Highlights
-
•
NxTAG chemistry offers an easy workflow, low hands-on-time and scalable throughput.
-
•
All post-extraction assay steps take place in a single, closed tube.
-
•
Clinical performance of NxTAG RPP is comparable to that of the xTAG RVP assay.
-
•
Analytical performance is also similar or better than xTAG and xTAG FAST assays.
Keywords: Luminex, Multiplex, NxTAG, Respiratory pathogen, Bead array, IVD
Abstract
The Luminex® NxTAG® Respiratory Pathogen Panel (NxTAG RPP) is an IVD-cleared assay for the simultaneous detection and identification of nucleic acids from 18 respiratory viruses and 2 (or 3 outside of the U.S.) atypical bacterial pathogens in nasopharyngeal swabs. Its scalability allows concurrent testing of up to 96 samples in a single batch. Nucleic acid extracted from 200 µL of raw specimen using the easyMAG® extractor is added directly to pre-plated, lyophilized bead reagents (LBRs), where multiplexed RT-PCR and hybridization to MagPlex-TAG™ microspheres occurs within a sealed reaction well using a single cycling program. Data acquisition is done on the MAGPIX® instrument which reads and sorts the reaction products directly from the sealed well following transfer of the assay plate from the thermal cycler. NxTAG is the newest innovation in bead-based nucleic acid chemistry developed by Luminex. Here we provide the detailed assay protocol and present data which describe the clinical and analytical performance characteristics of NxTAG RPP.
1. Introduction
Respiratory tract infection (RTI) involves a wide variety of etiologic agents and presents with overlapping signs and symptoms; therefore, prediction of the causative organism based on clinical findings alone is not reliable [1]. Among the various diagnostic tests available, multiplex nucleic acid amplification assays are capable of detecting and identifying multiple respiratory pathogens and are now commonly used in clinical microbiology laboratories. The xTAG® Respiratory Viral Panel (RVP) from Luminex® was the first multiplexed molecular assay for respiratory pathogens cleared for in vitro diagnostic use and was capable of detecting 12–19 common respiratory viruses and subtypes, depending on the geographical region [2]. xTAG RVP incorporated multiplex Reverse Transcription PCR (RT-PCR) and multiplex Target Specific Primer Extension (TSPE) with Luminex’s xTAG technology, a proprietary universal sequence tag sorting system that allows easy development and optimization of nucleic acid assays on xMAP® microspheres. In this assay system, samples are spiked with an internal control and subjected to nucleic acid extraction. Nucleic acid extracts are subjected to multiplex RT-PCR and treated with Shrimp Alkaline Phosphatase and Exonuclease I (EXO) to inactivate remaining nucleotides and degrade leftover primers. Multiplex TSPE is then used to detect and label any viral nucleic acids that were amplified by PCR, incorporating the unique sequence tag during extension. At the time of its introduction in 2007, it was a major innovation that revolutionized the approach to diagnosis of respiratory infections. While much faster than traditional culture and immunological tests, the entire xTAG RVP procedure still takes approximately 6–8 h to complete. In addition, as an open platform system, there is a potential risk for cross-contamination after specimen extraction and PCR amplification steps [3].
Subsequently, Luminex developed the xTAG FAST assay chemistry which employed tagged primers and labeled primers together in a target-specific multiplex RT-PCR assay in order to streamline the workflow and shorten the time to result to approximately 4 h. The FAST version 1 assay incorporated the original non-magnetic xTAG microspheres whereas the version 2 assay is detected on the magnetic MagPlex-TAG microspheres. Following the nucleic acid extraction and multiplex RT-PCR reaction, an aliquot of the amplified and tagged targets are simultaneously hybridized to the xTAG or MagPlex-TAG microspheres and labeled with reporter dye for detection on either the Luminex™ 200 or MAGPIX® analyzer. This format eliminated the post-PCR clean-up and TSPE steps from the workflow entirely, decreased the time to result by about 2–4 h, and reduced the tube opening/transfer steps down to one. xTAG RVP FAST (and/or xTAG RVP FAST v2) was IVD-cleared for 7–17 targets, depending on the geographical region.
NxTAG® is the newest generation of universal tag sorting chemistry from Luminex. The NxTAG® Respiratory Pathogen Panel (RPP) was launched in December 2015. NxTAG RPP incorporates multiplex RT-PCR with the universal tag system in the form of lyophilized reagents to provide a closed system for amplification and detection of 20–21 respiratory viral and bacterial targets. All of the PCR and bead hybridization components, including tagged primers, fluorescently-labeled primers, enzyme, dNTPs, buffers, and MagPlex-TAG microspheres, are combined in precise mixtures and lyophilized into small beads or Lyophilized Bead Reagents (LBRs). LBRs are pre-plated into individual wells of a 96-well plate that allows scalable batch testing for 1 to 96 reactions. Extracted total nucleic acid is added to pre-plated LBRs sealed in individual reaction wells by piercing the pipette tip through a foil seal. The reaction is mixed by pipette to resuspend the reaction components and resealed with a fresh foil seal. The reaction is then amplified via RT-PCR and the reaction product undergoes near simultaneous bead hybridization within the single sealed reaction well. No open procedures are needed after nucleic acid extraction and amplification, thereby virtually eliminating the risk for carryover contamination [4]. The hybridized, tagged beads are then sorted and read on the MAGPIX® instrument, and the signals are analyzed using the NxTAG RPP Assay File for SYNCT™ Software, providing a qualitative call for each of the targets and internal control within each reaction well.
The targets included in NxTAG RPP are: influenza A, influenza A H1, influenza A H3, influenza B, respiratory syncytial virus A (RSV A), respiratory syncytial virus B (RSV B), coronavirus 229E (CoV 229E), coronavirus OC43 (CoV OC43), coronavirus NL63 (CoV NL63), coronavirus HKU1 (CoV HKU1), human metapneumovirus (hMPV), rhinovirus/enterovirus, adenovirus, parainfluenza virus 1 (PIV-1), parainfluenza virus 2 (PIV-2), parainfluenza virus 3 (PIV-3), parainfluenza virus 4 (PIV-4), human bocavirus (hBoV), Chlamydophila pneumoniae, Mycoplasma pneumoniae and, outside of the U.S., Legionella pneumophila. The workflow of the NxTAG RPP assay is depicted in Fig. 1 .
Fig. 1.
NxTAG RPP Workflow.
2. Material and methods
2.1. Materials provided
-
1.
NxTAG Respiratory Pathogen Panel (RPP) Plate (One 96-well plate containing 2 Lyophilized Bead Reagents per well; Store at 2–8 °C in the resealable pouch provided; avoid exposure to light and moisture.)
