Abstract
Few studies have examined the epidemiology of respiratory viral infections in large tertiary centres over more than one season in the era of molecular diagnosis. Respiratory clinical specimens received between 1 January 2011 and 31 December 2012 were analysed. Respiratory virus testing was performed using a large panel of real‐time PCR or RT‐PCR. Results were analysed according to sample type (upper versus lower respiratory tract) and age group. In all, 2996 (2469 (82.4%) upper; 527 (17.6%) lower) specimens were analysed. Overall positivity rate was 47.4% and 23.7% for upper and lower respiratory samples, respectively. The highest positivity rate was observed in patients under 18 years old (p <0.001); picornaviruses were the most frequent viruses detected over the year. Influenza virus, respiratory syncytial virus, human metapneumovirus and coronaviruses showed a seasonal peak during the winter season, while picornaviruses and adenoviruses were less frequently detected in these periods. Multiple viral infections were identified in 12% of positive cases and were significantly more frequent in children (p <0.001). In conclusion, we observed significant differences in viral infection rates and virus types among age groups, clinical sample types and seasons. Follow‐up of viral detection over several seasons allows a better understanding of respiratory viral epidemiology.
Keywords: Adenovirus, coronavirus, epidemiology, influenza, picornavirus, respiratory syncytial virus, respiratory virus, seasonality
Introduction
Respiratory viral infections are a leading cause of morbidity and mortality, particularly in children, the elderly and immunocompromised persons. Rapid identification of viral aetiology is critical to avoid unnecessary antibiotics, to initiate antiviral treatment when available and to limit the spread of the infection 1.
Nucleic acid‐based amplification tests (NATs) allow sensitive detection of a broad panel of both conventional and emerging viruses in respiratory tract specimens. NATs are more sensitive than any other diagnostic method, including virus isolation in cell culture and antigen detection, and now form the backbone of clinical virology laboratory testing around the world. This advance has changed the landscape of virus detection and highlights the need to better understand the epidemiology of viruses ranging from rhinoviruses to influenza virus 2, 3.
Most of the available literature describing the epidemiology of respiratory viruses is focused on the paediatric population, other particular populations, or specific viral agents; studies are frequently limited to one season. A longitudinal examination of the epidemiology of viral respiratory agents among patients frequenting a large university centre is lacking; prevalence patterns in this group may differ from those in the community and may be subject to seasonal variation.
Our study aims to describe the general molecular epidemiology of viral respiratory infections in paediatric, adult and elderly populations admitted to or screened in a tertiary care centre over a 2‐year period, and, more specifically, to compare the epidemiological patterns of upper and lower respiratory specimens.
Methods
The University Hospital of Geneva is a tertiary care teaching hospital with 1600 acute‐care beds and >40 000 admissions each year. It is comprised of surgical and internal medicine services, as well as bone‐marrow and solid‐organ transplant units. The virology laboratory of the University Hospital of Geneva processes all clinical specimens from adult and paediatric inpatient and outpatient departments.
Clinical specimens
All respiratory specimens (nasopharyngeal swabs and aspirates, bronchial and/or tracheal aspirates and bronchoalveolar lavage (BAL) specimens) from adult and paediatric inpatients and outpatients received between 1 January 2011 and 31 December 2012 were included in the study. Nasopharyngeal swabs and aspirates were grouped as ‘upper respiratory samples’, while bronchial and tracheal aspirates, as well as BAL specimens, were considered ‘lower respiratory samples’. Further, specimens were considered ‘paediatric’ if they were from patients under 18 years of age, ‘adult’ if from patients between 18 and 65 years old, and ‘elderly’ if from patients over age 65. Collection of samples for analysis was not systematic but was based on clinical judgement according to local practice and guidelines that recommend viral screening in patients at risk of lower respiratory complications and systematically in transplant recipients, in hospitalized patients with an acute respiratory disease during the influenza season, and in those that do no respond to the usual empirical antibiotic treatment. However, detection of viral pathogens was systematic in BAL obtained from immunocompromised patients such as transplant recipients.
