Abstract
Ubiquitination is the vital system for controlling protein degradation and regulation of basic cellular processes. Deubiquitinases (DUBs) are emerging as an important regulator of several pathways related to cancer and other diseases. Their ability to detach ubiquitin from the target substrate and regulation of signaling makes it potential target to treat cancer and other fatal diseases. In the current review, we are trying to summarize deubiquitination, and their role in cancer and potential small molecules DUBs inhibitors which can be used as drugs for cancer treatment.
Keywords: Deubiquitinases, Ubiquitination, Cancer, Deubiquitination, DUBs, DUBs inhibitors
1. Introduction
Proteins are vital for the structure and function of the cells, and the regulation of protein synthesis is a prime aspect of cellular metabolism. About 30% of mammalian proteins are short lived, have very short half-life of less than 10 min and are rapidly degraded after translation (Schubert et al., 2000). Such a high level of protein degradation requires a dedicated system to regulate the selective unwanted protein degradation. Ubiquitin-proteasome system (UPS) has emerged as a key supervisor of protein function and stability. UPS has many vital roles in eukaryotic cellular processes including cell cycle progression, stress response, signal transduction, DNA repair, control of transcription factor activity and membrane trafficking (Coux et al., 1996, Hershko and Ciechanover, 1998, Ciechanover et al., 2000a, Ciechanover, 2006, Welchman et al., 2005). Ubiquitin plays an important role to degrade proteins through proteasome targeting as well as by direct sorting to the lysosome. Ubiquitin is a small eukaryotic polypeptide which marks unwanted or damaged proteins for degradation, and the proteasome, is a large molecule breaks down protein into smaller peptides, to be used in other anabolic processes (D'Arcy et al., 2015). More than 80% of proteins are degraded by UPS and that is why it has emerged as an important player in the regulation of various cellular processes (Rock et al., 1994). UPS plays a pivotal role in the pathogenesis of many human diseases like cancer and neurodegenerative disorders (Ciechanover et al., 2000b).
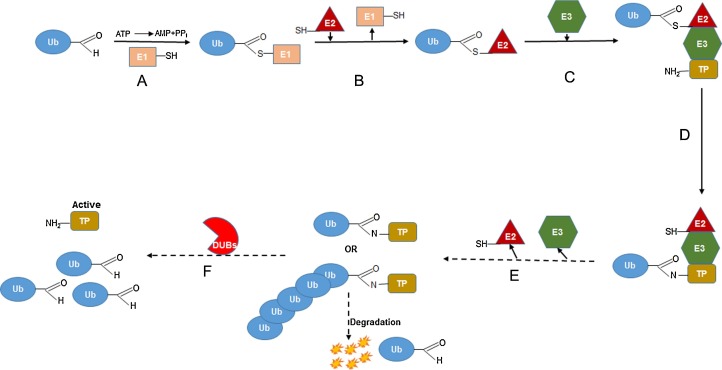
The process of ubiquitination is a multi-step process ultimately leading to the covalent modification of a protein substrate with small molecule ubiquitin. There are three types of ubiquitination: 1) mono-ubiquitination in which single ubiquitin is attached to target 2) multi ubiquitination or poly-mono-ubiquitination where several single ubiquitin are attached to target proteins 3) poly ubiquitination where substrate is attached with poly-ubiquitin chains (D'Arcy et al., 2015, Jentsch and Schlenker, 1995, Di Fiore et al., 2003, Lander et al., 2012). Ubiquitin has ≅76 conserved amino acid protein that covalently attached through a peptide bond between the carboxyl glycine residues at 76 position of ubiquitin to the amino groups of lysine residues in target proteins. The process of ubiquitination depends on the consecutive activity of three distinct enzymes, ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2) and ubiquitin-ligase or E3 ubiquitin ligase (E3) (fig. 1 ). In the first step, ubiquitin is activated by the E1 in the presence of ATP, forming a thio-ester bond between the carboxyl-terminal glycine residue of ubiquitin and the active site cysteine of the E1 enzyme. Once activated, ubiquitin is transferred from E1 to a cysteine residue of E2 ubiquitin carrier proteins. Substrate specificity is mediated by E3 ligases, which bind target substrates and co-ordinate the covalent attachment of ubiquitin. Two distinct families of E3 ligases exist, the HECT domain family that receives ubiquitin from the E2 ligase forming an ubiquitin-E3 intermediate, and the RING finger family of E3 ligases that form a molecular bridge between the E2 ligase and target proteins (D'Arcy et al., 2015, Ross et al., 2015, Voges et al., 1999, Pickart and Eddins, 2004). Once ubiquitin-E2, E3 and substrate complex is formed, ubiquitin binds to the substrate which is mediated by E3 ligase. In the subsequent process E2 and E3 are removed from complex and substrate protein will be either degraded or alternatively substrated could be free from ubiquitin through deubiquitinases and remain active (fig. 1).
Fig. 1.
