Abstract
Membrane fusion between virus and host cells is the key step for enveloped virus entry and is mediated by the viral envelope fusion protein. In murine coronavirus, mouse hepatitis virus (MHV), the spike (S) protein mediates this process. Recently, the formation of anti-parallel 6-helix bundle of the MHV S protein heptad repeat (HR) regions (HR1 and HR2) has been confirmed, implying coronavirus has a class I fusion protein. This bundle is also called fusion core. To facilitate the solution of the crystal structure of this fusion core, we deployed an Escherichia coli in vitro expression system to express the HR1 and HR2 regions linked together by a flexible linker as a single chain (named 2-helix). This 2-helix polypeptide subsequently assembled into a typical 6-helix bundle. This bundle has been analyzed by a series of biophysical and biochemical techniques and confirmed that the design technique can be used for coronavirus as we successfully used for members of paramyxoviruses.
Keywords: Mouse hepatitis virus, Coronavirus, Spike (S) protein, Fusion core, 6-Helix bundle, Heptad repeat regions (HR1 and HR2)
Mouse hepatitis virus (MHV) is a member of genus Coronavirus in the family Coronaviridae [1]. The family comprises a large and diverse group of enveloped viruses with a positive-stranded RNA genome of approximately 31 kb [2], [3]. The members of the family Coronaviridae exhibit broad host range, infecting many mammalian and avian species and causing upper respiratory, gastrointestinal, hepatic, and central nervous system diseases. In humans and fowl, coronaviruses primarily cause upper respiratory tract infections, while porcine and bovine coronaviruses establish enteric infections that result in severe economic loss [4]. The emergence of atypical pneumonia with severe acute respiratory syndrome (SARS) in 2003 and identification of the etiologic agent as a new coronavirus (SARS-CoV) have attracted the attention of the scientists and ordinary people for the importance of the family Coronaviridae [5], [6].
The coronavirus infection starts with fusion of the viral and cellular membranes [7]. This process is mediated by viral envelope spike (S) protein, which is thought to be a trimer on the virion. The MHV S protein is cleaved in the Golgi apparatus, by a host cell protease, into two similarly sized subunits: amino terminal S1 and carboxyl terminal S2. It is believed that the S1 subunit forms the globular head of the spike, whereas the S2 subunit forms the transmembrane stalk portion [8]. Sequence analysis suggests that the coronavirus spike protein has the structural features of a type I membrane protein, including a transmembrane domain near the carboxyl terminus of S2 and a hydrophobic signal peptide at the N-terminus of S1.
The S2 domain has an internal fusion peptide and two heptad repeat (HR) regions designated HR1 and HR2 [8], [9]. HR2 is located close to the transmembrane region, and HR1 is some 170 amino acids of its upstream. The HR regions are found in fusion proteins of many different viruses and form an important characteristic of class I viral fusion proteins (there are two classes of the viral fusion proteins under the current classification) [10], [11], [12], [13].
Typical class I viral fusion protein contains several notable features. It has a fusion peptide at the amino terminus that is thought to insert directly into the target membrane during the viral fusion process [12]. Carboxyl-terminal to the fusion peptide are two regions containing 4,3 hydrophobic (heptad) repeats, sequence motifs, which form a coiled-coil structure in the post-fusion state after series of the conformational changes of the fusion protein [12]. This coiled-coil structure is a stable, protease resistant core in the ectodomain of enveloped glycoprotein (or fusion protein), named as viral fusion core [12]. The fusion core complex consists of trimer of HR1/HR2 heterodimers, therefore, it is also called 6-helix bundle. The MHV fusion core complex has been confirmed biophysically and biochemically [14] but its crystal structure is yet to be elucidated. We previously developed a method to prepare this core complex for other viruses [15], [16], [17] by linking the HR1 and HR2 as a single chain. It was shown to be an effective method to prepare diffractable crystals for such a core complex. Here we deployed this method to MHV HR1/HR2 complex and indeed it shows that the single chain HR1/HR2 (detonated as 2-helix) could be prepared as a trimer of HR1/HR2 heterodimer suitable for good-quality crystal preparation.