Viral real‐time PCR detection
Each specimen was screened by nucleic acid detection for the presence of influenza A (both seasonal and 2009 pandemic H1N1 strains) or B virus, respiratory syncytial virus (RSV) A and B, parainfluenza virus 1–3, human metapneumovirus, rhinovirus A, B and C, enterovirus, adenovirus and, beginning on 23 August 2011, coronaviruses 229E, OC43, HKU1 and NL63. Screening was performed using individual one‐step real‐time Taqman©‐based PCR or RT‐PCR as described previously. Respiratory specimens were extracted using Easymag© (bioMérieux, Geneva, Switzerland) according to the manufacturer's recommendations. The viral real‐time PCR detection was performed as described for parainfluenza viruses 1 and 3 4, influenza viruses, RSV A and B, coronaviruses, parainfluenza virus 2, human metapneumovirus, coronaviruses, adenoviruses 5, 6 enteroviruses and rhinoviruses 7.
Extraction, presence of PCR inhibitors, and reverse transcription were controlled by spiking each specimen with a quantified standard of canine distemper virus; experiments were validated only if the resulting cycling threshold value was within the expected ranges. PCR detection was considered positive if the cycling threshold value was ≤39.
In addition to molecular tests, rapid tests for antigen detection (data not shown) are routinely used for influenza A and B viruses (Bionexia; bioMérieux, Lyon, France) and RSV (Quickvue; Quidel, San Diego, CA, USA) detection in the paediatric emergency wards.
Definition of clinical episodes
For patients with specimens that were repeatedly positive for the same viral agent in a period shorter than 3 weeks, only the first positive result was considered. If the interval between positive specimens was 3 weeks or longer, a different episode was assumed. A separate clinical episode was also assumed for patients with different viruses detected throughout their clinical course, independent of the time elapsed between sampling. If two upper or two lower respiratory samples were sent at the same time or within 3 days of one another, these were considered a single clinical episode if both were positive. Simultaneous detection of more than one virus in the same sample was considered as a single clinical episode with a multiple viral infection (MVI).
Statistical analysis
The independent t‐test was used to compare continuous data, while the chi‐squared test was used to compare categorical data. Associations with a p value <0.05 were considered significant. All data were analysed within IBM SPSS Statistics for Windows, Version 20.0 (IBM Corp, Armonk, NY, USA).
Results
During the study period, 2996 clinical respiratory specimens were analysed, with a total of 32 387 real‐time PCRs or RT‐PCRs performed. Upper respiratory samples predominated, representing 82.4% of all cases (2469); 17.6% (527) were lower respiratory samples. Among the 527 lower respiratory samples, 451 (85.6%) were collected via BAL. The overall positivity rate (PR) for any respiratory virus was 43.2%, while for upper and lower respiratory samples it was 47.4% and 23.7%, respectively (Fig. 1). Most (89%) nasopharyngeal aspirates were collected from paediatric patients, whereas most (87.9%) nasopharyngeal swabs were drawn from adult and elderly patients. For coronaviruses, 328 upper and 72 lower respiratory samples were analysed.
Figure 1.
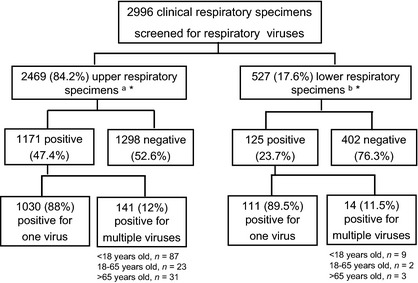
Clinical respiratory specimens analysed during the study period (1 January 2011 to 31 December 2012). aNasopharyngeal swabs and nasopharyngeal aspirates. bTracheal and/or bronchial aspirates and bronchoalveolar lavages *In 69 cases there were paired upper and lower respiratory specimens for the same episode.