Overview of ubiquitination and deubiquitination. TP; target protein, Ub; ubiquitin, E1, E2, and E3; different classes of ubiquitin, DUBs; deubiquinases.
2. Deubiquitination, deubiquitinases and cancer
Protein deubiquitination is reverse process of ubiquitination and performed by deubiquitinases or deubiquitinating enzymes (DUBs), which help in removal of ubiquitin from target proteins and involve in ubiquitin maturation, recycling and editing (Pfoh et al., 2015, Kim et al., 2003, Amerik and Hochstrasser, 2004, Nijman et al., 2005a, Reyes-Turcu and Wilkinson, 2009, Clague et al., 2013, Tyagi et al., 2015a). The function of deubiquitinases however, are rescuing the protein which is marked for degradation, and releasing ubiquitin from target proteins substrate. Dubs are playing an important role in cell growth, apoptosis and cancer (Pfoh et al., 2015, Hu et al., 2005, Avvakumov et al., 2006, Renatus et al., 2006) and are associated with 26S proteasome to rescue ubiquitin chain before the degradation of the substrate protein. These free polyubiquitin chains are processed by other DUBs to restore the ubiquitin in the cell (Pfoh et al., 2015). Approximately hundred human DUBs are discovered till date and these are divided into four classes; (1) ubiquitin specific proteases (USP), (2) ubiquitin C-terminal hydrolases (UCH), (3) ovarian tumor proteases (OTU), (4) Josephins and the Jab1/MPN/MOV34 metalloenzymes (JAMM). USPs are the major DUBs and are associated with most of the cancers and play an important role in regulation of various pathways, for example in Fanconi Anemia (FA). FA is a genomic instability syndrome characterized by bone marrow failure, developmental abnormalities and increased probability of cancers (Kee and D'Andrea, 2010). There are many reports, based on clinical investigation indicated that FA is a chromosomal instability disorder, and cells from FA patients accumulate DNA damage at an increased rate. Mono-ubiquitination of FANCD2 and FANCI is a vital event in FA pathway, downstream of this pathway, it interacts with FAND1, FANCN, FANCJ and BRCA1 (Knipscheer et al., 2009). USP1/UAF1 is known to deubiquitinases FANCD2 and FANCI. Disruption of USP1/UAF1 complex promote level of FANCD2/FANCI ubiquitination and DNA repair defects, suggesting a failure in the completion of FA pathway (Nijman et al., 2005b, Smogorzewska et al., 2007). ELG1 (Enhanced Levels of Genomic Instability) are recently found associated with USP1/USF1 and may play a role in the successful completion of the FA pathway (Lee et al., 2010, Yang et al., 2011, Shkedy et al., 2015).
Ubiquitin C-terminal hydrolase-L1 (UCTL1), is one of the most explored DUBs which is involved in neurodegenerative disorders such as Parkinson disease. It has been shown that expression of neurons of neuroendocrine system and gonads is unregulated in non-small cell lung cancer (Hibi et al., 1998, Hibi et al., 1999), oesophageal cancer (Takase et al., 2003) invasive colorectal cancer (Yamazaki et al., 2002) and pancreatic cancer (Tezel et al., 2000). Overexpression of the UCHL-1 has also been related with tumor progression, size, invasiveness and apoptosis in breast cancer (Wang et al., 2008).
Mechanism of action and potential role of different DUBs in the different cancers is well established (D'Arcy et al., 2015, Pfoh et al., 2015, Sacco et al., 2010) and shown in Table 1, Table 2 .
Table 1.
Deubiquitinases and their mechanism of action.
| DUBs | Pathway | DUBs | Pathway |
|---|---|---|---|
| USP1 | Fanconi anemia, DNA replication (Nijman et al., 2005b, Huang et al., 2006) | USP20 | Deubiquitinates and stabilizes HIF1-α (Li et al., 2005) |
| USP2 | Regulation through androgen synthesis and fatty acid synthatase (Graner et al., 2004) | USP21 | Deubiquitinates histone H2A, activating transcription (Nakagawa et al., 2008) |
| USP3 | Cell cycle regulation (Nicassio et al., 2007) | USP39, USP44 | Involve in mitotic spindle checkpoint (van Leuken et al., 2008, Stegmeier et al., 2007) |
| USP4 | Regulate WNT signaling athway (Zhao et al., 2009) | OTUB1 | Interacts with estrogen receptor alpha and negativily regulates ER-α-mediated transcription (Kee and D'Andrea, 2010) |
| USP5 | Cell cycle regulation (Dayal et al., 2009) | A20 | Negatively regulates NF-kB signaling (Sacco et al., 2010) |
| USP6 | Actin remodeling (Masuda-Robens et al., 2003) | Cezanne | Dowun-regulates NF-kB signaling (Enesa et al., 2008) |
| USP7 | Regulation of p53 (Sacco et al., 2010) | TRABID | Positive regulator of WNT signaling required for TCF-mediated transcription (Tran et al., 2008) |
| USP8 | Stabilization of ESCRT components (Huang et al., 2009, Schweitzer et al., 2007, Xu et al., 2009) | POH1 | Regulates ErbB2 ubiquitination (Liu et al., 2009) |
| USP9X | Regulation of AMPK kinase family | UCHL | Mechanism unknown |
| USP11 | Interact with BRCA2 and stabilizes IkB | UCHL5 | Regulates TGFβ signaling (Wicks et al., 2005) |
Table 2.
