Abstract
Infectious agents associated with community-acquired pneumonia (CAP) are under-studied. This study attempted to identify viruses from the upper respiratory tract in adults visiting emergency departments for clinically suspected CAP. Adults with suspected CAP enrolled in the ESCAPED study (impact of computed tomography on CAP diagnosis) had prospective nasopharyngeal (NP) samples studied by multiplex PCR (targeting 15 viruses and four intracellular bacteria). An adjudication committee composed of infectious disease specialists, pneumologists and radiologists blinded to PCR results reviewed patient records, including computed tomography and day 28 follow up, to categorize final diagnostic probability of CAP as definite, probable, possible, or excluded. Among the 254 patients enrolled, 78 (31%) had positive PCR, which detected viruses in 72/254 (28%) and intracellular bacteria in 8 (3%) patients. PCR was positive in 44/125 (35%) patients with definite CAP and 21/83 (25%) patients with excluded CAP. The most frequent organisms were influenza A/B virus in 27 (11%), rhinovirus in 20 (8%), coronavirus in seven (3%), respiratory syncytial virus in seven (3%) and Mycoplasma pneumoniae in eight (3%) of 254 patients. Proportion of rhinovirus was higher in patients with excluded CAP compared with other diagnostic categories (p = 0.01). No such difference was observed for influenza virus. Viruses seem common in adults attending emergency departments with suspected CAP. A concomitant clinical, radiological and biological analysis of the patient's chart can contribute to either confirm their role, or suggest upper respiratory tract infection or shedding. Their imputability and impact in early management of CAP deserve further studies.
Clinical Trials Registration. NCT01574066.
Keywords: Adjudication committee, community-acquired pneumonia, emergency departments, multiplex PCR, nasopharyngeal swabs, viruses
Introduction
Community-acquired pneumonia (CAP) is increasingly recognized as a leading cause of hospital admission and death among adults in industrialized countries [1] and one of the most common infectious diseases encountered in emergency departments (ED). Its annual incidence is estimated at 600 cases per 100 000 inhabitants, which represents in France around 300 000 cases per year causing 3500–11 000 annual deaths, particularly in the elderly population.
Currently, the diagnosis of CAP is heavily dependent on clinical and radiological findings, some of which are largely non-specific and of unknown aetiology [2], [3], [4], [5], and treatment remains in most cases empirical [6], [7], [8]. The conventional microbiological techniques, such as culture, serology, antigen detection, have practical challenges, notably, complex technology, poor sensitivity and prolonged turnaround time. Since the advent of PCR, the advances in these new molecular techniques have shown their applicability to improve the detection of agents causing various types of infections. The multiplex PCR method allows the potential amplification of many nucleic acid targets within a single reaction and has proven to be an attractive diagnostic method for multi-pathogen detection in respiratory tract infections [9], [10].
In ED, timely and accurate diagnosis is crucial for treatment decisions and the outcome of patients; it has been advocated that the prognosis of CAP depends on the immediate initiation of specific treatment no later than 8 h after diagnosis [11]. We present here the identification of infectious agents using multiplex PCR in adult patients with suspected CAP visiting EDs in France within the framework of the ESCAPED study, a prospective, open-label, multicentre impact measurement study of thoracic computed tomography (CT) scans.
Material and methods
The primary objective of the present microbiological study was to determine the prevalence of infectious agents (viruses and bacteria) in nasopharyngeal (NP) swabs using multiplex PCR in adults presenting with clinically suspected non-severe CAP. The study also aimed to describe detected pathogens according to the probability of diagnosis of CAP.
Context and design
The ESCAPED study was conducted in four university hospitals (Bichat, Cochin, Pitié-Salpêtrière and Tenon) in Paris, France. The study was approved by the ethics committee (Comité de Protection des Personnes—CPP 1, Paris, France) and patients gave written informed consent. Consecutive adult patients (≥18 years) presenting to the hospital EDs with a clinical suspicion of non-severe CAP (classes 1 and 2 on the CRB65 score) were prospectively enrolled. Clinical suspicion of CAP was based on investigator's own judgement and had to fulfil the following criteria: new onset of systemic features (at least one among sweat, fever, chills, aches and pain, temperature ≥38°C) and symptoms of an acute lower respiratory tract illness (at least one among: cough, sputum production, dyspnoea, chest pain, altered breathing sounds).