The proportion of specimens from male patients ranged from 47.4 to 64.7% depending on the population. The distribution of samples and the positivity rate according to age group are shown in Table 1. Median ages were 2 years (range 0–18 years), 41.5 years (range 18–65 years) and 84.5 years (65–103 years) for paediatric, adult and elderly populations, respectively. Lower respiratory samples were collected from both genders fairly equally among adult patients (47.4% male) but primarily from male patients among the elderly (63%). Overall, lower respiratory samples were collected chiefly from adults (51.2%), with only 26.2% and 22.6% from paediatric and elderly patients, respectively. Upper respiratory samples were somewhat more evenly distributed, with 26.1%, 36.6% and 37.3% from paediatric, adult and elderly populations, respectively. The highest positivity rate (78.6%) was found in upper respiratory samples collected from paediatric patients, the lowest (20.4%) in lower respiratory samples from adult patients. Paediatric patients had a significantly higher positivity rate (p <0.001).
Table 1.
Type, distribution and positivity rates of clinical respiratory specimens collected during the study period stratified by age
| Respiratory specimen | Paediatric | Adults | Elderly | |||
|---|---|---|---|---|---|---|
| n | PR (%) | n | PR (%) | n | PR (%) | |
|
Uppera n = 2469 |
645 | 78.6 | 904 | 44.6 | 920 | 46 |
|
Lowerb n = 527 |
138 | 40.6 | 270 | 20.4 | 119 | 23.5 |
PR, positivity rate.
Includes nasopharyngeal swabs and nasopharyngeal aspirates.
Includes tracheal/bronchial aspirates and bronchoalveolar lavages.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Of 527 lower respiratory samples, 69 (13.1%) had a paired upper respiratory sample analysed during the same episode. Among these, 20 (29%) were both positive, 39 (56.5%) were both negative, and ten (14.5%) had discordant results.
Fig. 2 describes the respective prevalence of different types of viruses. Picornaviruses were detected with the greatest frequency in all specimens and populations except upper respiratory samples collected from the elderly; in these, influenza viruses predominated. Indeed, across all specimens, influenza virus prevalence increased with patients' age. Parainfluenza viruses were the second most frequently found viruses in lower respiratory samples of the elderly population, whereas overall, adenoviruses were very infrequent in the non‐paediatric population. Among children, adenoviruses and RSV were the second and third most frequent viruses, both for upper and lower respiratory samples. In contrast, coronaviruses were rarely found in children, but were detected in 9–16% of combined specimens in adults and the elderly.
Figure 2.
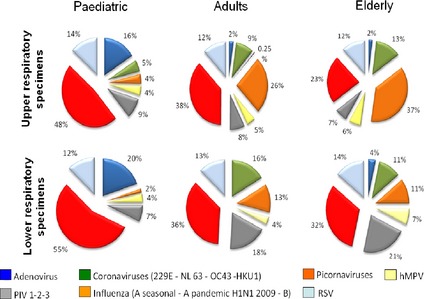
Viral prevalences according to age group (<18, 18–65 and >65 years old) in all respiratory specimens collected during the study period. hMPV, human metapneumovirus; PIV, parainfluenza virus; RSV, respiratory syncytial virus.
Multiple viral infections, defined as the simultaneous detection of more than one virus in one clinical specimen, were apparent in 141 (12%) and 14 (11.3%) of all positive upper and lower respiratory samples, respectively. Among the 141 positive upper respiratory samples, 123 (87.2%) were positive for two viruses, 16 (11.4%) for three viruses and two (1.4%) for four viruses, 14 lower respiratory samples were positive for two viruses. MVI diagnosed via upper respiratory samples were significantly more frequent among paediatric patients: 13.5% of paediatric samples revealed multiple viruses versus 2.5% and 3.4% of adult and elderly samples, respectively (p <0.001). Among upper respiratory samples from children, the most frequent co‐infections were picornaviruses with adenoviruses (22 cases, 15.4%) and adenoviruses with RSV (ten cases, 7%). The former was also the most frequent co‐infection detected among lower respiratory samples. Details of MVI are shown in Table 2.
Table 2.