Materials and methods
Prediction of the heptad repeat regions and construction of the expression vectors
The α-helix structure of the two HR regions of the S protein of MHV was predicted by LearnCoil-VMF program (http://nightingale.lcs.mit.edu/cgi-bin/vmf), which was specifically developed for identification of potential coiled-coil HR regions in viral fusion proteins by Kim and colleagues at Massachusetts Institute of Technology [18]. The proteinase K resistent regions of HR1 and HR2 have been identified by Bosch and colleagues [14]. The HR regions were chosen based on the prediction of LearnCoil-VMF and the previous published data [14]. The HR1 region used was derived from amino acids 968–1027 and HR2 from 1216 to 1253 as shown in Fig. 1 .
Fig. 1.
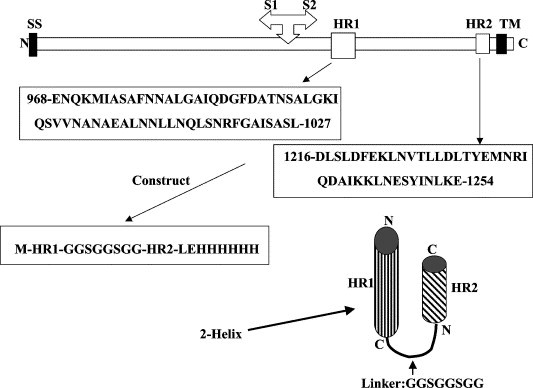
Schematic representations of MHV S protein and the construct used in this study. SS, signal sequence; TM, transmembrane domain; HR1 and HR2, heptad repeats 1 and 2. Cleavage site of S1 and S2 domains is shown. Amino acid sequences of the HR1 and HR2 regions are shown and the 2-helix construct is schematically represented.
The genes encoding the HR1 and HR2 regions of MHV were synthesized by PCR using eight synthesized primers according to the published sequences of murine coronavirus MHV strain A59 (Gene Bank No. M18379). The codon usage was optimized according to Escherichia coli expression preferences. HR1 and HR2 were connected by an eight amino acid linker (GGSGGSGG, single letter abbreviation of amino acids) and a 6-His tag was introduced by PCR primers at the C-terminus to facilitate the protein purification. The choice of the eight amino acids as linker was based on our previous successful work on other viral fusion core [15], [16], [17]. The constructs were cloned into the NdeI and XhoI sites (introduced by synthetic PCR primers) of pET22b expression vector (Novagen). Under this construction strategy, two extra amino acids were created between the C-terminal end of the HR2 and 6-His tag. They are “Leu–Glu” for XhoI restriction cloning site (CTCGAG for nucleotides). This construct was named as MHV 2-helix (Fig. 1). The construction was verified by direct DNA sequencing.
The primers overlapped with each other for 12–18 nucleotide bases. The NdeI and XhoI restriction sites were introduced by primer F and P8 (as shown below in capitalized/italic denominators). The gene of MHV 2-helix was obtained by two-step PCR. Firstly, the eight synthetic primers (0.2 OD/μl) were mixed and the PCR reactions were run with the following conditions: denatured at 94 °C for 5 min and then 15 cycles of 94 °C, 0.5 min; 55 °C, 1 min; and 72 °C, 1 min, For the second PCR, the first PCR product was used as template and the new PCR product was amplified by using primers F and P8 at the following conditions: denatured at 94 °C for 5 min and then 32 cycles of 94 °C, 0.5 min; 55 °C, 1 min; and 72 °C, 1 min.