-
2.
MS2 bacteriophage internal control (1 × 1.5 mL vial; Store at −25 to 8 °C.)
-
3.
Foil Seals (8 pieces; Store at 2–30 °C. Store at 15–30 °C after first use.)
-
4.
NxTAG RPP Files (An assay file, data acquisition protocol, and package insert provided on a USB.)
-
5.
SYNCT™ Software (data analysis software.)
2.2. Materials required but not provided
2.2.1. Equipment
-
1.
Computer with Operating System Microsoft® Windows® 7 or higher, 64-bit, specifications as stated in the SYNCT™ Software User Manual.
-
2.
Luminex® MAGPIX® instrument, including xPONENT® Software, calibrators, verifiers, and controls.
-
3.
Multichannel pipette or single channel pipette (10–200 μL).
-
4.
Sonicator bath (Ultrasonic Cleaner, Cole-Parmer®, A-08849-00 or equivalent).
-
5.
PCR cooler rack (Eppendorf® 022510509 or equivalent).
-
6.
Pierceable TPE Capmat-96 for ABI 9700 thermal cycler systems or other thermal cyclers without adjustable lids (E&K Scientific, Cat. No. EK-64088).
-
7.
Nucleic acid extraction system (e.g. NucliSENS® easyMAG® System with Generic protocol 2.0.1).
-
8.
Thermal Cycler.
2.2.2. Consumables
-
1.
xMAP Drive Fluid (Cat # MPXDF-4PK-1)
-
2.
0.1 N NaOH
-
3.
DNase/RNase-Free Water
-
4.
NxTAG Probe Adjustment Strip (Cat # C000Z0452)
-
5.
Non-Skirted Plate (Cat # C000Z0453) (96-well in clear frame, for ABI 9700 thermal cycler system or other thermal cyclers that are not compatible with fully-skirted plate)
-
6.
Skirted Plate (Cat # C000Z0455) (96-well in white frame)
2.3. Specimen collection and nucleic acid extraction
NOTES:
-
•
Standard precautions should be taken with regard to sample collection, handling, and storage prior to extraction (refer to CLSI MM13-A and Farkas et al.) [5], [6].
-
•
Specimens and external controls should be extracted using the bioMérieux® NucliSENS® easyMAG® system.
-
•
Specimens can be stored at 2–8 °C for up to 7 days after collection in Universal Transport Media (UTM™) or equivalent. If the specimen is not going to be tested within 7 days of collection, it may be stored at ≤−70 °C for up to 12 months.
-
•
Follow your institutional or CLSI guidelines for appropriate run and QC controls.
-
1.
Spike 10 μL of MS2 (Internal Control) into 200 μL of the specimen sample.
-
2.Use the extraction method recommended for the NxTAG RPP assay on the bioMérieux® NucliSENS® easyMAG®:
-
a.Set the Protocol to Generic 2.0.1.
-
b.Set the Type for Primary.
-
c.Set Matrix for Other.
-
d.Set Elution volume for 110 μL.
-
e.Set the Sample volume to 0.200 mL.
-
a.
-
3.
Proceed with “Multiplex RT-PCR, Bead Hybridization, and Detection” or store extracted nucleic acid at ≤−70 °C for up to one month until testing with NxTAG RPP.
2.4. Multiplex RT-PCR, bead hybridization and detection
NOTE: Perform PCR setup in the pre-PCR area.
-
1.Program the following PCR protocol into a thermal cycler with a heated lid (105 °C), and pre-heat thermal cycler to 42 °C prior to plate setup:
-
a.1 cycle at 42 °C, 20 min
-
b.1 cycle at 95 °C, 2 min 30 s
-
c.15 cycles at 95 °C, 20 s; 65 °C, 60 s; 72 °C, 10 s
-
d.24 cycles at 95 °C, 20 s; 56 °C, 60 s; 72 °C, 10 s
-
e.1 cycle at 37 °C, 45 min
-
f.Hold at 37 °C
-
a.
NOTE: The total thermal cycling run time should be approximately 2 h 15 min.
-
2.
If frozen, thaw the extracted nucleic acid samples. Briefly vortex the samples followed by a quick spin to collect the samples to the bottom of the tube.
-
3.
Place samples on a chilled PCR cooler block or equivalent.
-
4.Remove the assay plate from its storage pouch. Place the required number of wells into the appropriate PCR set-up plate (e.g., skirted Plate for Eppendorf® and Bio-Rad® thermal cyclers and non-skirted Plate for ABI thermal cycler).
-
a.Firmly press down on the strips to snap into place, ensuring they are flush with the plate surface.
-
b.Return unused wells to the pouch, seal and store at recommended storage conditions.
-
a.
NOTE: Protect the assay plate from prolonged light exposure.
-
5.
Tap the plate on the benchtop to ensure the LBRs are at the bottom of the well.
-
6.
Place the plate on a chilled PCR cooler block or equivalent.
-
7.
Use the end-tabs to peel the clear release liner. Do not touch the black adhesive mask that surrounds each well. This adhesive is used to seal the plate following sample addition.
-
8.Dispense 35 μL of sample or control to each PCR well by using the pipette tip to pierce the foil at an angle:
-
a.Insert the tip a third to halfway down into the well.
-
b.Dispense the sample into the well and wait 1 to 2 s while maintaining the pipette tip inside the well.
-
c.Push the tips all the way to the bottom of the well and pipette up and down at least three times to reconstitute the LBRs.
-
a.
NOTE: When piercing the foil be sure to NOT touch the black adhesive.
-
9.
Reseal the plate after the sample addition using the precut strips of foil provided. Apply the foil(s) directly on top of the plate and press firmly on and around the wells to ensure a tight seal. Ensure the foil covers the wells and surrounding black adhesive. Do not vortex and spin down the plate.
-
10.
Place the foil-sealed plate in the thermal cycler and run the protocol as described in Step 1. Place a TPE Capmat-96 on top of the sealed plate if using a thermal cycler without an adjustable lid. Ensure the thermal cycler is pre-heated to 42 °C.
-
11.
While waiting for the cycling to complete, prepare the MAGPIX instrument. Refer to “Instrument Preparation for Data Acquisition”.
-
12.
After the cycling program is complete, do not remove the seal. Transfer the foil-sealed plate directly to the pre-heated MAGPIX. The plate should be read by the MAGPIX instrument immediately after the end of cycling.
2.5. Instrument preparation for data acquisition
NOTES:
-
•
Refer to the xPONENT® for MAGPIX Software User Manual for more information.
-
•
Refer to the “Initial Instrument Setup” section of the product insert the first time the instrument is used for NxTAG RPP.
-
1.