An expert adjudication committee comprising a senior infectious disease specialist, pneumologist and radiologist reviewed patient records using all clinical data including day 28 follow up, radiological (thoracic X-ray and CT scan performed at ED admission), and laboratory findings to assess the diagnosis of CAP a posteriori. They jointly established the diagnosis of CAP and a final diagnostic probability according to four categories—definite, probable, possible, and excluded CAP. The committee was blinded to PCR results.
Microbiological samples
Systematic NP swabs were collected at enrolment from the nose, and also the throat in a sub-population of patients, and placed in a Middle Virocult MWE (Sigma, St Louis, MO, USA) transport medium. Samples were kept at room temperature and sent to the virology laboratory of Bichat-Claude Bernard Hospital as soon as possible after collection. The NP samples used in this study were not frozen and thawed. The other samples such as blood, urine and sputum were collected for bacteriological examinations within the scope of routine care as indicated by the attending physician.
Multiplex PCR
The RespiFinder-19 assay (Pathofinder, Maastricht, the Netherlands) was used to detect 15 respiratory viruses—coronaviruses 229E, NL63, OC43; human metapneumovirus; influenza A, A(H1N1)pdm2009 and B viruses, parainfluenza viruses 1, 2, 3 and 4; respiratory syncytial viruses A and B; rhinovirus; adenovirus; and four intracellular bacteria—Bordetella pertussis, Chlamydophila pneumoniae, Legionella pneumophila, Mycoplasma pneumoniae—in one reaction. The RespiFinder-19 analysed the amplified PCR products by capillary electrophoresis using a DNA analyser (ABI 3130; Applied Biosystems, Darmststadt, Germany) and provided diagnosis within 6 h. The multiplex PCR method used in the present study was validated by the virology lab that participates every year to the European quality control program for each respiratory virus.
Statistical analysis
Data collected included demographic variables such as gender and age, clinical signs of CAP, coexisting conditions, severity score, as assessed by Fine score (12) and biological data, in particular inflammatory biomarkers.
Continuous data were compared using Student t test or analysis of variance. The Mann-Whitney U test or Kruskal-Wallis method were used for non-parametric comparisons. For categorical variables, proportions were compared using chi-squared or Fisher's exact test. Statistical analyses were performed using Stata software, version 11.2 (StataCorp, College Station, TX, USA).
Results
Patients' characteristics
In the ESCAPED study, 319 adult patients with suspected CAP were enrolled between November 2011 and December 2012. Of these, 254 (80%) patients with NP swabs available were included in the study, of whom, 51% were female, and with a mean (SD) age of 65 ± 19 years. The baseline characteristics of the 254 PCR study patients are shown in Table 1 . One third (34%) of patients belonged to the Class IV or Class V of the FINE score (12). The baseline clinical characteristics of the 65 patients without swab (Non PCR Group) were not statistically different from those of patients who underwent PCR assay (PCR Study Group) except headache (more frequent in patients in PCR Study Group) and impaired consciousness (more frequent in patients in Non PCR Group).
Table 1.