Primers used for making the MHV 2-helix construct:
-
F:
5′-atgcgaattcCATATGgaaaaccagaaaatgatcgc-3′
-
P1:
5′-gaaaaccagaaaatgatcgcttctgctttcaacaacgctctgggtgctatccaggacg-3′
-
P2:
5′-gactggattttgcccagagcagagttagtagcgtcgaagccgtcctggatagcacccag-3′
-
P3:
5′-ctctgggcaaaatccagtctgttgttaacgctaacgctgaagctctgaacaacctgctg-3′
-
P4:
5′-cagagaagcagagatagcaccgaaacggttagacagctggttcagcaggttgttcagagc-3′
-
P5:
5′-gctatctctgcttctctgggtggttctggtggttctggtggtgacctgtctctggacttc-3′
-
P6:
5′-catttcgtaagtcaggtccagcagagtaacgttcagtttttcgaagtccagagacaggtc-3′
-
P7:
5′-gacctgacttacgaaatgaaccgtatccaggacgctatcaaaaaactgaacgaatc-3′
-
P8:
5′-ctagCTCGAGttctttcaggttgatgtaagattcgttcagttttttgatagcg-3′
Protein expression, purification, and gel-filtration analysis
The recombinant pET22b plasmids (MHV 2-helix) were transformed into E. coli strain BL21 (DE3). A single colony of the E. coli BL21 (DE3) transformed with the positive plasmid was grown in 2× YT medium containing 100-μg/mL of ampicillin in a 37 °C incubator. When the culture density (OD600) reached 0.6–0.8, the culture was induced with 0.2 mM IPTG and continuously grown for an additional 10 h at 16 °C before the cells were harvested. Bacterial cells were homogenized by sonication in phosphate-buffered saline (PBS, 10 mM sodium phosphate, pH 7.4; 150 mM NaCl). The lysates were clarified by centrifugation at 18,000g for 20 min at 4 °C. The supernatants were loaded on Ni-NTA column (QIAGEN) that was equilibrated with PBS. The contaminated protein was washed with washing buffer (1× PBS, 60 mM imidazole) over 10 column volumes and the target protein was eluted with elution buffer (1× PBS, 500 mM imidazole) for five column volumes. The resultant protein 2-helix was concentrated to a proper concentration by ultra-filtration (10 kDa cut-off). Proteins were analyzed by 10% Tricine SDS–PAGE. For further purification, the concentrated 2-helix protein was loaded onto a Superdex 75 HR10/30 (Pharmacia Biotech) column with an Akta Purifier System (Pharmacia Biotech). The fractions of the peak were collected and analyzed by a 10% Tricine SDS–PAGE. The peak molecular mass was estimated by comparison with the protein standards run on the same column (see Table 1 ).
Table 1.
Purification process of the 2-helix protein
Chemical cross-linking of the complex
The purified 2-helix protein after gel-filtration was dialyzed against cross-linking buffer (50 mM Hepes, pH 8.3; 100 mM NaCl) and concentrated to approximately 2 mg/mL by ultra-filtration (10 kDa cut-off). Protein concentrations were determined by absorbance at 280 nm, assuming an A280 of 0.22 for a 1.0 mg/mL. The proteins were cross-linked with ethyleneglycol bis(succinimidylsuccinate) (EGS) (Sigma). The reactions were incubated for 1 h on ice at different concentrations of EGS (0, 0.1, 0.5, and 1.5 mM EGS) and quenched with 50 mM glycine. Cross-linked samples were analyzed by 10% Tricine SDS–PAGE under reducing conditions.
Circular dichroism spectroscopic analysis
Circular dichroism (CD) spectra were performed on a Jasco J-715 spectrophotometer. The working buffer was PBS (10 mM sodium phosphate, pH 7.3; 150 mM NaCl). Wavelength spectra were recorded at 25 °C using a 0.1-cm path-length cuvette. Thermodynamic stability was measured at 222 nm by recording the CD signals in the temperature range of 25–90 °C with a scan rate of 5 °C/min.
Expression and preparation of selenomethionine derivative of the MHV 2-helix
The selenomethionine derivative of the MHV 2-helix was expressed using E. coli BL21 (DE3) cultured in minimal media M9 containing 60 mg/L l-SetMet. When the culture optical density (OD600) reached 0.6, the further six amino acids (lysine, threonine, phenylalanine, leucine, isoleucine, and valine) were added to the culture. The culture was then induced with 0.2 mM IPTG and grown for an additional 10 h at 16 °C. Purification of the selenomethionene-incorporated MHV 2-helix was performed as that used for the native MHV 2-helix.
