔN) IFITM1. (A) IFITM1 (EGFP, green) could colocalized (Merge, yellow) with the endocytic markers (CTxB, CAV1; Red), whereas IFITM1 ΔN (EGFP, green) could not colocalized with these proteins (Red). (B) IFITM1 (EGFP, green) were distributed in cytoplasmic and concentrated in the perinuclear region, which was localized to Golgi (Red) but excluded from mitochondria (Mito, Red) or endoplasmic reticulum (ER, Red). Green fluorescence of the truncation (IFITM1 ΔN) was distributed diffusely in the cells and absent from Golgi (Red), which indicated that the N-terminal region of IFITM1 was responsible for subcelluar localization. Blue fluorescence showed the nuclei stained by Hoechst 33342. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
We further characterized the cellular localization of PoIFITM1 through cotransfection with organelle marker plasmids. PoIFITM1 was distributed into punctuate clusters throughout the cytoplasm and also appeared concentrated in the perinuclear region (Fig. 8B). The images of EGFP-tagged PoIFITM1 merged well with the red fluorescent protein DsRed that targeted to the Golgi, but not with those targeted to mitochondria or endoplasmic reticulum (Fig. 8B), suggesting that PoIFITM1 is mainly located in the Golgi apparatus. The ΔN mutant was found diffusely distributed throughout the cell with absent from the Golgi, which demonstrates that the N-terminal region is responsible for localizing PoIFITM1 to the Golgi.
4. Discussion
We report a characterization of Paralichthys olivaceus IFITM1 and demonstrated that it exhibited strong antiviral activity against DNA and RNA viruses of aquatic animals. PoIFITM1 inhibited viral entry into host cells, with the N-terminal region targeting to Golgi. This study firstly reveals that IFITM-mediated viral restriction is conserved from lower vertebrates to mammals.
One striking finding in our study is that PoIFITM1 elicits an ability to prevent against DNA virus. RGV is a large double-strand DNA virus of the Iridoviridae family. Ectopic expression of PoIFITM1 restricted RGV replication, whereas RNA interference knockdown impaired the restriction. So far, nothing is known about the IFITMs activity against DNA viruses, although the inhibition of retroviruses, positive and negative-strand RNA viruses has been extensively described in mammals [1]. Therefore, this finding expands the spectrum of viruses inhibited by the IFITM family.
Emerging lines of evidence have demonstrated that IFITMs disrupt the entry step of virus infection [12], [13]. The following observations suggest that action mechanism of PoIFITM1 is similar to that of mammals. First, the virus binding and entry experiment revealed that viral particles entering into the cells to proceed replication were greatly reduced, in the presence of PoIFITM1. Second, PoIFITM1 colocalized with endocytosis marker proteins. The association between IFITM2, 3 and endocytosed transferrin has been observed in mammals, whereas IFITM1 barely exhibited colocalization with transferrin [8]. This is most likely because transferrin is a clathrin-mediated endocytic marker. Indeed, when we incubated cells with CTxB, a caveola-mediated endocytosis marker [34], an extensive colocalization was seen. Moreover, PoIFITM1 colocalized with CAV1, which is an essential component of caveolae and functions in the endocytosis [35]. IFITM1 has been suggested to interact and form a complex with CAV1 to function in some processes including endocytosis [36]. These findings indicate that PoIFITM1 associates with endocytic compartments. Finally, PoIFITM1 displayed a unique characteristic not observed in other known IFITM family members: subcellular distribution within the Golgi apparatus. The localization of IFITM proteins correlates with their antiviral function. Mammalian IFITMs have been described to reside in late endosomes or lysosome, where they prevent viruses fuse with host cell membranes, and thereby restrict virus replication [12]. Our recent studies have discovered that RGV entered FEC cells via a pH-dependent caveola-mediated endocytosis (J. Wang, R. Zhu and Q. Y. Zhang, unpublished data). In this entry route, the endocytosed virus requires access to acidic organelle to undergo viral-host membrane fusion. The Golgi complex is a potential fusion site for these viruses [37]. It raises the possibility that PoIFITM1 may target to the Golgi, and it is thus able to block viral membrane fusion process. Further investigation should be needed to elucidate the mechanism by which PoIFITM1 restricts virus infection with the Golgi targeting capability.
A marked structure difference between PoIFITM1 and the identified mammalian IFITM members is the length of the N-terminal region. PoIFITM1 has a relatively longer N-terminal region. Deletion mutagenesis revealed that this region was crucial for the antiviral function of IFITM1, as in the case of other viruses studied [10], [16]. Due to the removing of the N-terminal region, the colocalization of PoIFITM1 with endocytosis markers as well as with Golgi was abolished; therefore PoIFITM1 lost the chance to encounter the endocytosed viral particles. This finding is consistent with a recent study that N-terminal region of IFITM3 modulates its antiviral activity by controlling the cellular localization [16].