Deubiquitinases involve in different types of cancer.
| Cancer | Deubiquitinase involved |
|---|---|
| Lung | USP2, USP4, USP5, USP8, USP18, A20, USP33, CYLD, OTUB1 (Sacco et al., 2010, Komander et al., 2009, Dikic et al., 2009) |
| Prostate | USP1, USP33, BRCC36 (Sacco et al., 2010, Dikic et al., 2009) |
| Brain | USP1, USP2, USP3, USP4, USP5, USP6, USP8, A20, USP18, USP22, USP33, USP39, USP44, CYLD, AMSH-LP, MYSM, POH1, UCHL, UCHL5, OUTB1, TRABID, A20 (Renatus et al., 2006, Kee and D'Andrea, 2010, Dikic et al., 2009) |
| Leukemia | USP1, USP3, USP5, USP8, USP9X, USP20, USP21, USP33, USP44, CYLD, AMSH, POH1 (Renatus et al., 2006, Dikic et al., 2009) |
| Colon | USP2, USP5, USP7, USP9X, USP21, USP22, AMSH, BRCC36, POH1, UCHL1 (Renatus et al., 2006, Dikic et al., 2009) |
| Ovarian | USP1, USP5, USP18, USP39, CYLD, BAP1, UCHL1, UCH25, A20 (Renatus et al., 2006, Dikic et al., 2009) |
| Breast | USP7, USP15, USP33, POH1, UCHL5 (Sacco et al., 2010) |
| Pancreas | USP11, USP39 (Sacco et al., 2010) |
| Kidney | BAP1 (Dikic et al., 2009) |
| Gastric | USP1, USP44, UCHL1 (Sacco et al., 2010) |
| Fanconi Anemia | USP1, UAF1 (Dikic et al., 2009) |
| Liver | USP1, USP4, USP5, USP15, USP21, AMSH, AMSH-LP, CEZANNE, TRABD (Sacco et al., 2010) |
3. Therapeutic potential of DUBs for the treatment of cancer
Accumulation of genetic mutations and aberrant signaling of various growth and survival related pathways in cancer cells (Tyagi et al., 2015b, Tyagi et al., 2015c, Srivastava et al., 2015, Arora et al., 2015, Tyagi et al., 2014) leads to the clinical diversity and therapeutic resistance. However, advance understanding of the complex biology of cancer cells (Tyagi and Ghosh, 2011, Tyagi et al., 2011, Bhardwaj et al., 2014, Deshmukh et al., 2015) and the involvement of deubiquitinating enzymes in cancer (Schubert et al., 2000, Ciechanover, 2006, Pfoh et al., 2015) reveals the therapeutic potential of DUBs for the treatment of cancer. As mentioned in Table 2, DUBs are playing an important role in different type of cancers. More extensive studies are needed in this area of research to explore the detailed mechanism and target of DUBs. Study and designing the targeted small molecule DUB inhibitors, will most probably a new therapeutics for different types of cancers. DUBs have been identified in most of the cancer (Table 2) and targeting them can be an effective way to treat, diagnose and prevent the disease.
DUBs are involved in cell cycle regulation and DNA damage pathways and since, in cancer and different stress conditions all the proteins which regulate cell cycle and DNA damage/repair are either up/down regulated (Singh et al., 2013, Singh et al., 2011, Kumar and de Massy, 2010, Gupta et al., 2010), it would be interesting to investigate the potential role of DUBs in cell cycle and DNA damage/repair. In most of the cancers, tumor suppresser genes are degraded by ubiquitin which can be rescued by application of controlled mechanism of enhancing deubiquitinases in the tumor cells, to prevent degradation of tumor suppresser proteins. Development of small molecules which inhibits DUBs would be a great idea to target cancer cells and it will be facilitated by the development of suitable high throughput screening. Two molecules were identified that inhibit the SARS coronavirus DUBs papain like protease (PLpro) (Ratia et al., 2008, Ghosh et al., 2009). Some small molecules inhibit USP7 and USP8 (Daviet and Colland, 2008). Compound like G5 and F6 were reported as total DUBs inhibitor (Aleo et al., 2006) along with USP2, USP7 and SENP2 deSUMOylase (Aleo et al., 2006, Fontanini et al., 2009, Nicholson et al., 2008). b-AP15 was identified as an USP14 and UCHL5 inhibitor at a concentration of about 5 μM (D'Arcy et al., 2011). AM146, RA-14, RAMB1, RA-9 and WP1130 are other molecules which inhibit many DUBs including USPs and UCH and induce accumulation of polyubiquitinated proteins (Anchoori et al., 2011, Issaenko and Amerik, 2012, Kapuria et al., 2010). WP1130 in combination with bortezomid had showed antitumor activity in mantle cell lymphoma (Pham et al., 2010). Several other small molecules have been reported as DUBs inhibitors like Eeyarestatin (Fiebiger et al., 2004), Velcade (Bold, 2004) and Kyprolis (Steele, 2013).