Baseline characteristics of the 254 patients enrolled in PCR study according to PCR results
| PCR study group (254 patients), n (%) | PCR-positive (78 patients), n (%) | PCR-negative (176 patients), (%) | p Value (PCR-pos. versus PCR-neg.) | |
|---|---|---|---|---|
| Demographic characteristics | ||||
| Sex, female | 130 (51) | 37 (47) | 93 (53) | 0.43 |
| Age, years, mean (SD) | 65 (19) | 60 (21) | 66 (18) | 0.06 |
| Clinical features | ||||
| Temperature >38 or <36°C | 88 (35) | 28 (36) | 60 (34) | 0.44 |
| Cough | 191 (75) | 72 (92) | 119 (68) | <0.001 |
| Chest pain | 88 (35) | 19 (24) | 69 (39) | 0.03 |
| Sputum production | 120 (47) | 48 (62) | 72 (41) | 0.001 |
| Dyspnoea | 182 (72) | 58 (74) | 124 (71) | 0.31 |
| Chills | 83 (33) | 24 (31) | 59 (34) | 0.34 |
| Headache | 47 (19) | 20 (26) | 27 (15) | 0.03 |
| Aches and pain | 50 (20) | 24 (31) | 26 (15) | 0.001 |
| Crepitation (unilateral) | 87 (34) | 32 (41) | 55 (31) | 0.17 |
| Heart rate >90/min | 172 (68) | 54 (69) | 118 (67) | 0.42 |
| Respiratory rate >20/min | 121 (60) | 43 (68) | 78 (56) | 0.07 |
| Coexisting conditions | ||||
| At least one comorbidity | 114 (45) | 32 (41) | 82 (47) | 0.41 |
| Neoplastic disease | 28 (11) | 3 (4) | 25 (14) | 0.02 |
| Congestive cardiac disease | 28 (11) | 8 (10) | 20 (11) | 1 |
| Chronic lung disease | 71 (28) | 21 (27) | 50 (28) | 0.88 |
| Before ED characteristics | ||||
| Duration of symptoms before ED,(hours, mean (SD) | 173.3 (240.5) | 171.6 (245.8) | 174.0 (238.8) | 0.9 |
| Antibiotics before ED | 89 (35) | 34 (44) | 55 (31) | 0.04 |
| FINE classificationa | ||||
| Class I | 38 (15) | 17 (22) | 21 (12) | |
| Class II | 73 (29) | 22 (28) | 51 (29) | |
| Class III | 56 (22) | 21 (27) | 35 (20) | 0.06 |
| Class IV | 68 (27) | 15 (19) | 53 (30) | |
| Class V | 19 (7) | 3 (4) | 16 (9) | |
Abbreviations: ED, emergency departments; Neg., negative; Pos., positive. Numbers in bold are p values <0.05.
The p value was calculated with the use of t test, otherwise chi-squared test was used.
A prediction rule to assess the severity of community-acquired pneumonia [12].
Multiplex PCR findings
Among the 254 patients, 78 (31%) patients had at least one infectious agent detected (Fig. 1 ). The baseline characteristics of the 78 patients were not different from those with negative PCR results (Table 1) except neoplastic disease was more frequent in the negative PCR group.
Fig. 1.
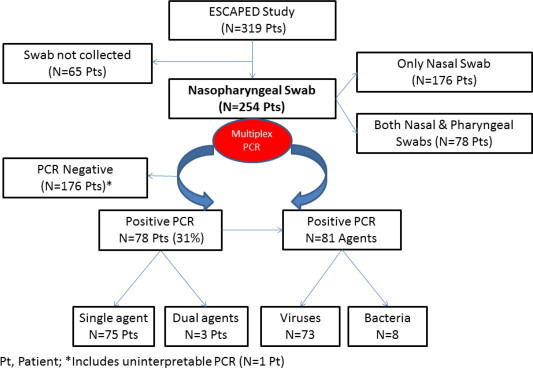
Flow chart of detection of infectious agents in nasopharyngeal swabs by multiplex PCR in the 319 patients included in the ESCAPED study.
The vast majority of patients (n = 75, 96%) had a single infectious agent, with a total of 81 pathogens detected in these 78 patients. Of the 254 study patients, 72 (28%), had a virus detected (Table 2 ). In the 78 patients with positive PCR, the most prevalent viral pathogens were influenza A/B virus in 35%, rhinovirus in 26%, respiratory syncytial virus A/B in 9% and coronavirus 229E/NL63/OC43 in 9% of patients. Other viruses identified were human metapneumovirus, parainfluenza viruses 3/4 and adenovirus. The only bacterium detected was M. pneumoniae in eight, 3% of patients. The seasonal distribution of the detected pathogens is shown in Fig. 2 .
Table 2.