Collectively, the data presented in the current study demonstrate that a functional IFITM1 ortholog exits in fish. RGV is a pathogen that causes lethal disease in aquaculture animals [24], [38]. Strategies that effectively induce IFITM1 activity could serve as potent therapeutic approaches for treatment against RGV infection. Therefore, our results also pave the way for future development of antiviral agents for controlling infection by pathogenic viruses in fish.
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (2009CB118704, 2010CB126303), and the National Natural Science Foundation of China (30800854, 31072231).
References
- 1.Siegrist F., Ebeling M., Certa U. The small interferon-induced transmembrane genes and proteins. J Interferon Cytokine Res. 2011;31:183–197. doi: 10.1089/jir.2010.0112. [DOI] [PubMed] [Google Scholar]
- 2.Sällman Almén M., Bringeland N., Fredriksson R., Schiöth H.B. The dispanins: a novel gene family of ancient origin that contains 14 human members. PLoS One. 2012;7:e31961. doi: 10.1371/journal.pone.0031961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka S.S., Matsui Y. Developmentally regulated expression of mil-1 and mil-2, mouse interferon-induced transmembrane protein like genes, during formation and differentiation of primordial germ cells. Mech Dev. 2002;119:261–267. doi: 10.1016/s0925-4773(03)00126-6. [DOI] [PubMed] [Google Scholar]
- 4.Martensen P.M., Justesen J. Small ISGs coming forward. J Interferon Cytokine Res. 2004;24:1–19. doi: 10.1089/107999004772719864. [DOI] [PubMed] [Google Scholar]
- 5.Klymiuk I., Kenner L., Adler T., Busch D.H., Boersma A., Irmler M. In vivo functional requirement of the mouse ifitm1 gene for germ cell development, interferon mediated immune response and somitogenesis. PLoS One. 2012;7:e44609. doi: 10.1371/journal.pone.0044609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brass A.L., Huang I.C., Benita Y., John S.P., Krishnan M.N., Feeley E.M. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang I.C., Bailey C.C., Weyer J.L., Radoshitzky S.R., Becker M.M., Chiang J.J. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7:e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J., Pan Q., Rong L., Liu S.L., Liang C. The IFITM proteins inhibit HIV-1 infection. J Virol. 2011;85:2126–2137. doi: 10.1128/JVI.01531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raychoudhuri A., Shrivastava S., Steele R., Kim H., Ray R., Ray R.B. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J Virol. 2011;85:12881–12889. doi: 10.1128/JVI.05633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weidner J.M., Jiang D., Pan X.B., Chang J., Block T.M., Guo J.T. Interferon induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J Virol. 2010;84:12646–12657. doi: 10.1128/JVI.01328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diamond M.S., Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2012;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feeley E.M., Sims J.S., John S.P., Chin C.R., Pertel T., Chen L.M. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7:e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li K., Markosyan R.M., Zheng Y.M., Golfetto O., Bungart B., Li M. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013;9:e1003124. doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everitt A.R., Clare S., Pertel T., John S.P., Wash R.S., Smith S.E. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y.H., Zhao Y., Li N., Peng Y.C., Giannoulatou E., Jin R.H. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat Commun. 2013;4:1418. doi: 10.1038/ncomms2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia R., Pan Q., Ding S., Rong L., Liu S.L., Geng Y. The N-terminal region of IFITM3 modulates its antiviral activity through regulating IFITM3 cellular localization. J Virol. 2012;86:13697–13707. doi: 10.1128/JVI.01828-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsen R., Rokenes T.P., Robertsen B. Inhibition of infectious pancreatic necrosis virus replication by atlantic salmon Mx1 protein. J Virol. 2004;78:7938–7944. doi: 10.1128/JVI.78.15.7938-7944.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu R., Zhang Y.B., Zhang Q.Y., Gui J.F. Functional domains and the antiviral effect of the dsRNA-dependent protein kinase PKR from Paralichthys olivaceus. J Virol. 2008;82:6889–6901. doi: 10.1128/JVI.02385-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou J., Secombes C.J. Teleost fish interferons and their role in immunity. Dev Comp Immunol. 2011;35:1376–1387. doi: 10.1016/j.dci.