Multiple respiratory viral infections detected during the study period
| Upper respiratory samplesa | Lower respiratory samples | ||||||
|---|---|---|---|---|---|---|---|
| <18 yearsb | n | Positive for two viruses | n | Positive for two viruses | |||
| 22 | Picornavirus | Adenovirus | 6 | Picornavirus | Adenovirus | ||
| 10 | Adenovirus | RSV | 1 | Picornavirus | RSV | ||
| 9 | Picornavirus | RSV | 1 | Picornavirus | PIV | ||
| 5 | Picornavirus | PIV | 1 | RSV | PIV | ||
| 4 | Adenovirus | hMPV | |||||
| 4 | Adenovirus | PIV | |||||
| 3 | Adenovirus | PIV | |||||
| 3 | Adenovirus | Coronavirus | |||||
| 3 | RSV | Coronavirus | |||||
| n | Positive for three viruses | ||||||
| 3 | Picornavirus | PIV | Adenovirus | ||||
| 2 | Picornavirus | RSV | Adenovirus | ||||
| 2 | Picornavirus | RSV | Coronavirus | ||||
| 18–65 years | n | Positive for two viruses | n | Positive for two viruses | |||
| 3 | Picornavirus | RSV | 1 | Picornavirus | Coronavirus | ||
| 2 | Picornavirus | Influenza virus | 1 | Picornavirus | PIV | ||
| 2 | Picornavirus | Coronavirus | |||||
| n | Positive for three viruses | ||||||
| 2 | Picornavirus | RSV | Influenza virus | ||||
| >65 years | n | Positive for three viruses | n | Positive for two viruses | |||
| 7 | Influenza virus | Coronavirus | 1 | Picornavirus | Adenovirus | ||
| 4 | Picornavirus | Influenza virus | 1 | Coronavirus | Influenza virus | ||
| 2 | RSV | Influenza virus | |||||
hMPV, human metapneumovirus ; PIV, parainfluenza virus; RSV, respiratory syncytial virus.
Only the multiple infections with associations found in more than one case are shown.
There were two cases of multiple infections with four viruses detected in this group (upper respiratory specimens, <18 years, one case of picornavirus–adenovirus–PIV2–PIV3 co‐infection and one case of adenovirus–hMPV–PIV–influenza virus co‐infection).
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
The number of screened specimens increased in winter periods but the positivity rate, although also slightly higher in winter months, remained relatively stable (Fig. 3a). The highest positivity rate was obtained in February 2012 (76.5%) and the lowest in August 2012 (25.4%). Regarding the distribution of respiratory viruses over the year, nfluenza virus, RSV, human metapneumovirus and coronaviruses showed a clear seasonal pattern, with a marked increase in PR throughout the winter months, while picornavirus and adenovirus prevalence remained relatively stable over the year, with a decrease in the PR during winter months. Seasonal prevalence patterns are depicted in Fig. 3b. For multiple infections, the cases with simultaneous detection of picornaviruses and adenoviruses were found throughout the year, whereas those involving influenza viruses and coronaviruses were more frequent in winter.
Figure 3.
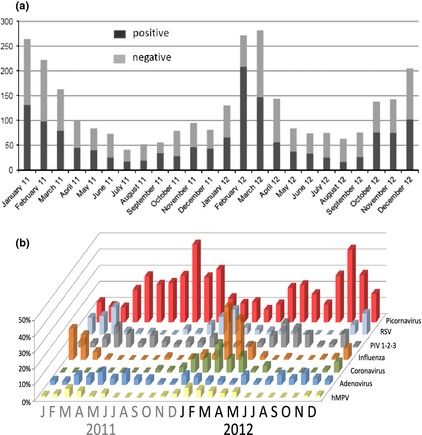
(a) Number of positive and negative respiratory clinical specimens received and analysed at the Laboratory of Virology of the University of Geneva Hospitals during the study. (b) Positivity rate for each group of viruses from clinical specimens analysed at the Laboratory of Virology of the University of Geneva Hospitals during the study period. hMPV, human metapneumovirus; PIV, parainfluenza virus; RSV, respiratory syncytial virus.