4. DUBs in immunotherapy
Nowadays immunotherapy is widely used to treat many diseases including cancer. Immunotherapy is the treatment that uses person’s immune system to fight diseases and this can be either stimulate the immune system to work harder and cleverer or providing immune system machineries to escalation of immunity. DUBs widely involve in the regulation of cell signaling and these signaling pathways are frequently altered in most of the cancers.
P53 is a well known tumor repressor protein involves in cell cycle control and frequently mutated in tumor cells (Harris and Levine, 2005). Many of the USPs involve in P53 regulation. USP7 and USP15 involve in the control of P53-MDM2 pathway by regulating the stability of both P53 and MDM2 (Kon et al., 2010, Zou et al., 2014). USP2, USP4, USP5, USP10 and USP29 also involve in the regulation of P53 activity (D'Arcy et al., 2015). TNF-κB is another vital player in an immune response system which frequently deregulated and constitutively activated in cancer cells. Many DUBs like A20 and CYLD act as tumor repressor through their ability to downregulate TNF-κB signaling by acting on several components of the pathway (Harhaj and Dixit, 2012). USP21 and Cezanne inhibit TNF-κB activation by regulating ubiquitin level of RIPK1 (D'Arcy et al., 2015). Dr. Greenberg group from University of Pennsylvania showed that BRCC36 containing deubiquitinating complex BRISC which is also a sister protein complex of nuclear RAP80-BRCA1 complex, deubiquitinates type I interferon receptor (IFNAR1), resulting in its delayed lysosomal dependent degradation. They showed that BRISC deficient cells shows reduced inflammatory gene expression and BRISC deficient mice have an attenuated interferon response with a survival advantage from a LPS dependent septic shock (Zheng et al., 2013). All the above mentioned DUSs could be the potential candidate for immunotherapy.
5. Future perspectives
DUBs can be the potential target for treatment of many cancer or other fatal diseases but it needs to be investigated and explored. Small molecules inhibitors against DUBs could enhance the probability to treat various vital diseases. DUBs emerge as a promising target in the development of new disease specific treatment.
Conflict of interest
None.
Funding
None.
Acknowledgement
The authors tried to cover all relevant paper but afraid if some publications were not included.
References
- Aleo E., Henderson C.J., Fontanini A., Solazzo B., Brancolini C. Identification of new compounds that trigger apoptosome-independent caspase activation and apoptosis. Cancer Res. 2006;66:9235–9244. doi: 10.1158/0008-5472.CAN-06-0702. [DOI] [PubMed] [Google Scholar]
- Amerik A.Y., Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Anchoori R.K., Khan S.R., Sueblinvong T., Felthauser A., Iizuka Y., Gavioli R., Destro F., Isaksson V.R., Peng S., Roden R.B., Bazzaro M. Stressing the ubiquitin-proteasome system without 20S proteolytic inhibition selectively kills cervical cancer cells. PLoS. One. 2011;6:e23888. doi: 10.1371/journal.pone.0023888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S., Tyagi N., Bhardwaj A., Rusu L., Palanki R., Vig K., Singh S.R., Singh A.P., Palanki S., Miller M.E., Carter J.E., Singh S. Silver nanoparticles protect human keratinocytes against UVB radiation-induced DNA damage and apoptosis: potential for prevention of skin carcinogenesis. Nanomedicine. 2015;11:1265–1275. doi: 10.1016/j.nano.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvakumov G.V., Walker J.R., Xue S., Finerty P.J., Jr., Mackenzie F., Newman E.M., Dhe-Paganon S. Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8) J. Biol. Chem. 2006;281:38061–38070. doi: 10.1074/jbc.M606704200. [DOI] [PubMed] [Google Scholar]
- Bhardwaj A., Srivastava S.K., Singh S., Arora S., Tyagi N., Andrews J., McClellan S., Carter J.E., Singh A.P. CXCL12/CXCR4 signaling counteracts docetaxel-induced microtubule stabilization via p21-activated kinase 4-dependent activation of LIM domain kinase 1. Oncotarget. 2014;5:11490–11500. doi: 10.18632/oncotarget.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bold R. Development of the proteasome inhibitor velcade (Bortezomib) by julian adams Ph.D., and michael kauffman, M.D., Ph.D. Cancer Invest. 2004;22:328–329. doi: 10.1081/cnv-120030223. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Orian A., Schwartz A.L. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Orian A., Schwartz A.L. The ubiquitin-mediated proteolytic pathway: mode of action and clinical implications. J. Cell Biochem. 2000;34:40–51. doi: 10.1002/(sici)1097-4644(2000)77:34+<40::aid-jcb9>3.0.co;2-6. (40–51) [DOI] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin proteolytic system: from an idea to the patient bed. Proc. Am. Thorac. Soc. 2006;3:21–31. doi: 10.1513/pats.200510-106JH. [DOI] [PubMed] [Google Scholar]