Open the xPONENT Software.
-
2.
Perform the probe height adjustment at least once a week to ensure optimal performance of this assay. Perform the Enhanced Startup Routine at least once a week. Adjust the probe height, as needed, using a skirted or non-skirted plate with the NxTAG Probe Adjustment Strip and one alignment sphere. For more information on adjusting the sample probe height, refer to the xPONENT for MAGPIX Software User Manual. The plate name MUST be saved as NxTAG RPP Assay Plate.
-
3.
Select ON under Plate Heater and enter 37 in the Set Temperature field to heat the MAGPIX heater plate to 37 °C. Click Apply.
-
4.
Navigate to the Batches page > Batches tab > click Create a New Batch from an Existing Protocol. Choose the NxTAG RPP T-A protocol for this batch, click Next. If a Run and Orders were created in the SYNCT Software, the batch should have the same name as the Run name created in the SYNCT Software.
-
5.
Select the appropriate wells where the samples will be analyzed and then click Unknown. The selected wells are highlighted.
-
6.
Import the sample list by clicking Import List or enter the appropriate Sample ID for each well. Dilution should be left on the default. If orders were created in the SYNCT Software, ensure that the Sample and Control IDs entered match the IDs used when creating the Order in SYNCT Software. SYNCT software also provides the option to export the Sample and Control IDs for import into xPONENT through the Import List function. The Sample ID name cannot be duplicated within a Run. Each sample MUST have a unique ID. If you are running replicates or running the same control sample more than once, please make sure you assign a unique Sample ID for each replicate.
-
7.
Click Save. The batch is now saved as a pending batch and ready to run.
-
8.
Navigate to Maintenance page > Cmds & Routines tab. Add appropriate reagents to the off-plate reagent reservoirs as specified by the Post-Batch Routine indicated in the software. Click Retract.
-
9.
Upon completion of the cycling, place the plate on the preheated MAGPIX heater block. Click Eject to place the plate on the heated block. If the probe height was adjusted with the skirted plate, ensure you put the reaction on the skirted plate before placing on the heater block. When placing the plate on the heater block, ensure that the numbers are on the left side and the letters are closest to you. Be sure to leave the foil seal in place. Click Retract to retract the holder.
-
10.
Navigate to the Batches page > Batches tab > choose the batch from the pending batches list and click Run to start data acquisition. Verify information in warning dialog boxes and click OK.
-
11.
After the last sample is read, navigate to Home > Probe and Heater > turn off the heater and click Eject to remove the plate from the heater block and turn OFF the heater.
-
12.
Carefully discard the test wells into a biohazard bag, sealing the bag to avoid aerosolization of the amplicons. If re-using the Skirted Plate (or Non-Skirted Plate), clean by soaking in a 10% bleach solution for 15 min. Rinse the Skirted Plate under running tap water to remove bleach, and air dry on paper towels or wipe with 70% alcohol for fast drying, if necessary.
2.6. Obtaining test results
-
1.
Once all the sample Orders in the Run have been reviewed and edited (if necessary), click Process Run. A dialog box displays, “Confirm all orders are correct before proceeding. You cannot change the sample type or expected control results after the Run has been analyzed. Do you want to continue?”
-
2.
Click Yes to continue, or No to cancel.
-
3.
Once the Run has completed processing, the Run is removed from the NxTAG Run view. The results of the Run can be found by clicking the Results icon from the System Navigation Menu and locating the processed Run from the list.
3. Results
3.1. Prospective evaluation study
The clinical performance of the NxTAG RPP Assay was evaluated in a multicenter study conducted at four clinical sites in North America. Nasopharyngeal swab specimens from pediatric or adult subjects who were hospitalized, admitted to a hospital emergency department, visited an outpatient clinic, or resided at a long-term care facility, and exhibited clinical signs and symptoms of respiratory tract infection were eligible for inclusion in the prospective study. A total of 2132 clinical specimens that met the study inclusion criteria were prospectively collected from pediatric and adult subjects suspected of having respiratory tract infection. Of these, 934 (43.8%) were collected from January to April 2014 and the remaining 1198 (56.2%) were enrolled between January and March 2015. Table 1 provides a summary of the general demographic information of the prospectively collected nasopharyngeal swabs that were included in the prospective data analysis. Among the 2132 specimens tested by comparator assays (xTAG RVP and bidirectional sequencing), 14.1% were infected with rhinovirus, 12.8% were positive for influenza A, 9.6% were positive for RSV A or B, 6.8% were positive for hMPV (N = 144) and 4.3% were positive for influenza B (N = 91). The prevalence of adenovirus, the coronaviruses (CoV 229E, CoV NL63, CoV 229E, CoV HKU1), parainfluenza viruses (PIV-1, PIV-2, PIV-2, PIV-3 and PIV-4) and human bocavirus (hBoV) in the study population ranged from 0.1% to 3.0%. The majority of hBoV (96.4%), adenovirus (90.0%), rhinovirus/enterovirus (80.0%), CoV NL63 (80.0%), RSV (72.8%), parainfluenza viruses (71.9%) and hMPV (71.5%) infections were reported in pediatric subjects while other respiratory viruses, including influenza, were generally evenly distributed between age groups. Most influenza A positive specimens were typed as influenza A H3 (75.4%). C. pneumoniae and M. pneumoniae represented <0.5% of the infections in the prospective study.
Table 1.
General demographic details for the prospective dataset.