Discussion
We describe the epidemiological patterns of respiratory viruses among hospitalized and ambulatory patients of all ages screened at a tertiary care centre across all seasons of a 2‐year period. Overall, the positivity rate for any respiratory virus was 43.2% and it was significantly higher in the paediatric population. Picornaviruses had the highest prevalence. Co‐infections with more than one virus were relatively uncommon; they were detected most frequently in paediatric samples (13.5% of positive cases). The main strengths of our investigation are the high number of specimens analysed as well as the use of ‘real‐life’ data; these results are likely to reflect the epidemiology of respiratory viruses as observed by clinicians delivering routine care in a tertiary care centre and for a wide age range. The increased sensitivity of molecular diagnostic methods, provides a more faithful depiction of the epidemiology of respiratory viruses at the ‘front line’, where patients with a higher risk of complications are hospitalized. Our results suggest that viruses circulating in large hospitals parallel those observed in the community.
We observed the highest positivity rate in the paediatric population, with overall prevalence reaching nearly 80%. This could partially be explained by the frequent collection of nasopharyngeal aspirates rather than swabs, as the former may be more sensitive 8. However, it has also been established that the viral load in children is significantly higher than in adults, and our results are comparable to other studies in children that used swabs. Khamis et al. reported a PR of 50% in nasopharyngeal aspirates from the paediatric patients of an Omani hospital using multiplex PCR. In this study, the most prevalent virus was RSV, but the study population was limited to children under 5 years old 9. Another large, prospective study following paediatric patients for 2 years and employing real‐time RT‐PCR also detected RSV most frequently, but again patients were young (<2 years) with suspected pneumonia 10. In our study, picornaviruses were most frequently detected in the paediatric population. Varying outcomes among studies reflect their differences in diagnostic technique, the clinical criteria applied to the study population, and the panel of detected viruses. These heterogeneities render comparisons among published studies difficult, if not altogether misleading.
In our study, multiple viral infections were observed in >10% of positive cases overall, and almost 15% of positive paediatric cases. Other studies, however, have shown even higher rates of co‐infections. In the study by Khamis et al. 9, MVI were found in 18% of cases. Zhang et al. 11 reported a rate of 29.5% in children younger than 3 years old, while Kouni et al. 12 reported an MVI rate of 42.5% in children between 1 month and 14 years old, as detected by DNA/RNA microarray assay. Although the clinical impact of multiple infections is unknown, a more severe clinical course was observed in the latter study 12. It is also unclear whether infection with the first virus may facilitate or prevent infection with other viruses. In the present study, picornaviruses were those most frequently involved in co‐infections, which is certainly related to the fact that picornaviruses are detected throughout the year and are the most prevalent. It is also possible, however, that picornavirus serotypes vary over the year; some of these might manifest a seasonal pattern, variable virulence and/or the capacity to predispose their host to additional viral infections. The most frequent co‐infection was with picornaviruses and adenoviruses. Not surprisingly, this association was consistent throughout the year, whereas the MVI involving influenza virus, RSV or coronaviruses were more frequently found in winter.
Moreover, a positive detection by NATs does not necessarily signify active replication, because it might be the remnant traces of a recent infection. Hence, an infection following a previous one with still‐detectable nucleic acid cannot be excluded. Alternatively, detection of multiple respiratory viruses might be a surrogate marker of immunosuppression, as persistent infection has been described for several respiratory viruses in immunocompromised patients 13. Finally, it seems that co‐detection of different respiratory viruses is not random. A recently published study showed that there were positive and negative associations between some viruses. For example, rhinoviruses and adenoviruses were positively associated whereas influenza A virus and rhinoviruses showed a negative association 14. Of note, in our study influenza virus was almost completely absent among the viruses involved in co‐infections in the paediatric population, in which picornaviruses were very prevalent.
Seasonality is well described for several viral respiratory pathogens. Existing evidence suggests that the seasonality of some pathogens may be driven by enhanced wintertime survival, and also by increased host susceptibility resulting from relative ‘wintertime immune suppression’ 15. A cold environment decreases innate defence mechanisms such as mucociliary clearance, so increasing susceptibility to viral infections 16. In a large Malaysian study performed over 27 years and with >10 000 respiratory samples, while RSV and other, less frequent viruses produced peaks throughout the year, they spiked more notably from September to December, correlating with the rainy season 17. For those viruses showing seasonal patterns, the months in which the peaks occur may be determined by local topography and latitude, making comparisons among studies performed in different countries difficult 18.