Mass spectroscopy
The purified and desalted 2-helix proteins (both native and SeMet-incorporated 2-helix) were exchanged into the buffer of 20 mM Tris–HCl, pH 8.0, and then analyzed by using Bruker Daltonics Biflex III MALDI-TOF mass spectrometer for the molecular mass of the polypeptide.
Results and discussion
2-Helix polypeptide is expressed as a soluble protein
Based on the prediction of the LearnCoil-VMF program and the work by Bosch et al. [14], the HR1 and HR2 regions of MHV-A59 have been chosen in this study (Fig. 1). The single-chain construct of the fusion core HR1/HR2 was made as 2-helix as described under Materials and methods. The schematic representation of this construct is shown in Fig. 1. As expected from our previous work on paramyxovirus fusion core 2-helix proteins [15], [16], [17], the MHV 2-helix polypeptide preparation from the E. coli expression system in this study was expressed in high level and soluble in the physiological condition, i.e., in the buffers of either PBS or pH 8.0, Tris (Fig. 2 ). This implies that the protein has most likely been folded correctly. The protein has been shown to be soluble in the concentration of up to 40 mg/mL.
Fig. 2.
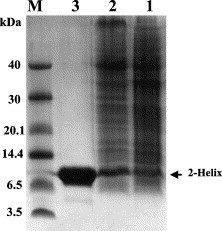
SDS–PAGE gel profile showing the 2-helix expression and purification. M, Molecular mass standards in kilo-dalton. Lane 1: non-induced. Lane 2: induced by IPTG. Lane 3: purified by affinity chromatography. Samples run on 10% Tricine SDS–PAGE.
2-Helix polypeptide forms a stable coiled-coil complex: the 6-helix bundle
The purified proteins were concentrated to about 10 mg/mL in PBS and analyzed by gel filtration and chemical cross-linking for estimation of the molecular mass. The MHV 2-helix protein was eluted between the corresponding positions of 52 and 14 kDa (Fig. 3 ). In comparison, the computed molecular mass of 2-helix (with 6× Histidine) is about 13 kDa, which indicates that the 2-helix might form oligomers (about 40 kDa). MALDI-TOFF results showed that the 2-helix monomer is about 12.6 kDa with two possible charges (Fig. 4 A). Subsequently, chemical cross-linking demonstrated that the 2-helix protein oligomer was, in fact, a trimer (Fig. 5 ) from the estimated molecular mass of the cross-linked products. In addition, the content of trimer increased with the concentration increase of the chemical cross-linker. Clear trimer band with monomer could also be seen if the samples (reduced by SDS loading buffer) were not heated at 100 °C (Lane 2 in Fig. 5A). All this indicates that the 2-helix protein forms a stable trimeric complex.
Fig. 3.
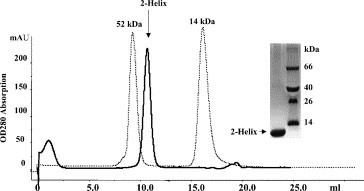
Gel-filtration profile of the 2-helix protein. The inset picture shows the existence of the pure 2-helix protein in the peak. The protein standards run on the same column and are shown by dashed lines.
Fig. 4.
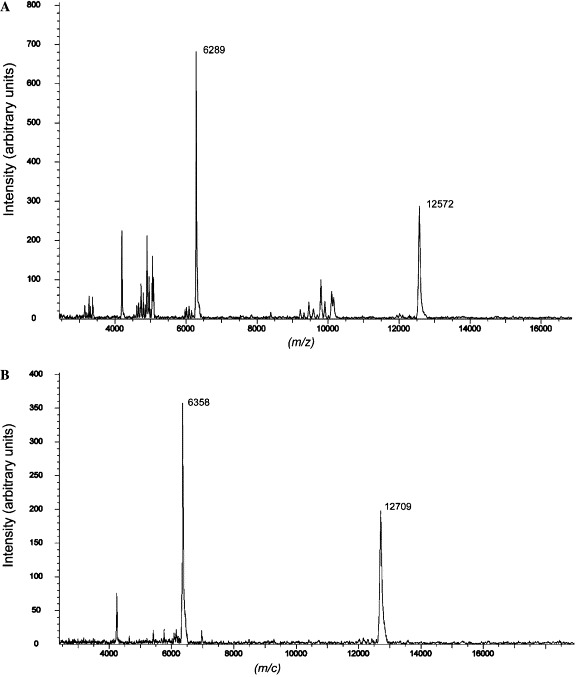
MALDI-TOF mass spectra of native and SeMet-labeled 2-helix proteins. The molecular mass of native 2-helix protein was 12,572 Da (A). The molecular mass of SeMet 2-helix protein was 12,709 Da (B). The molecular mass difference between (A) and (B) shows three Se molecules were incorporated. The two peaks in both cases indicate that the proteins carry two charges.