Multiplex PCR detection of virus and bacteria in nasopharyngeal swabs collected from 78 of the 254 adult patients enrolled in the PCR study
| Type | Patients with positive PCR (n = 78) n (%) |
|---|---|
| Single agent | 75 (96) |
| Influenza A/B | 27 (35) |
| Rhinovirus | 20 (26) |
| Respiratory syncytial virus A/B | 7 (9) |
| Coronavirus 229E/NL63/OC43 | 7 (9) |
| Mycoplasma pneumoniae | 6 (8) |
| Parainfluenza virus 3/4 | 4 (5) |
| Human metapneumovirus (hMPV) | 3 (4) |
| Adenovirus | 1 (1) |
| Multiple agents | 3 (4) |
| Co229E—Mycoplasma pneumoniae | 1 (1) |
| Coronavirus OC43—hMPV | 1 (1) |
| Rhinovirus—Mycoplasma pneumoniae | 1 (1) |
Fig. 2.
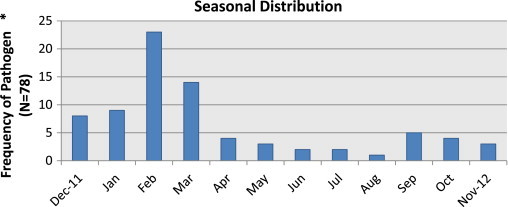
Seasonal distribution of detected pathogens in nasopharyngeal swabs by multiplex PCR in the 319 patients included in the ESCAPED study. * As compared with the total number of samples.
The multiplex PCR detected viruses in both NP samples in 72 (92% concordant results) of the 78 patients with both NP swabs taken. The central virology laboratory received and processed 91% (232/254) of NP swabs within 24 h. The probability of PCR positivity was not influenced by the time intervals between NP swab collection and PCR testing. The PCR positivity rate for viral infection and the agents found were comparable between NP (20/78) and only nasal (58/176) samples.
Probability of diagnosis of CAP and PCR results
Among the 254 patients, 49% were classified as definite CAP, 7% as probable, 11% as possible and 33% were excluded by the adjudication committee (Table 3 ). Although the PCR findings were not different according to the diagnosis probability, among the 125 patients with definite CAP, 44 (35%) had positive PCR results and 81 (65%) were PCR negative. Among the 83 patients with excluded CAP, 21 (25%) were PCR positive. The probability of PCR detection of rhinoviruses was significantly higher in excluded CAP than in the other three groups combined (10/21 (48%) versus 10/57 (18%), respectively, p 0.01), whereas there was no difference in detection of influenza viruses (9/21 (43%) versus 18/57 (32%), respectively). Referring to the category of diagnosis, the sensitivity and specificity of multiplex PCR were 35% and 74%, and the positive and negative predictive values were 56% and 54%, respectively.
Table 3.
PCR findings according to the adjudication committee diagnostic probability of community-acquired pneumonia (CAP) in the 254 adult patients enrolled in the PCR study
| Adjudication committee diagnosis of CAP | PCR results, n (%) |
|||
|---|---|---|---|---|
| Positive |
Negative | Total | ||
| Types of agents | (n) | |||
| Definite | 44 (35)a | 81 (65) | 125 | |
| Influenza A/B | 12 | |||
| Rhinovirus | 6 | |||
| Respiratory syncytial virus A/B | 5 | |||
| Coronavirus 229E/NL63/OC43 | 4 | |||
| Mycoplasma pneumoniaa | 6 | |||
| Parainfluenza virus 3/4 | 4 | |||
| Human metapneumovirus | 3 | |||
| Adenovirus | 1 | |||
| Multiple agentsa | 3 | |||
| Probable | 4 (22) | 14 (78) | 18 | |
| Influenza A/B | 1 | |||
| Rhinovirus | 1 | |||
| Respiratory syncytial virus A/B | 1 | |||
| Coronavirus 229E/NL63/OC43 | 1 | |||
| Possible | 9 (32) | 19 (68) | 28 | |
| Influenza A/B | 5 | |||
| Rhinovirus | 3 | |||
| Coronavirus 229E/NL63/OC43 | 1 | |||
| Excluded | 21 (25) | 62 (75) | 83 | |
| Influenza A/B | 9 | |||
| Rhinovirus | 10 | |||
| Respiratory syncytial virus A/B | 1 | |||
| Coronavirus 229E/NL63/OC43 | 1 | |||
| Total | 78 (31) | 176 (69) | 254 | |
Includes the eight patients with positive PCR for Mycoplasma pneumoniae.