| Site 1 | Site 2 | Site 3 | Site 4 | All Sites | |
|---|---|---|---|---|---|
| Patients tested | 675 | 375 | 498 | 584 | 2132 |
| Gender | |||||
| Male | 322 (47.7%) | 155 (41.3%) | 264 (53.0%) | 281 (48.1%) | 1022 (47.9%) |
| Female | 353 (52.3%) | 220 (58.7%) | 234 (47.0%) | 303 (51.9%) | 1110 (52.1%) |
| Age (yrs) | |||||
| 0–1 | 105 (15.6%) | 25 (6.7%) | 155 (31.1%) | 168 (28.8%) | 453 (21.2%) |
| >1–5 | 66 (9.8%) | 22 (5.9%) | 92 (18.5%) | 70 (12.0%) | 250 (11.7%) |
| >5–21 | 87 (12.9%) | 73 (19.5%) | 101 (20.3%) | 92 (15.8%) | 353 (16.6%) |
| >21–65 | 174 (25.8%) | 152 (40.5%) | 111 (22.3%) | 147 (25.2%) | 584 (27.4%) |
| >65 | 243 (36.0%) | 103 (27.5%) | 39 (7.8%) | 107 (18.3%) | 492 (23.1%) |
| Subject Status | |||||
| Outpatients | 309 (45.8%) | 157 (41.9%) | 49 (9.8%) | 39 (6.7%) | 554 (26.0%) |
| Hospitalized | 255 (37.8%) | 144 (38.4%) | 332 (66.7%) | 329 (56.3%) | 1060 (49.7%) |
| Emergency Department | 111 (16.4%) | 74 (19.7%) | 117 (23.5%) | 216 (37.0%) | 518 (24.3%) |
| Immune Status | |||||
| Immunocompromised | 116 (17.2%) | 89 (23.7%) | 0 (0.0%) | 59 (10.1%) | 264 (12.4%) |
| Immunocompetent | 506 (75.0%) | 285 (76.0%) | 0 (0.0%) | 525 (89.9%) | 1316 (61.7%) |
| Not Determined | 53 (7.9%) | 1 (0.3%) | 498 (100%) | 0 (0.0%) | 552 (25.9%) |
3.1.1. Prospective clinical performance
Of the 2132 prospective specimens (95.3%), 2031 generated valid NxTAG RPP results for all analytes on the first attempt. Invalid results were generated for one or more analytes in 101 specimens tested. Invalid results were primarily due to external control failure or non-specific signals in external controls (3.8% of invalids). Other causes for invalid results included low bead count (0.5%), internal control failure (0.3%), and inconclusive results due to abnormal signals (0.1%). The frequency of invalid results was evenly distributed among the clinical sites. All available residual specimens were re-run with NxTAG RPP and generated valid results upon re-test. Table 2 summarizes the performance characteristics of NxTAG RPP using xTAG RVP or bidirectional sequencing as comparator. Clinical sensitivities (before discrepant analysis) for all viral pathogens identified by NxTAG RPP, with the exception of PIV-2 and PIV-4 ranged from 92.9% to 100%. For PIV-2 and PIV-4, sensitivity values were derived from very few positive specimens and were 50% (1/2; 95% Confidence Interval [CI], 1.3% to 98.7%) and 60% (3/5; 95% CI, 14.7% to 94.7%), respectively. The combined sensitivity value for the bacterial pathogens (C. pneumoniae and M. pneumoniae) was 70.0% (7/10; 95% CI, 34.8% to 93.3%). Clinical specificity (before discrepant analysis) ranged from 96.3% to 99.9% for the viruses and 99.9% to 100% for the bacterial pathogens.
Table 2.
NxTAG RPP clinical performance before discrepant analysis (prospective sample Set).
| Target | Positive Agreement (Sensitivity) |
Negative Agreement (Specificity) |
# “No Call” by Comparator | ||||
|---|---|---|---|---|---|---|---|
| TP/TP + FN | % | 95% CI | TN/TN + FP | % | 95% CI | ||
| Influenza A (matrix) | 259/273 | 94.9 | 91.5–97.2 | 1822/1859 | 98.0 | 97.3–98.6 | 0 |
| Influenza A H1 | 21/21 | 100 | 83.9–100 | 2091/2111 | 99.1 | 98.5–99.4 | 0 |
| Influenza A H3 | 203/206 | 98.5 | 95.8–99.7 | 1872/1917 | 97.7 | 96.9–98.3 | 9 |
| Influenza B | 87/91 | 95.6 | 89.1–98.8 | 2019/2033 | 99.3 | 98.8–99.6 | 8 |
| Respiratory syncytial virus A | 73/73 | 100 | 95.1–100 | 2037/2052 | 99.3 | 98.8–99.6 | 7 |
| Respiratory syncytial virus B | 131/133 | 98.5 | 94.7–99.8 | 1978/1990 | 99.4 | 98.9–99.7 | 9 |
| Respiratory syncytial virus A & B (combined) | 204/206 | 99.0 | 96.5–99.7 | 4015/4042 | 99.3 | 99.0–99.5 | 16 |
| Coronavirus 229E | 21/21 | 100 | 83.9–100 | 2098/2111 | 99.4 | 98.9–99.7 | 0 |
| Coronavirus OC43 | 30/31 | 96.8 | 83.3–99.9 | 2092/2101 | 99.6 | 99.2–99.8 | 0 |
| Coronavirus NL63 | 62/65 | 95.4 | 87.1–99.0 | 2053/2065 | 99.4 | 99.0–99.7 | 2 |
| Coronavirus HKU1 | 13/14 | 92.9 | 66.1–99.8 | 2113/2118 | 99.8 | 99.4–99.9 | 0 |
| All coronaviruses (combined) | 126/131 | 96.2 | 91.4–98.4 | 8356/8395 | 99.5 | 99.4–9.7 | 2 |
| Human metapneumovirus | 135/144 | 93.8 | 88.5–97.1 | 1958/1976 | 99.1 | 98.6–99.5 | 12 |
| Enterovirus/Rhinovirus | 286/300 | 95.3 | 92.3–97.4 | 1764/1832 | 96.3 | 95.3–97.1 | 0 |
| Adenovirus | 20/20 | 100 | 83.2–100 | 2078/2112 | 98.4 | 97.8–98.9 | 0 |
| Parainfluenza 1 | 5/5 | 100 | 47.8–100 | 2115/2116 | 99.9 | 99.7–100 | 11 |
| Parainfluenza 2 | 1/2 | 50.0 | 1.3–98.7 | 2121/2122 | 99.9 | 99.7–100 | 8 |
| Parainfluenza 3 | 20/21 | 95.2 | 76.2–99.9 | 2086/2103 | 99.2 | 98.7–99.5 | 8 |
| Parainfluenza 4 | 3/5 | 60.0 | 14.7–94.7 | 2116/2127 | 99.5 | 99.1–99.7 | 0 |
| Human bocavirus | 27/28 | 96.4 | 81.7–99.9 | 2081/2104 | 98.9 | 98.4–99.3 | 0 |
| Chlamydophila pneumoniae | 0/1 | 0.0 | 0.0–97.5 | 2131/2131 | 100 | 99.8–100 | 0 |
| Mycoplasma pneumoniae | 7/9 | 77.8 | 40.0–97.2 | 2121/2123 | 99.9 | 99.7–100 | 0 |
3.1.2. Discrepant analysis for prospective specimens
There were 59 specimens identified as positive by reference method but negative by NxTAG RPP (i.e. RPP false negative). Of these, 35 (59.3%) were confirmed as negative (i.e. reference false positive) by either a FDA-cleared RT-PCR assay routinely used at the clinical sites (BioFire FilmArray RP or xTAG RVP) or by bidirectional sequencing using analytically validated primers that targeted genomic regions distinct from those targeted by NxTAG RPP. The 35 reference false positive specimens included: 14 influenza A, 2 H3 subtype of influenza A, 3 influenza B, 6 hMPV, 1 PIV-2, 1 PIV-3 and 8 rhinovirus/enterovirus. There were 356 specimens identified as negative by reference method but positive by NxTAG RPP (i.e. RPP false positive). Of these, 96 (27.0%) were confirmed as positive (i.e. reference false negative) by either the FDA-cleared RT-PCR assay routinely used at the clinical sites or discrepant sequencing analysis. These reference false negative specimens included: 12 influenza A, 11 H1 subtype of influenza A, 34 H3 subtype of influenza A, 4 influenza B, 8 RSV-A, 3 RSV-B, 2 hMPV, 2 PIV-3, 18 rhinovirus/enterovirus and 2 adenovirus. Site testing results were not available for the coronaviruses, hBoV, C. pneumoniae or M. pneumoniae. While it has not been shown definitively, it remains possible that some of the 260 unconfirmed RPP false positives may be due to increased sensitivity of RPP compared to the reference methods. When considering the total number of tests performed (i.e. 20 tests per reaction × 2132 reactions = 42,640 tests), the percentage of unconfirmed RPP false positives calls is only 0.6%. Table 3 shows the resolved performance values following discrepant analysis.