- Clague M.J., Barsukov I., Coulson J.M., Liu H., Rigden D.J., Urbe S. Deubiquitylases from genes to organism. Physiol. Rev. 2013;93:1289–1315. doi: 10.1152/physrev.00002.2013. [DOI] [PubMed] [Google Scholar]
- Coux O., Tanaka K., Goldberg A.L. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. 801–847. [DOI] [PubMed] [Google Scholar]
- D'Arcy P., Brnjic S., Olofsson M.H., Fryknas M., Lindsten K., De C.M., Perego P., Sadeghi B., Hassan M., Larsson R., Linder S. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat. Med. 2011;17:1636–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- D'Arcy P., Wang X., Linder S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol. Ther. 2015;147:32–54. doi: 10.1016/j.pharmthera.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Daviet L., Colland F. Targeting ubiquitin specific proteases for drug discovery. Biochimie. 2008;90:270–283. doi: 10.1016/j.biochi.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Dayal S., Sparks A., Jacob J., Allende-Vega N., Lane D.P., Saville M.K. Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J. Biol. Chem. 2009;284:5030–5041. doi: 10.1074/jbc.M805871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh S.K., Srivastava S.K., Bhardwaj A., Singh A.P., Tyagi N., Marimuthu S., Dyess D.L., Dal Z.V., Carter J.E., Singh S. Resistin and interleukin-6 exhibit racially-disparate expression in breast cancer patients, display molecular association and promote growth and aggressiveness of tumor cells through STAT3 activation. Oncotarget. 2015;6:11231–11241. doi: 10.18632/oncotarget.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore P.P., Polo S., Hofmann K. When ubiquitin meets ubiquitin receptors: a signalling connection. Nat. Rev. Mol. Cell Biol. 2003;4:491–497. doi: 10.1038/nrm1124. [DOI] [PubMed] [Google Scholar]
- Dikic I., Wakatsuki S., Walters K.J. Ubiquitin-binding domains − from structures to functions. Nat. Rev. Mol. Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enesa K., Zakkar M., Chaudhury H., Luong l.A., Rawlinson L., Mason J.C., Haskard D.O., Dean J.L., Evans P.C. NF-kappaB suppression by the deubiquitinating enzyme Cezanne: a novel negative feedback loop in pro-inflammatory signaling. J. Biol. Chem. 2008;283:7036–7045. doi: 10.1074/jbc.M708690200. [DOI] [PubMed] [Google Scholar]
- Fiebiger E., Hirsch C., Vyas J.M., Gordon E., Ploegh H.L., Tortorella D. Dissection of the dislocation pathway for type I membrane proteins with a new small molecule inhibitor eeyarestatin. Mol. Biol. Cell. 2004;15:1635–1646. doi: 10.1091/mbc.E03-07-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanini A., Foti C., Potu H., Crivellato E., Maestro R., Bernardi P., Demarchi F., Brancolini C. The isopeptidase inhibitor G5 triggers a caspase-independent necrotic death in cells resistant to apoptosis: a COMPARATIVE STUDY WITH THE PROTEASOME INHIBITOR BORTEZOMIB. J. Biol. Chem. 2009;284:8369–8381. doi: 10.1074/jbc.M806113200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A.K., Takayama J., Aubin Y., Ratia K., Chaudhuri R., Baez Y., Sleeman K., Coughlin M., Nichols D.B., Mulhearn D.C., Prabhakar B.S., Baker S.C., Johnson M.E., Mesecar A.D. Structure-based design synthesis, and biological evaluation of a series of novel and reversible inhibitors for the severe acute respiratory syndrome-coronavirus papain-like protease. J. Med. Chem. 2009;52:5228–5240. doi: 10.1021/jm900611t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graner E., Tang D., Rossi S., Baron A., Migita T., Weinstein L.J., Lechpammer M., Huesken D., Zimmermann J., Signoretti S., Loda M. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell. 2004;5:253–261. doi: 10.1016/s1535-6108(04)00055-8. [DOI] [PubMed] [Google Scholar]
- Gupta V., Kalaiarasan P., Faheem M., Singh N., Iqbal M.A., Bamezai R.N. Dominant negative mutations affect oligomerization of human pyruvate kinase M2 isozyme and promote cellular growth and polyploidy. J. Biol. Chem. 2010;285:16864–16873. doi: 10.1074/jbc.M109.065029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harhaj E.W., Dixit V.M. Regulation of NF-kappaB deubiquitinases. Immunol. Rev. 2012;246:107–124. doi: 10.1111/j.1600-065X.2012.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S.L., Levine A.J. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. 425–479. [DOI] [PubMed] [Google Scholar]