Routine bacteriological findings
Routine bacteriological examinations were available for 190 (75%) of the 254 patients. Bacterial agents were detected in 28 patients, of whom 11 (39%) had a probably non-pulmonary infection, and 17 (61%) had detection of bacteria with ‘lung tropism’, possibly associated with CAP (Table 4 ). They were: Streptococcus pneumoniae in nine patients; Legionella pneumophila and other intracellular agents in three patients; and Haemophilus influenzae, Branhamella catarrhalis, a Gram-positive coccus, Mycobacterium tuberculosis, and an Enterobacteriaceae, each in one patient. Of the 17 patients with bacteria with “lung tropism”, 11 had definite and two had probable or possible CAP according to the experts.
Table 4.
Positive bacteriological data from routine assessment in 28 of the 254 adult patients with suspected CAP enrolled in of the study
| PCR detection of viruses | Type of bacteriologial pathogen | Modality of detection |
Diagnosis of CAPa | |||
|---|---|---|---|---|---|---|
| Respiratory sample | Blood culture | Urine antigen | Others | |||
| PCR positive | ||||||
| Influenza A virus | Streptococcus pneumoniae | Blood culture | Definite | |||
| Influenza A virus | Escherichia coli; Staphylococcus coagulase negative | Ascitic fluid | Excluded | |||
| Influenza A virus | Gram-positive cocci | Sputum | Possible | |||
| Influenza A virus | Group B streptococcus | Urine culture | Definite | |||
| Rhino virus | Branhamella catarrhalis | Sputum | Excluded | |||
| Rhino virus | Escherichia coli | Urine culture | Definite | |||
| Rhino virus | Streptococcus pneumoniae (sputum and urine antigen); Escherichia coli (sputum culture) | Sputum | Urine antigen | Definite | ||
| Rhino virus | Streptococcus pneumoniae | Sputum | Excluded | |||
| PCR negative | ||||||
| Negative | Streptococcus pneumoniae | Blood culture | Urine antigen | Definite | ||
| Negative | Other intracellular | Serology | Definite | |||
| Negative | Other intracellular | Serology | Probable | |||
| Negative | Mycobacterium tuberculosis | Sputum | Excluded | |||
| Negative | Streptococcus pneumoniae | Urine antigen | Definite | |||
| Negative | Escherichia coli | Vertebral puncture culture | Excluded | |||
| Negative | Legionella pneumophila | Urine antigen | Definite | |||
| Negative | Other pyogen | Blood culture | Possible | |||
| Negative | Enterobacteriaceae | Sputum | Excluded | |||
| Negative | Enterobacteriaceae | Urine culture | Definite | |||
| Negative | Other pyogen | Urine culture | Excluded | |||
| Negative | Enterobacteriaceae | Blood culture | Urine culture | Excluded | ||
| Negative | Enteroccoccus sp.; Citrobacter | Urine culture | Excluded | |||
| Negative | Streptococcus pneumoniae | Sputum | Urine antigen | Definite | ||
| Negative | Haemophilus influenzae | Blood culture | Definite | |||
| Negative | Streptococcus pyogenes (blood); Escherichia coli (urine) | Blood culture | Urine culture | Possible | ||
| Negative | Enterobacteriaceae | Blood culture | Excluded | |||
| Negative | Streptococcus pneumoniae | Urine antigen | Definite | |||
| Negative | Streptococcus pneumoniae | Urine antigen | Definite | |||
| Streptococcus pneumoniae | Blood culture | Urine antigen | Definite | |||
Adjudication committee diagnosis of CAP.