Table 3.
NxTAG RPP performance after discrepant analysis (prospective sample set).
| Target | Positive Agreement (Sensitivity) |
Negative Agreement (Specificity) |
# “No Call” by Comparator | ||||
|---|---|---|---|---|---|---|---|
| TP/TP + FN | % | 95% CI | TN/TN + FP | % | 95% CI | ||
| Influenza A (matrix) | 271/271 | 100 | 98.6–100 | 1836/1860 | 98.0 | 98.1–99.1 | 0 |
| Influenza A H1 | 21/21 | 100 | 83.9–100 | 2091/2111* | 99.1 | 98.5–99.4 | 0 |
| Influenza A H3 | 237/238 | 96.6 | 97.7–99.9 | 1874/1885 | 99.4 | 98.9–99.7 | 9 |
| Influenza B | 91/92 | 98.9 | 94.1–99.8 | 2022/2032 | 99.5 | 99.1–99.7 | 8 |
| Respiratory syncytial virus A | 81/81 | 100 | 95.1–100 | 2037/2044 | 99.7 | 99.3–99.8 | 7 |
| Respiratory syncytial virus B | 134/136 | 98.5 | 94.8–99.6 | 1978/1987 | 99.5 | 99.2–99.8 | 9 |
| Coronavirus 229E | 21/21 | 100 | 83.9–100 | 2098/2111 | 99.4 | 98.9–99.7 | 0 |
| Coronavirus OC43 | 30/31 | 96.8 | 83.3–99.9 | 2092/2101 | 99.6 | 99.2–99.8 | 0 |
| Coronavirus NL63 | 62/65 | 95.4 | 87.1–99.0 | 2053/2065 | 99.4 | 99.0–99.7 | 2 |
| Coronavirus HKU1 | 13/14 | 92.9 | 66.1–99.8 | 2113/2118 | 99.8 | 99.4–99.9 | 0 |
| Human metapneumovirus | 137/140 | 97.9 | 93.9–99.3 | 1964/1980 | 99.2 | 98.7–99.5 | 12 |
| Enterovirus/Rhinovirus | 304/310 | 98.1 | 95.8–99.1 | 1772/1822 | 97.3 | 96.4–97.9 | 0 |
| Adenovirus | 22/22 | 100 | 85.1–100 | 2078/2110 | 98.5 | 97.9–98.9 | 0 |
| Parainfluenza 1 | 5/5 | 100 | 47.8–100 | 2115/2116 | 99.9 | 99.7–100 | 11 |
| Parainfluenza 2 | 1/1 | 100 | 20.7–100 | 2122/2123 | 99.9 | 99.7–100 | 8 |
| Parainfluenza 3 | 22/22 | 100 | 85.1–100 | 2087/2102 | 99.3 | 98.8–99.6 | 8 |
| Parainfluenza 4 | 3/5 | 60.0 | 14.7–94.7 | 2116/2127 | 99.5 | 99.1–99.7 | 0 |
| Human bocavirus† | 27/28 | 96.4 | 81.7–99.9 | 2081/2104 | 98.9 | 98.4–99.3 | 0 |
| Chlamydophila pneumoniae† | 0/1 | 0.0 | 0.0–97.5 | 2131/2131 | 100 | 99.8–100 | 0 |
| Mycoplasma pneumoniae† | 7/9 | 77.8 | 40.0–97.2 | 2121/2123 | 99.9 | 99.7–100 | 0 |
11/20 NxTAG RPP false positive specimens were un-subtypeable by the comparator. xTAG RVP does not detect influenza A(H1N1)pdm09 whereas NxTAG RPP does.
Discrepant analysis not available.
3.1.3. Co-infections in prospective specimens
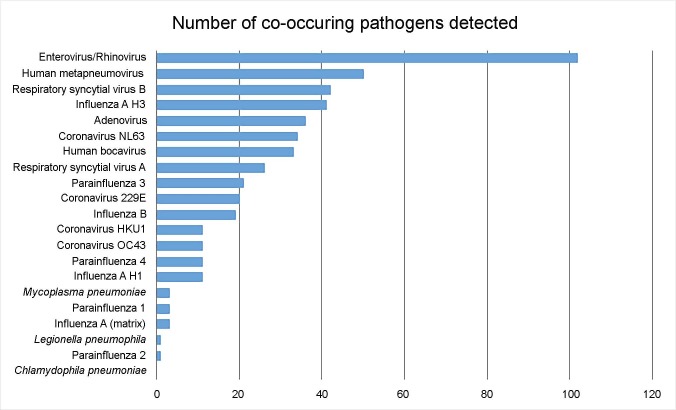
NxTAG RPP detected a total of 217 mixed infections in the prospective clinical evaluation. This represented 17.9% (217/1211) of NxTAG RPP positive specimens during the study period. 182 (83.9%) were dual infections, 23 (10.6%) were triple infections, 11 (5.1%) were positive for four targets and 1 (0.5%) was positive for five targets. All respiratory pathogens probed by NxTAG RPP (with the exception of C. pneumoniae) were implicated in co-infections. The most prevalent virus involved in mixed infections was enterovirus/rhinovirus (47%), followed by RSV (31.3%), influenza A H1 or H3 (24%), hMPV (23%), adenovirus (16.6%) and CoV-NL63 (15.7%) (Fig. 2 ).
Fig. 2.
Number of co-occurring pathogens in 217 mixed infections detected by NxTAG RPP (prospective sample set).