- Hibi K., Liu Q., Beaudry G.A., Madden S.L., Westra W.H., Wehage S.L., Yang S.C., Heitmiller R.F., Bertelsen A.H., Sidransky D., Jen J. Serial analysis of gene expression in non-small cell lung cancer. Cancer Res. 1998;58:5690–5694. [PubMed] [Google Scholar]
- Hibi K., Westra W.H., Borges M., Goodman S., Sidransky D., Jen J. PGP9.5 as a candidate tumor marker for non-small-cell lung cancer. Am. J. Pathol. 1999;155:711–715. doi: 10.1016/S0002-9440(10)65169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Li P., Song L., Jeffrey P.D., Chenova T.A., Wilkinson K.D., Cohen R.E., Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T.T., Nijman S.M., Mirchandani K.D., Galardy P.J., Cohn M.A., Haas W., Gygi S.P., Ploegh H.L., Bernards R. and A.D. D'Andrea: regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- Huang X., Langelotz C., Hetfeld-Pechoc B.K., Schwenk W., Dubiel W. The COP9 signalosome mediates beta-catenin degradation by deneddylation and blocks adenomatous polyposis coli destruction via USP15. J. Mol. Biol. 2009;391:691–702. doi: 10.1016/j.jmb.2009.06.066. [DOI] [PubMed] [Google Scholar]
- Issaenko O.A., Amerik A.Y. Chalcone-based small-molecule inhibitors attenuate malignant phenotype via targeting deubiquitinating enzymes. Cell Cycle. 2012;11:1804–1817. doi: 10.4161/cc.20174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S., Schlenker S. Selective protein degradation: a journey's end within the proteasome. Cell. 1995;82:881–884. doi: 10.1016/0092-8674(95)90021-7. [DOI] [PubMed] [Google Scholar]
- Kapuria V., Peterson L.F., Fang D., Bornmann W.G., Talpaz M., Donato N.J. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010;70:9265–9276. doi: 10.1158/0008-5472.CAN-10-1530. [DOI] [PubMed] [Google Scholar]
- Kee Y., D'Andrea A.D. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010;24:1680–1694. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Park K.C., Chung S.S., Bang O., Chung C.H. Deubiquitinating enzymes as cellular regulators. J. Biochem. 2003;134:9–18. doi: 10.1093/jb/mvg107. [DOI] [PubMed] [Google Scholar]
- Knipscheer P., Raschle M., Smogorzewska A., Enoiu M., Ho T.V., Scharer O.D., Elledge S.J., Walter J.C. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Clague M.J., Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Kon N., Kobayashi Y., Li M., Brooks C.L., Ludwig T., Gu W. Inactivation of HAUSP invivo modulates p53 function. Oncogene. 2010;29:1270–1279. doi: 10.1038/onc.2009.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., de Massy Bernard. Initiation of meiotic recombination in mammals. Genes. 2010;1:521–549. doi: 10.3390/genes1030521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander G.C., Estrin E., Matyskiela M.E., Bashore C., Nogales E., Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.Y., Yang K., Cohn M.A., Sikdar N., D'Andrea A.D., Myung K. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through Its interactions with PCNA and USP1. J. Biol. Chem. 2010;285:10362–10369. doi: 10.1074/jbc.M109.092544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wang D., Messing E.M., Wu G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1alpha. EMBO Rep. 2005;6:373–378. doi: 10.1038/sj.embor.7400377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Buus R., Clague M.J., Urbe S. Regulation of ErbB2 receptor status by the proteasomal DUB POH1. PLoS One. 2009;4:e5544. doi: 10.1371/journal.pone.0005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda-Robens J.M., Kutney S.N., Qi H., Chou M.M. The TRE17 oncogene encodes a component of a novel effector pathway for Rho GTPases Cdc42 and Rac1 and stimulates actin remodeling. Mol. Cell Biol. 2003;23:2151–2161. doi: 10.1128/MCB.23.6.2151-2161.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kajitani T., Togo S., Masuko N., Ohdan H., Hishikawa Y., Koji T., Matsuyama T., Ikura T., Muramatsu M., Ito T. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008;22:37–49. doi: 10.1101/gad.1609708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicassio F., Corrado N., Vissers J.H., Areces L.B., Bergink S., Marteijn J.A., Geverts B., Houtsmuller A.B., Vermeulen W., Di Fiore P.P., Citterio E. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr. Biol. 2007;17:1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- Nicholson B., Leach C.A., Goldenberg S.J., Francis D.M., Kodrasov M.P., Tian X., Shanks J., Sterner D.E., Bernal A., Mattern M.R., Wilkinson K.D., Butt T.R. Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein Sci. 2008;17:1035–1043. doi: 10.1110/ps.083450408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman S.M., Luna-Vargas M.P., Velds A., Brummelkamp T.R., Dirac A.M., Sixma T.K., Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Nijman S.M., Huang T.T., Dirac A.M., Brummelkamp T.R., Kerkhoven R.M., D'Andrea A.D., Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Pfoh R., Lacdao I.K., Saridakis V. Deubiquitinases and the new therapeutic opportunities offered to cancer. Endocr. Relat. Cancer. 2015;22:T35–T54. doi: 10.1530/ERC-14-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham L.V., Tamayo A.T., Li C., Bornmann W., Priebe W., Ford R.J. Degrasyn potentiates the antitumor effects of bortezomib in mantle cell lymphoma cells in vitro and in vivo: therapeutic implications. Mol. Cancer Ther. 2010;9:2026–2036. doi: 10.1158/1535-7163.MCT-10-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M., Eddins M.J. Ubiquitin: structures functions, mechanisms. Biochim. Biophys. Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliji S., Chaudhuri R., Fu W., Prabhakar B.S., Johnson M.E., Baker S.C., Ghosh A.K., Mesecar A.D. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renatus M., Parrado S.G., D'Arcy A., Eidhoff U., Gerhartz B., Hassiepen U., Pierrat B., Riedl R., Vinzenz D., Worpenberg S., Kroemer M. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14:1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu F.E., Wilkinson K.D. Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chem. Rev. 2009;109:1495–1508. doi: 10.1021/cr800470j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock K.L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A.L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Ross J.M., Olson L., Coppotelli G. Mitochondrial and ubiquitin proteasome system dysfunction in ageing and disease: two sides of the same coin? Int. J. Mol. Sci. 2015;1716(8):19458–19476. doi: 10.3390/ijms160819458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco J.J., Coulson J.M., Clague M.J., Urbe S. Emerging roles of deubiquitinases in cancer-associated pathways. IUBMB Life. 2010;62:140–157. doi: 10.1002/iub.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U., Anton L.C., Gibbs J., Norbury C.C., Yewdell J.W., Bennink J.R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Schweitzer K., Bozko P.M., Dubiel W., Naumann M. CSN controls NF-kappaB by deubiquitinylation of IkappaBalpha. EMBO J. 2007;26:1532–1541. doi: 10.1038/sj.emboj.7601600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkedy D., Singh N., Shemesh K., Amir A., Geiger T., Liefshitz B., Harari Y., Kupiec M. Regulation of Elg1 activity by phosphorylation. Cell Cycle. 2015;0 doi: 10.1080/15384101.2015.1068475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Bhattacharya S., Paul J. Entamoeba invadens: dynamics of DNA synthesis during differentiation from trophozoite to cyst. Exp. Parasitol. 2011;127:329–333. doi: 10.1016/j.exppara.2010.08.013. [DOI] [PubMed] [Google Scholar]
- Singh N., Bhattacharya A., Bhattacharya S. Homologous recombination occurs in Entamoeba and is enhanced during growth stress and stage conversion. PLoS One. 2013;8:e74465. doi: 10.1371/journal.pone.0074465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E.R., III, Hurov K.E., Luo J., Ballif S.P., Hofmann K., D'Andrea A.D., Elledge S.J. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S.K., Bhardwaj A., Arora S., Tyagi N., Singh S., Andrews J., McClellan S., Wang B., Singh A.P. MicroRNA-345 induces apoptosis in pancreatic cancer cells through potentiation of caspase-dependent and −independent pathways. Br. J. Cancer. 2015;113:660–668. doi: 10.1038/bjc.2015.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele J.M. Carfilzomib: a new proteasome inhibitor for relapsed or refractory multiple myeloma. J. Oncol. Pharm. Pract. 2013;19:348–354. doi: 10.1177/1078155212470388. [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Rape M., Draviam V.M., Nalepa G., Sowa M.E., Ang X.L., McDonald E.R., III, Li M.Z., Hannon G.J., Sorger P.K., Kirschner M.W., Harper J.W., Elledge S.J. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- Takase T., Hibi K., Yamazaki T., Nakayama H., Taguchi M., Kasai Y., Ito K., Akiyama S., Nagasaka T., Nakao A. PGP9.5 overexpression in esophageal squamous cell carcinoma. Hepatogastroenterology. 2003;50:1278–1280. [PubMed] [Google Scholar]
- Tezel K., Nagasaka T., Nakao A. PGP9.5 as a prognostic factor in pancreatic cancer. Clin. Cancer Res. 2000;6:4764–4767. [PubMed] [Google Scholar]