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Hickford D.E., Frankenberg S., Shaw G., Renfree M.B. Evolution of vertebrate interferon inducible transmembrane proteins. BMC Genomics. 2012;13:155. doi: 10.1186/1471-2164-13-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson M.C., Sangrador-vegas A., Smith T.J., Carirns M.T. Cloning and characterization of two genes encoding rainbow trouthomologues of the IFITM protein family. Vet Immunol Immunopathol. 2006;110:357–362. doi: 10.1016/j.vetimm.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 22.Wang X., Chen X.H. Molecular cloning and expression analysis of interferon-inducible transmembrane protein 1 in large yellow croaker Pseudosciaena crocea. Vet Immunol Immunopathol. 2008;124:99–106. doi: 10.1016/j.vetimm.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Chen S.L., Ren G.C., Sha Z.X., Shi C.Y. Establishment of a continuous embryonic cell line from Japanese flounder Paralichthys olivaceus for virus isolation. Dis Aquat Org. 2005;60:241–246. doi: 10.3354/dao060241. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q.Y., Xiao F., Li Z.Q., Gui J.F., Mao J.H., Chinchar G.V. Characterization of an iridovirus form the cultured pig frog (Rana grylio) with lethal syndrome. Dis Aquat Org. 2001;48:27–36. doi: 10.3354/dao048027. [DOI] [PubMed] [Google Scholar]
- 25.Zhu R.L., Lei X.Y., Ke F., Yuan X.P., Zhang Q.Y. Genome of turbot rhabdovirus exhibits unusual non-coding regions and a novel additional ORF that could be expressed in fish cell. Virus Res. 2011;155:495–505. doi: 10.1016/j.virusres.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y.D., Zhang Y.B., Zhu R., Jiang J., Zhang Q.Y., Gui J.F. Construction of a subtractive cDNA library from the Paralichthys olivaceus embryonic cells induced by a double-stranded RNA virus. Virol Sin. 2005;20:168–172. [In Chinese] [Google Scholar]
- 27.Zhu R., Zhang Y.B., Chen Y.D., Dong C.W., Zhang F.T., Zhang Q.Y. Molecular cloning and stress-induced expression of Paralichthys olivaceus heme-regulated initiation factor 2α kinase. Dev Comp Immunol. 2006;30:1047–1059. doi: 10.1016/j.dci.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 28.Lei X.Y., Ou T., Zhang Q.Y. Rana grylio virus (RGV) 50L is associated with viral matrix and exhibited two distribution patterns. PLoS One. 2012;7:e43033. doi: 10.1371/journal.pone.0043033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Yount J.S., Moltedo B., Yang Y.Y., Charron G., Moran T.M., López C.B. Palmitoylome profiling reveals S-palmitoylation dependent antiviral activity of IFITM3. Nat Chem Biol. 2010;6:610–614. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yount J.S., Karssemeijer R.A., Hang H.C. S-palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. J Biol Chem. 2012;287:19631–19641. doi: 10.1074/jbc.M112.362095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y.B., Li Q., Gui J.F. Differential expression of two Carassius auratus Mx genes in cultured CAB cells induced by grass carp hemorrhage virus and interferon. Immunogenetics. 2004;56:68–75. doi: 10.1007/s00251-004-0658-1. [DOI] [PubMed] [Google Scholar]
- 33.Habjan M., Penski N., Wagner V., Spiegel M., Overby A.K., Kochs G. Efficient production of Rift Valley fever virus-like particles: the antiviral protein MxA can inhibit primary transcription of bunyaviruses. Virology. 2009;385:400–408. doi: 10.1016/j.virol.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 34.Blagojevic G., Mahmutefendic H., Kucic N., Tomas M.I., Lucin P. Endocytic Trafficking of cholera toxin in Balb 3T3 cells. Croat Chem Acta. 2008;81:191–202. [Google Scholar]
- 35.Luetterforst R., Stang E., Zorzi N., Carozzi A., Way M., Parton R.G. Molecular characterization of caveolin association with the Golgi complex: identification of a cis-Golgi targeting domain in the caveolin molecule. J Cell Biol. 1999;145:1443–1459. doi: 10.1083/jcb.145.7.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y., Yang G., Hu G. Binding of IFITM1 enhances the inhibiting effect of caveolin-1 on ERK activation. Acta Biochim Biophys Sin. 2009;41:488–494. doi: 10.1093/abbs/gmp034. [DOI] [PubMed] [Google Scholar]
- 37.Guo C.J., Whu Y.Y., Yang X.B., Yang L.S., Mi S., Huang Y.X. Tiger frog virus (an iridovirus) entry into HepG2 cells via a pH-dependent, atypical caveola-mediated endocytosis pathway. J Virol. 2011;85:6416–6426. doi: 10.1128/JVI.01500-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gui J.F., Zhu Z.Y. Molecular basis and genetic improvement of economically important traits in aquaculture animals. Chin Sci Bull. 2012;57:1751–1760. [Google Scholar]