In our study, picornaviruses had the highest prevalence over all seasons, although other viruses such as influenza virus and RSV increased their prevalence in winter. The reasons why picornaviruses and adenoviruses display no discernible seasonal patterns are poorly understood. As expected, we found that influenza viruses and other viruses increase in prevalence with a seasonal distribution in winter (see Fig. 3b). In these periods, picornaviruses and adenoviruses were less frequently detected. Whether picornaviruses delay influenza circulation or whether the initiation of influenza circulation decreases the prevalence of picornaviruses is not clear either. It has been suggested that rhinovirus infection might protect against influenza virus infection either through viral interference at the cellular level or via interferon or other cytokines. Indeed, it has been hypothesized that in some countries such as France and Norway, rhinovirus circulation delayed the spread of pandemic H1N1 influenza in 2009 19, 20. As noted above, a recent study showed a negative association between influenza A virus and rhinovirus co‐infection 14.
On a practical level, whether a complete respiratory panel needs to be employed for viral detection in summer months is unknown. The approach should probably be adapted to the local epidemiology of each particular centre. In Geneva, a city with a continental climate and inhabitants who travel internationally with relative frequency, employing a complete panel year‐round appears justified, despite a low detection of some viruses. However, as shown in Fig. 3a, despite a slight decrease of PR in summer months, overall PR for respiratory viruses remained relatively stable throughout the year. A complete respiratory viral panel would also be appropriate for immunocompromised patients, particularly lung transplant recipients or children with chronic pulmonary diseases. The additional costs incurred by the use of a complete panel would likely be offset by the attendant reduction in unnecessary hospitalizations and antibiotic therapy.
Our study has some limitations. First, the analysis is limited to the data available in our laboratory; no clinical end‐points were included. A positive result from an upper respiratory sample (e.g. nasopharyngeal aspirate) may represent an upper respiratory infection or a lower one (bronchiolitis for example), hence upper respiratory sampling does not guarantee an upper respiratory infection. Second, pooled data from bacterial and fungal testing were not available; these would have been particularly illuminating among immunocompromised patients. In addition, the prevalence of RSV may be underestimated in our study: because a rapid test for RSV antigen detection is available in our centre in the paediatric emergency wards: not every child with suspected RSV infection was tested by NATs. This may explain, in part, why picornaviruses were detected more frequently in the present study, whereas most other studies report RSV as the most frequent virus in children. Finally, the proportion of immunocompetent and immunocompromised patients is not known; we offer an analysis of the whole population (ambulatory and hospitalized, immunocompetent and immunocompromised).
Our study provides an overview of viral respiratory infections in a large tertiary centre. Picornaviruses are the most frequently detected respiratory viruses among paediatric, adult and geriatric populations, although influenza virus predominated in the latter. Influenza virus, RSV and human metapneumovirus display a seasonal pattern, whereas picornaviruses and adenoviruses are detected with regularity throughout the year. Multiple infections are commonly observed in children, although their overall prevalence is relatively low. A clearer picture of respiratory virus prevalence patterns across all seasons and age groups aids in the design of diagnostic strategies and can help to reduce unwarranted antimicrobial consumption.
Transparency Declaration
This study was supported by the Swiss National Science Foundation (grant 32003B‐1Z7160 awarded to LK). No conflicts of interest.