- Tran H., Hamada F., Schwarz-Romond T., Bienz M. Trabid: a new positive regulator of Wnt-induced transcription with preference for binding and cleaving K63-linked ubiquitin chains. Genes Dev. 2008;22:528–542. doi: 10.1101/gad.463208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi N., Ghosh P.C. Folate receptor mediated targeted delivery of ricin entrapped into sterically stabilized liposomes to human epidermoid carcinoma (KB) cells: effect of monensin intercalated into folate-tagged liposomes. Eur. J. Pharm. Sci. 2011;43:343–353. doi: 10.1016/j.ejps.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Tyagi N., Rathore S.S., Ghosh P.C. Enhanced killing of human epidermoid carcinoma (KB) cells by treatment with ricin encapsulated into sterically stabilized liposomes in combination with monensin. Drug Deliv. 2011;18:394–404. doi: 10.3109/10717544.2011.567309. [DOI] [PubMed] [Google Scholar]
- Tyagi N., Bhardwaj A., Singh A.P., McClellan S., Carter J.E., Singh S. p-21 activated kinase 4 promotes proliferation and survival of pancreatic cancer cells through AKT- and ERK-dependent activation of NF-kappaB pathway. Oncotarget. 2014;5:8778–8789. doi: 10.18632/oncotarget.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi N., Tyagi M., Pachauri M., Ghosh P.C. Potential therapeutic applications of plant toxin-ricin in cancer: challenges and advances. Tumour. Biol. 2015 doi: 10.1007/s13277-015-4028-4. [DOI] [PubMed] [Google Scholar]
- Tyagi N., Marimuthu S., Bhardwaj A., Deshmukh S.K., Srivastava S.K., Singh A.P., McClellan S., Carter J.E., Singh S. p-21 activated kinase 4 (PAK4) maintains stem cell-like phenotypes in pancreatic cancer cells through activation of STAT3 signaling. Cancer Lett. 2015;10 doi: 10.1016/j.canlet.2015.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi A., Bhardwaj S.K., Srivastava S., Arora S., Marimuthu S.K., Deshmukh A.P., Carter J.E., Singh S. Development and characterization of a novel in vitro progression model for UVB-Induced skin carcinogenesis. Sci. Rep. 2015;5:13894. doi: 10.1038/srep13894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges D., Zwickl P., Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. 1015–68. [DOI] [PubMed] [Google Scholar]
- Wang W.J., Li Q.Q., Xu J.D., Cao X.X., Li H.X., Tang F., Chen Q., Yang J.M., Xu Z.D., Liu X.P. Over-expression of ubiquitin carboxy terminal hydrolase-L1 induces apoptosis in breast cancer cells. Int. J. Oncol. 2008;33:1037–1045. doi: 10.3892/ijo_00000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchman R.L., Gordon C., Mayer R.J. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- Wicks S.J., Haros K., Maillard M., Song L., Cohen R.E., Dijke P.T., Chantry A. The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF-beta signalling. Oncogene. 2005;24:8080–8084. doi: 10.1038/sj.onc.1208944. [DOI] [PubMed] [Google Scholar]
- Xu M., Takanashi M., Oikawa K., Tanaka M., Nishi H., Isaka K., Kudo M., Kuroda M. USP15 plays an essential role for caspase-3 activation during Paclitaxel-induced apoptosis. Biochem. Biophys. Res. Commun. 2009;388:366–371. doi: 10.1016/j.bbrc.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Yamazaki T., Hibi K., Takase T., Tezel E., Nakayama H., Kasai Y., Ito K., Akiyama S., Nagasaka T., Nakao A. PGP9.5 as a marker for invasive colorectal cancer. Clin. Cancer Res. 2002;8:192–195. [PubMed] [Google Scholar]
- Yang K., Moldovan G.L., Vinciguerra P., Murai J., Takeda S., D'Andrea A.D. Regulation of the Fanconi anemia pathway by a SUMO-like delivery network. Genes Dev. 2011;25:1847–1858. doi: 10.1101/gad.17020911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Schlesiger C., Masucci M.G., Lindsten K. The ubiquitin specific protease 4 (USP4) is a new player in the Wnt signalling pathway. J. Cell Mol. Med. 2009;13:1886–1895. doi: 10.1111/j.1582-4934.2008.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q., Jin J., Hu H., Li H.S., Romano S., Xiao Y., Nakaya M., Zhou X., Cheng X., Yang P., Lozano G., Zhu C., Watowich S.S., Ullrich S.E., Sun S.C. USP15 stabilizes MDM2 to mediate cancer cell survival and inhibit antitumor T cell responses. Nat. Immunol. 2014;15(6):562–570. doi: 10.1038/ni.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leuken R.J., Luna-Vargas M.P., Sixma T.K., Wolthuis R.M., Medema R.H. Usp39 is essential for mitotic spindle checkpoint integrity and controls mRNA-levels of aurora B. Cell Cycle. 2008;7:2710–2719. doi: 10.4161/cc.7.17.6553. [DOI] [PubMed] [Google Scholar]
- Zheng H., Gupta V., Patterson-Fortin J., Bhattacharya S., Katlinski K., Wu J., Varghese B., Carbone C.J., Aressy B., Fuchs S.Y., Greenberg R.A. A BRISC-SHMT complex deubiquitinates IFNAR1 and regulates interferon responses. Cell Rep. 2013;5:180–193. doi: 10.1016/j.celrep.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]