Fig. 5.
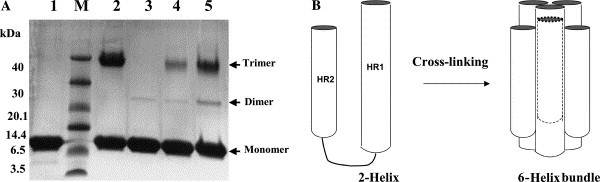
(A) Chemical cross-linking of the 2-helix protein. Cross-linking products were separated on 10% Tricine SDS–PAGE followed by Coomassie brilliant blue staining. Protein markers are shown in Lane M. Lane 1: MHV 2-helix boiled in 100 °C. Lane 2: MHV 2-helix not boiled in 100 °C. Lanes 3–5 indicate the increasing concentrations of the EGS used (0.1, 0.5, and 1.5 mM, respectively). (B) Schematic picture to show the possible cross-linking 6-helix bundle.
The secondary structure of the 2-helix protein was examined by CD spectroscopy as described under Materials and methods. The absorption curve showed that the 2-helix protein had double minima at 208 and 222 nm (Fig. 6 A), consistent with typical α-helix structure. The thermal stability test by CD spectroscopy indicated that trimer formation of the 2-helix was very stable in PBS (Fig. 6B), indicating that the 2-helix trimer represents the core structure of the post-fusion state of the coiled-coil bundle, as thoroughly studied for other viral fusion proteins [10], [11], [12], [19], [20], [21].
Fig. 6.
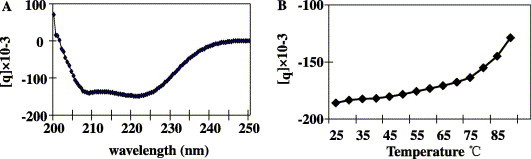
The CD spectra and thermal stability measurement of the MHV 2-helix protein in PBS. (A) Typical α-helix structure was seen with double minima at 208 and 222 nm. (B) The thermal stability was recorded at 222 nm with a Tm of 86 °C.
To facilitate the phase-determination of the 2-helix crystals, selenomethionine derivative preparations were made and the incorporation of the selenomethionine into the 2-helix preparation was confirmed by MALDI-TOFF mass spectrometry (Fig. 4B). Moreover, it seems that the start-codon ATG-encoded methionine remains in the expressed 2-helix protein as the molecular increase between the native and SeMet derivative indicates that three methionines were replaced by SeMet (molecular mass increases by 3 × (79 − 32) = 141 Da). The crystal structure has now been solved at 2.4 Å and has been reported elsewhere [22].
Conclusion
Our results do show that the strategy we used for other enveloped viruses, such as paramyxoviruses [15], [16], [17], to analyze the fusion core in the form of single chain HR1 and HR2, can be used for coronavirus. Coronavirus does most likely adapt to the similar fusion mechanism to other viruses with a class I fusion protein. A clear crystal structure of this fusion core from a member of coronavirus is now known and has answered some important questions [22].
Acknowledgments
This work was supported by Project 973 of the Ministry of Science and Technology of China (Grant No. 2003CB514116). GFG’s stay at the Tsinghua University was supported by Chunhui Project Scheme of the Ministry of Education, China.