3.2. Retrospective and contrived evaluation study
The retrospective evaluation included a total of 326 unique, preselected, anonymized, remnant nasopharyngeal swab specimens from pediatric and adult subjects who were confirmed as infected with influenza A H1 subtype, adenovirus, parainfluenza, coronaviruses, C. pneumoniae, M. pneumoniae or enterovirus D68. NxTAG RPP accurately detected 323 out of 329 (98.2%) viral and bacterial pathogens present in the 326 clinical specimens tested (Table 4 ). Due to the limited number of samples positive for the atypical bacterial pathogens, contrived samples were prepared by spiking varying clinically-relevant concentrations of six strains of C. pneumoniae (1 × 102 to 2 × 106 copies/mL) or six strains of M. pneumoniae (1 × 102 to 2 × 106 copies/mL) into negative clinical specimens [7], [8], [9]. Fifty contrived specimens for each of the two atypical bacteria were prepared and tested along with 50 negative clinical specimens in a randomized, blinded fashion at 3 testing sites. NxTAG RPP was able to detect all concentrations of C. pneumoniae and M. pneumoniae in the contrived specimens (100%; 50/50; 95% CI, 92.9% to 100%).
Table 4.
Percent agreement of NxTAG RPP in the pre-selected sample set.
| Target | Positive Agreement |
95% CI | |
|---|---|---|---|
| TP/(TP + FN) | % | ||
| Adenovirus | 30/30 | 100 | 88.4–100 |
| Influenza A H1 | 35/35 | 100 | 90.0–100 |
| Parainfluenza 1 | 38/38 | 100 | 90.8–100 |
| Parainfluenza 2 | 33/33 | 100 | 89.4–100 |
| Parainfluenza 3 | 34/34 | 100 | 89.7–100 |
| Parainfluenza 4 | 41/42 | 97.6 | 87.4–99.6 |
| Coronavirus 229E | 17/17 | 100 | 80.5–100 |
| Coronavirus OC43 | 16/16 | 100 | 79.4–100 |
| Coronavirus NL63 | 15/15 | 100 | 78.2–100 |
| Coronavirus HKU1 | 44/49 | 89.8 | 77.8–96.6 |
| Enterovirus D68 | 14/14 | 100 | 76.8–100 |
| Chlamydophila pneumoniae | 2/2 | 100 | 15.8–100 |
| Mycoplasma pneumoniae | 4/4 | 100 | 39.8–100 |
| Total Agreement | 323/329 | 98.2 | 96.1–99.3 |
3.3. Analytical performance
3.3.1. Analytical limit of detection
The limit of detection (LoD) for each NxTAG RPP target was assessed by analyzing serial dilutions of simulated samples made from high-titer pathogen stocks from commercial suppliers, or clinical specimens when the target pathogen was not commercially available. All sample dilutions were prepared using Universal Transport Medium (UTM). The LoD titer for each target was defined as the lowest concentration at which ≥95% (≥19/20) of samples tested generated positive calls. The LoD concentrations varied between targets but the majority was in the range of 3.37 × 10−3 to 2.82 × 101 TCID50/mL [10], [11]. Three stocks samples were provided in alternate unit measurements and were determined to have the following LoDs: 3.91 × 102 copies/mL for hBoV, 1.57 × 104 copies/mL for CoV HKU1 and 1.42 × 102 CCU/mL (Color Changing Units) [12].
3.3.2. Analytical specificity
3.3.2.1. Reactivity and cross-reactivity
Potential cross-reactivity of the assay was assessed using pathogens that cause respiratory infections but are not probed by the assay, pathogens that may be found in respiratory specimens, as well as pathogens that the assay is designed to detect. Cross-reactivity was evaluated in simulated specimens prepared by spiking cultured organisms into UTM. Viral and bacterial targets were prepared at 1 × 105 TCID50/mL or 1 × 106 CFU/mL, respectively, or at the highest concentration possible based on the organism stock concentration. Three replicates of each pathogen were extracted and then analyzed with NxTAG RPP. A total of 107 pathogens were tested, of which 80 are not and 27 (21 targets plus additional strains of influenza A H1) are probed by the assay [10], [11]. None of the pathogens included in this study were found to cross-react, with the exception of three strains of non-pandemic influenza A H1 (A/Brisbane/59/07, A/Solomon Islands/3/2006 and A/Singapore/63/04) (data not shown). These strains cross-reacted with CoV 229E when the titer of the influenza A H1 was above 1 × 104 TCID50/mL. Based on both laboratory testing and in silico prediction, high titers of these three non-pandemic influenza A H1 strains may result in a false positive call for CoV 229E. Based on in silico analysis, there is also a potential that the presence of CoV 229E may cause a false positive influenza H1 call and the presence of PIV-2 may cause a false positive influenza H3 call. Nevertheless no false positive calls were observed with these targets in either the prospective or pre-selected sample sets in the clinical study.
Laboratory testing was supplemented with in silico data where prediction rules were used to predict both reactivity and cross-reactivity of specific strains. GenBank sequences for 77 avian, human and swine influenza A strains, 39 adenovirus serotypes, 3 human poliovirus strains and the SARS CoV, were aligned with all primer sequences in the NxTAG RPP. Reactivity and cross-reactivity was predicted based on thermodynamic analysis of mismatches between the primers and genomic sequences. With the exception of one influenza A H5N1 swine strain (A/swine/East Java/UT6010/2007(H5N1)), all strains analyzed are predicted to react with the correct primers but show no cross-reactivity with the other analyte primers in the assay. The H5N1 swine strain is not expected to react with the influenza A primers or any other primers in the assay.
3.3.2.2. Interference testing
The accuracy of NxTAG RPP was determined by testing the analytes in the presence of potential interfering substances and organisms that are not probed by the assay. Competitive interference due to pathogens that are probed by NxTAG RPP was also evaluated to assess the effects that clinically relevant co-infections may have on the assay. The NxTAG RPP assay was able to successfully detect all analytes at a concentration of three times the LoD (3x LoD) in the presence of 25 potential interfering substances and organisms (data not shown). Although FluMist did not interfere with the assay’s ability to identify other analytes, NxTAG RPP was able to recognize and make a positive call for the attenuated viruses present in the FluMist vaccine as expected (i.e., influenza A, influenza A(H1N1)pdm09, influenza A H3 and influenza B). In addition, no interference was seen when analytes that are a part of NxTAG RPP were evaluated for competitive interference with one pathogen present at a high titer (1.00 × 105 TCID50/mL), and a second pathogen at a low titer (3x LoD).