Clin Microbiol Infect 2014; 20: O578–O584
References
- 1. Sanghavi SK, Bullotta A, Husain S, Rinaldo CR. Clinical evaluation of multiplex real‐time PCR panels for rapid detection of respiratory viral infections. J Med Virol 2012; 84: 162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mahony JB, Petrich A, Smieja M. Molecular diagnosis of respiratory virus infections. Crit Rev Clin Lab Sci 2011; 48: 217–249. [DOI] [PubMed] [Google Scholar]
- 3. Yan Y, Zhang S, Tang YW. Molecular assays for the detection and characterization of respiratory viruses. Semin Respir Crit Care Med 2011; 32: 512–526. [DOI] [PubMed] [Google Scholar]
- 4. Cordey S, Thomas Y, Cherpillod P, van BS, Tapparel C, Kaiser L. Simultaneous detection of parainfluenza viruses 1 and 3 by real‐time reverse transcription‐polymerase chain reaction. J Virol Methods 2009; 156: 166–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garbino J, Inoubli S, Mossdorf E et al Respiratory viruses in HIV‐infected patients with suspected respiratory opportunistic infection. AIDS 2008; 22: 701–705. [DOI] [PubMed] [Google Scholar]
- 6. Garbino J, Soccal PM, Aubert JD et al Respiratory viruses in bronchoalveolar lavage: a hospital‐based cohort study in adults. Thorax 2009; 64: 399–404. [DOI] [PubMed] [Google Scholar]
- 7. Tapparel C, Cordey S, van BS et al New molecular detection tools adapted to emerging rhinoviruses and enteroviruses. J Clin Microbiol 2009; 47: 1742–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sung RY, Chan PK, Choi KC et al Comparative study of nasopharyngeal aspirate and nasal swab specimens for diagnosis of acute viral respiratory infection. J Clin Microbiol 2008; 46: 3073–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khamis FA, Al‐Kobaisi MF, Al‐Areimi WS, Al‐Kindi H, Al‐Zakwani I. Epidemiology of respiratory virus infections among infants and young children admitted to hospital in Oman. J Med Virol 2012; 84: 1323–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Homaira N, Luby SP, Petri WA et al Incidence of respiratory virus‐associated pneumonia in urban poor young children of Dhaka, Bangladesh, 2009–2011. PLoS ONE 2012; 7: e32056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang G, Hu Y, Wang H, Zhang L, Bao Y, Zhou X. High incidence of multiple viral infections identified in upper respiratory tract infected children under three years of age in Shanghai, China. PLoS ONE 2012; 7: e44568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kouni S, Karakitsos P, Chranioti A, Theodoridou M, Chrousos G, Michos A. Evaluation of viral co‐infections in hospitalized and non‐hospitalized children with respiratory infections using microarrays. Clin Microbiol Infect 2013; 19: 772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tapparel C, Cordey S, Junier T et al Rhinovirus genome variation during chronic upper and lower respiratory tract infections. PLoS ONE 2011; 6: e21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tanner H, Boxall E, Osman H. Respiratory viral infections during the 2009–2010 winter season in Central England, UK: incidence and patterns of multiple virus co‐infections. Eur J Clin Microbiol Infect Dis 2012; 31: 3001–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fisman D. Seasonality of viral infections: mechanisms and unknowns. Clin Microbiol Infect 2012; 18: 946–954. [DOI] [PubMed] [Google Scholar]
- 16. Giesbrecht GG. The respiratory system in a cold environment. Aviat Space Environ Med 1995; 66: 890–902. [PubMed] [Google Scholar]
- 17. Khor CS, Sam IC, Hooi PS, Quek KF, Chan YF. Epidemiology and seasonality of respiratory viral infections in hospitalized children in Kuala Lumpur, Malaysia: a retrospective study of 27 years. BMC Pediatr 2012; 12: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bloom‐Feshbach K, Alonso WJ, Charu V et al Latitudinal variations in seasonal activity of influenza and respiratory syncytial virus (RSV): a global comparative review. PLoS ONE 2013; 8: e54445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Casalegno JS, Ottmann M, Duchamp MB et al Rhinoviruses delayed the circulation of the pandemic influenza A (H1N1) 2009 virus in France. Clin Microbiol Infect 2010; 16: 326–329. [DOI] [PubMed] [Google Scholar]
- 20. Anestad G, Nordbo SA. Virus interference. Did rhinoviruses activity hamper the progress of the 2009 influenza A (H1N1) pandemic in Norway? Med Hypotheses 2011; 77: 1132–1134. [DOI] [PubMed] [Google Scholar]