References
- 1.Siddell S., Wege H., Ter Meulen V. The biology of coronaviruses. J. Gen. Virol. 1983;64(Pt 4):761–776. doi: 10.1099/0022-1317-64-4-761. [DOI] [PubMed] [Google Scholar]
- 2.Lee H.J., Shieh C.K., Gorbalenya A.E., Koonin E.V., La Monica N., Tuler J., Bagdzhadzhyan A., Lai M.M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180(2):567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spaan W., Cavanagh D., Horzinek M.C. Coronaviruses: structure and genome expression. J. Gen. Virol. 1988;69(Pt 12):2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- 4.S.G. Siddell, Plenum Press, New York, 1995, pp. 1–10
- 5.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 6.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 7.Gallagher T.M., Buchmeier M.J. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279(2):371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Groot R.J., Luytjes W., Horzinek M.C., van der Zeijst B.A., Spaan W.J., Lenstra J.A. Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J. Mol. Biol. 1987;196(4):963–966. doi: 10.1016/0022-2836(87)90422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers P., Pringle C.R., Easton A.J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J. Gen. Virol. 1990;71(Pt 12):3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- 10.Skehel J.J., Wiley D.C. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell. 1998;95(7):871–874. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- 11.Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 12.Eckert D.M., Kim P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 13.Gibbons D.L., Vaney M.C., Roussel A., Vigouroux A., Reilly B., Lepault J., Kielian M., Rey F.A. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature. 2004;427(6972):320–325. doi: 10.1038/nature02239. [DOI] [PubMed] [Google Scholar]
- 14.Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu J.Q., Zhang C.W., Rao Z., Tien P., Gao G.F. Biochemical and biophysical analysis of heptad repeat regions from the fusion protein of Menangle virus, a newly emergent paramyxovirus. Arch. Virol. 2003;148(7):1301–1316. doi: 10.1007/s00705-003-0105-x. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y., Lou Z., Liu Y., Cole D.K., Su N., Qin L., Li X., Bai Z., Rao Z., Gao G.F. Crystallization and preliminary crystallographic analysis of the fusion core from two new zoonotic paramyxoviruses, Nipah virus and Hendra virus. Acta. Crystallogr. D Biol. Crystallogr. 2004;60(Pt 6):1161–1164. doi: 10.1107/S0907444904009515. [DOI] [PubMed] [Google Scholar]
- 17.Zhu J., Zhang C.W., Qi Y., Tien P., Gao G.F. The fusion protein core of measles virus forms stable coiled-coil trimer. Biochem. Biophys. Res. Commun. 2002;299(5):897–902. doi: 10.1016/s0006-291x(02)02761-4. [DOI] [PubMed] [Google Scholar]
- 18.Singh M., Berger B., Kim P.S. LearnCoil-VMF: computational evidence for coiled-coil-like motifs in many viral membrane-fusion proteins. J. Mol. Biol. 1999;290(5):1031–1041. doi: 10.1006/jmbi.1999.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamb R.A., Joshi S.B., Dutch R.E. The paramyxovirus fusion protein forms an extremely stable core trimer: structural parallels to influenza virus haemagglutinin and HIV-1 gp41. Mol. Membr. Biol. 1999;16(1):11–19. doi: 10.1080/096876899294715. [DOI] [PubMed] [Google Scholar]
- 20.Weissenhorn W., Dessen A., Calder L.J., Harrison S.C., Skehel J.J., Wiley D.C. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 1999;16(1):3–9. doi: 10.1080/096876899294706. [DOI] [PubMed] [Google Scholar]
- 21.Bentz J. Membrane fusion mediated by coiled coils: a hypothesis. Biophys. J. 2000;78(2):886–900. doi: 10.1016/S0006-3495(00)76646-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y., Liu Y., Lou Z., Qin L., Li X., Bai Z., Pang H., Tien P., Gao G.F., Rao Z. Structural basis for coronavirus-mediated membrane fusion: Crystal structure of MHV spike protein fusion core. J. Biol. Chem. 2004;279:30514–30522. doi: 10.1074/jbc.M403760200. [DOI] [PMC free article] [PubMed] [Google Scholar]