3.3.3. Repeatability and reproducibility
Repeatability was assessed for each NxTAG RPP target using multi-analyte samples comprised of two to four analytes prepared by spiking cultured organisms into UTM. Nasopharyngeal swabs collected from anonymous asymptomatic volunteers were used as negative samples. The spiked samples were assessed at two concentrations: low positive at the LoD (1x LoD) and moderate positive (3x LoD). Each sample set was tested in 20 replicates starting from sample extraction, and assessed in a single run by the same operator using the same set of instruments. For all strains tested, 95–100% of replicates (19 or 20 out of 20) were detected at both the low positive and moderate positive concentrations and the negative samples were negative for all targets.
A reproducibility study was performed to assess the total variability of the NxTAG RPP assay across operators, study sites, testing days and instruments. The assay was evaluated by two operators at each of three sites by testing a 17-member reproducibility panel in triplicate on 5 non-consecutive days, for a total of 30 batch runs (2 operators × 5 days × 3 sites). The reproducibility panel was comprised of one negative sample, 8 low positive multi-analyte samples (1x LoD) and 8 medium positive multi-analyte samples (3x LoD) prepared in UTM. For each member of the panel, a total of 90 data points (30 batch runs × 3 replicates per batch run) were generated. Agreement with the expected result ranged from 94.4 to 100% per analyte and was 99.5% overall.
3.3.4. Carryover and cross-contamination
To assess the potential for carryover and cross-contamination between reaction wells, a study was conducted using negative samples (UTM) alternating with replicates of a high titer purified nucleic acid sample in a checkerboard pattern in the NxTAG RPP plate. Two representative viral (PIV-3) and bacterial (M. pneumoniae) analytes were examined in separate runs. High titer purified viral nucleic acid samples (1.98 × 105 TCID50/mL for PIV-3 and 1.0 × 106 CCU/mL for M. pneumoniae) were prepared in UTM, in order to obtain positive calls 100.0% of the time and maximize the potential for cross-contamination. No carryover or cross-contamination was observed.
4. Discussion
NxTAG is the newest innovation in universal tag sorting chemistry from Luminex and is the basis of the NxTAG RPP assay. This technology retains the advantages of Luminex’s xTAG universal array technology while providing several enhancements over its predecessors. As with xTAG technology, NxTAG allows for scalable testing which makes it amenable to both low- and high-throughput applications, providing flexibility for routine daily testing of low specimen volumes but also the ability to handle unpredicted increases in sample volume that may occur. Key advancements with NxTAG technology include: a significantly reduced hands-on-time (∼7 min for 8 samples), reduced overall turnaround time (approximately 5 h per batch, including extraction), and most notably, the convenience of all RT-PCR and bead hybridization reagents lyophilized and pre-plated into individual wells. This allows all steps of the assay process (post-extraction) to be carried out in a single, closed tube to both maximize the ease-of-use and minimize the risk of cross-contamination. In addition, to accommodate large sample volumes, it would be possible for the user to automate or semi-automate the post-extraction steps through use of standard automated 96-well plate handlers and pipetting robots. Manual set-up, just using a 96-well plate format for the extracted samples and a multi-channel pipettor, allows an entire 96-well NxTAG plate to be set up in less than 10 min. The new assay chemistry also leverages new and updated primer designs for the assay targets, in addition to the workflow enhancements.
As presented here, the clinical performance is comparable to that of the original xTAG RVP assay, with sensitivities of >92.9% and specificities of ≥96.3% for most pathogens. Exceptions are noted for PIV-2 and PIV-4, and the bacterial pathogens, where sensitivity values were derived from very few positive specimens, and ranged from 50 to 70%. These two targets exhibited 100% and 97.6% positive agreement in the pre-selected sample set, respectively, where data were generated from 33 and 42 pre-selected positive specimens. A particular strength of a multiplex panel, such as NxTAG RPP, is the ability to detect coinfections. In this study, coinfections were seen in 17.9% of clinical samples. While many of the causative organisms are treated similarly, the ability to accurately detect the organisms present and the most likely pathogen is very important when confronted with several organisms in a sample which are of varying risk to the patient. Furthermore, ability to detect co-infections is most important for organisms that can be treated, such as influenza, RSV, adenovirus, or M. pneumoniae where infection control measures are applied and if the patient population is predominantly immunocompromised. Analytical performance is also similar to somewhat better than the preceding xTAG and xTAG FAST assays, with LoDs ranging from 3.37 × 10−3 to 2.82 × 101 TCID50/mL as compared to LoDs of 3 × 10−2 to 8 × 103 TCID50/mL for the previous assays [10], [13], [14], [15].
Previously published studies have reported the performance of NxTAG RPP as compared to other multiplex molecular respiratory panels. These studies demonstrate excellent performance of the NxTAG RPP assay and confirm a diagnostic accuracy that is comparable to other commercially available multiplex molecular panels on a variety of assay chemistries, as well as in-house developed tests. Several of these studies are summarized below.
Beckmann and Hirsch compared NxTAG RPP to the RespiFinder-22 (RP-22) multiplex ligation assay in parallel with 282 respiratory specimens [4]. Concordant results were obtained in 93.3% (2 6 3) of samples, with concordant positives in 167 (59.2%) and concordant negatives in 96 (34%). NxTAG RPP identified more co-infections (10.3% vs. 5.9%) and more viral pathogens overall, with most additional positive results involved in dual rhinovirus/RSV infections. Discordant samples were retested by quantitative PCR and showed that NxTAG RPP positive/RP-22 negative samples were mainly due to low pathogen levels, suggesting a higher sensitivity of NxTAG-RPP, and also when detecting multiple infections. Hands-on time after extraction was less for NxTAG RPP (0.25–0.5 vs. 2–4 h), as was total turnaround time (5–7 vs. 8–16 h).
Chen and coworkers evaluated the performance of NxTAG RPP in comparison to the FilmArray Respiratory Panel (FA-RP) or singleplex real-time PCR as reference [16]. NxTAG RPP demonstrated overall diagnostic sensitivity and specificity of 98.9 and 99.0%, respectively. Influenza A H7N9 was also detected by the influenza A virus matrix gene target while the influenza A subtyping gene targets in the panel remained negative. Complete concordance between NxTAG-RPP and FA-RP was observed in 98.8% (318/322) of positive samples with substantial agreement observed for most targets. The authors concluded that with the good diagnostic performance, high sample throughput and reasonable turnaround time, NxTAG RPP is a suitable multiplex platform for routine screening of respiratory specimens in hospital-based laboratories.
Similarly, Tang et al. evaluated NxTAG RPP using 404 clinical respiratory specimens previously tested by the FA-RP assay [17]. Clinical sensitivities and specificities ranged from 80.0 to 100.0% and 98.9–100.0%, respectively with 95.5% agreement on influenza A genotype. They suggest that the scalability of 96 reactions in a batch makes the assay a potential solution when high-throughput testing is needed during burdensome nosocomial outbreaks, influenza seasons, and pandemics.
Brotons et al. compared the diagnostic accuracy of NxTAG RPP to the Anyplex II RV16 assay with 319 prospectively collected nasopharyngeal aspirates [18]. A total of 268 of 319 (84.0%) specimens yielded concordant results. Positive percent agreement values ranged from 83.3 to 100%, while the negative percent agreement was >99% for all targets except for enterovirus/rhinovirus (94.4%). In addition, NxTAG RPP detected single bacterial and mixed viral-bacterial infections in seven samples.
Sails and colleagues evaluated NxTAG RPP in comparison to an in-house laboratory developed multiplex real-time PCR panel on 314 clinical samples [19]. The overall agreement between the assays was very high, as indicated by Kappa coefficients ranging from 0.85 for hMPV up to 0.96 for RSV A and 96.2% agreement for rhinovirus/enterovirus and 100% for influenza A, influenza B, PIV-4 and RSV B. The high sample throughput and low hands-on time make NxTAG RPP suitable for screening clinical samples for respiratory pathogens.
5. Conclusions
The NxTAG Respiratory Pathogen Panel is the newest generation of bead-based multiplexing for respiratory pathogen detection available from Luminex. Based on NxTAG chemistry, it provides several advantages over the previous xTAG universal array chemistry. All reaction components are provided lyophilized and pre-plated into individual, sealed reaction wells. Once the extracted nucleic acid sample is added and the well is resealed, there is no further hands-on manipulation. This workflow both maximizes ease-of-use and minimizes risk of cross-contamination. As described in this protocol, after set-up, the reactions are processed on a thermal cycler and then read in the MAGPIX analyzer. The prospective multi-center study and several published studies described herein demonstrate the high accuracy of NxTAG RPP as compared to its xTAG predecessors and other multiplex assays, combined with a simplified workflow, low hands-on-time and scalable throughput that can meet the needs of a variety of laboratory settings.
References
- 1.Caliendo A.M. Multiplex PCR and emerging technologies for the detection of respiratory pathogens. Clin. Infect. Dis. 2011;52(Suppl. 4):S326–S330. doi: 10.1093/cid/cir047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krunic N., Yager T.D., Himsworth D., Merante F., Yaghoubian S., Janeczko R. xTAG RVP assay: analytical and clinical performance. J. Clin. Virol. 2007;40(Suppl. 1):S39–S46. doi: 10.1016/S1386-6532(07)70009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rand K.H., Rampersaud H., Houck H.J. Comparison of two multiplex methods for detection of respiratory viruses: FilmArray RP and xTAG RVP. J. Clin. Microbiol. 2011;49:2449–2453. doi: 10.1128/JCM.02582-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beckmann C., Hirsch H.H. Comparing Luminex NxTAG-respiratory pathogen panel and RespiFinder-22 for multiplex detection of respiratory pathogens. J. Med. Virol. 2016;88:1319–1324. doi: 10.1002/jmv.24492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CLSI, Collection, Transport, Preparation, and Storage of Specimens for Molecular Methods, first ed., Approved Guideline. Wayne, PA, 2005.
- 6.Farkas D.H., Kaul K.L., Wiedbrauk D.L., Kiechle F.L. Specimen collection and storage for diagnostic molecular pathology investigation. Arch. Pathol. Lab. Med. 1996;120:591–596. [PubMed] [Google Scholar]
- 7.Senn L., Jaton K., Fitting J.W., Greub G. Does respiratory infection due to Chlamydia pneumoniae still exist? Clin. Infect. Dis. 2011;53:847–848. doi: 10.1093/cid/cir515. [DOI] [PubMed] [Google Scholar]
- 8.Nilsson A.C., Bjorkman P., Persson K. Polymerase chain reaction is superior to serology for the diagnosis of acute Mycoplasma pneumoniae infection and reveals a high rate of persistent infection. BMC Microbiol. 2008;8:93. doi: 10.1186/1471-2180-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyashita N., Saito A., Kohno S. Multiplex PCR for the simultaneous detection of Chlamydia pneumoniae, Mycoplasma pneumoniae and Legionella pneumophila in community-acquired pneumonia. Respir. Med. 2004;98:542–550. doi: 10.1016/j.rmed.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 10.Luminex Molecular Diagnostics Inc., NxTAG®, Respiratory Pathogen Panel Package Insert, Rev C, Toronto, ON, Canada, 2018.
- 11.FDA, 510(k) Substantial Equivalence Determination for the NxTAG® Respiratory Pathogen Panel, K152386, 2017.
- 12.Bernet C., Garret M., de Barbeyrac B., Bebear C., Bonnet J. Detection of Mycoplasma pneumoniae by using the polymerase chain reaction. J. Clin. Microbiol. 1989;27:2492–2496. doi: 10.1128/jcm.27.11.2492-2496.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luminex Molecular Diagnostics Inc., xTAG® Respiratory Viral Panel (RVP), Rev P, Toronto, ON, Canada, 2016.
- 14.Luminex Molecular Diagnostics Inc., xTAG® Respiratory Viral Panel Fast, Rev C, Toronto, ON, Canada, 2017.
- 15.Luminex Molecular Diagnostics Inc., xTAG® RVP FAST v2, Rev G, Toronto, ON, Canada, 2018.
- 16.Chen J.H.K., Lam H.Y., Yip C.C.Y. Clinical evaluation of the new high-throughput Luminex NxTAG respiratory pathogen panel assay for multiplex respiratory pathogen detection. J. Clin. Microbiol. 2016;54:1820–1825. doi: 10.1128/JCM.00517-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Y.W., Gonsalves S., Sun J.Y. Clinical evaluation of the Luminex NxTAG respiratory pathogen panel. J. Clin. Microbiol. 2016;54:1912–1914. doi: 10.1128/JCM.00482-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brotons P., Henares D., Latorre I., Cepillo A., Launes C., Munoz-Almagro C. Comparison of NxTAG respiratory pathogen panel and anyplex II RV16 tests for multiplex detection of respiratory pathogens in hospitalized children. J. Clin. Microbiol. 2016;54:2900–2904. doi: 10.1128/JCM.01243-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sails A.D., Eltringham G., Valappil M., Waugh S., Saunders D. Comparison of the Luminex NxTAG respiratory pathogen panel and a multiplex in-house real-time PCR panel for the detection of respiratory viruses in symptomatic patients. J. Med. Microbiol. 2017 doi: 10.1099/jmm.0.000562. [DOI] [PubMed] [Google Scholar]