A bacterial–viral co-infection was present in seven (3.6%) of the 190 patients. The proportion of bacterial documentation was not different according to antibiotic administration before admission to ED. Overall, 95 (37%) CAP patients had detection of a viral or bacterial agent that was possibly responsible for the CAP.
Discussion
The present study found that approximately one-third of adult ED patients presenting with suspected CAP had PCR detection of viruses in the upper respiratory tract, mainly influenza A/B virus and rhinoviruses. For bacteria, multiplex PCR detected only M. pneumoniae, although one patient was diagnosed with CAP due to L. pneumophila by antigenuria detection.
To our knowledge, this is the first prospective clinical trial in ED describing the viruses associated with suspected CAP in adult patients taking into account an adjudication committee diagnosis of CAP as ‘reference’ (due to the inadequate accuracy of conventional diagnostic tests). Of note, the adjudication committee based its judgement not only on follow-up data, but also on systematic day 0 thoracic CT scan, which is unique. The value of 35% PCR-positive patients with definite CAP is in agreement with several studies without an adjudication committee, which reported detection of viruses in a substantial proportion of patients with CAP. Jennings et al. and Johansson et al. both reported a viral diagnosis in 29% of hospitalized adult patients with CAP of similar age group (61–70 years) by PCR, whereas Liu et al. detected 36% viruses in adult outpatients with CAP and Templeton et al. detected viruses in 56% of adult patients from outpatient and inpatient departments, of whom half of the patients were <60 years old [2], [3], [4], [13]. The most frequently detected viruses in our study were influenza A/B virus and rhinoviruses, as reported in previous studies [14]. Like our study, these reports addressed patients with an approximately similar rate, 28–32%, of coexisting conditions. Paediatric studies identified viral infections by real-time PCR in 28% of hospitalized patients, increasing to 50% in those with asthma [14], [15]. The frequency and distribution of viruses detected were slightly different in two recent studies of patients in intensive care units and of elderly adults with moderate to severe illness [16], [17]. These differences may partly be due to differences in viruses circulating in the community during the study period or to the sensitivity and specificity of diagnostic technology used. The seasonal distribution of detected pathogens in our study was similar to that observed in the community French surveillance network of patients with influenza-like illness in 2012–2013 (V Enouf, Institut Pasteur, personal communication). All of these findings confirm the major changes induced by molecular techniques in describing CAP-associated microorganisms; in comparison, a review of 26 prospective studies in ten European countries (approximately 6000 patients), published in 1998 before the wide use of PCR, found respiratory viruses accounted for only 8% of patients with CAP [18].
In patients with a definite CAP diagnosis, the 35% rate of PCR-positive results has to be balanced with the still high rate of negative results, which could be explained by other viral, bacterial or fungal agents that are not included as targets in the RespiFinder-19 assay. The fact that NP samples are only a proxy of the infectious agents present deeper in the lung has to be considered. Whether lower respiratory samples would have facilitated imputability and given fewer negative results has to be studied in patients with more severe conditions that allow for invasive diagnostic investigations [16]. It is important to admit that multiplex PCR may have different analytical sensitivities for different viruses, which would affect the diagnostic yield. It is interesting to underline that the eight patients with PCR-detected M. pneumoniae had definite CAP, reinforcing the interest of this test in the context of M. pneumoniae epidemics.
The 25% PCR-positive patients among those with an excluded diagnosis of CAP might be explained by the fact that the infection was localized in the bronchi and the upper respiratory tract and did not affect the pulmonary parenchyma. This could also be explained by the PCR detection of viral shedding in patients with other diseases mimicking CAP symptoms [19]. In our study, the higher proportion of rhinoviruses in patients with excluded CAP suggests that at least part of these rhinoviruses were either viral shedding or could be associated with limited upper respiratory tract infection, as suggested previously in symptomatic and asymptomatic children and adults [20], [21], [22]. The seasonal distribution of influenza A/B virus detection and the similar proportion in excluded and non-excluded CAP suggest that pneumonia is not the primary manifestation of influenza, as already known [23]. Another hypothesis for this high rate of positive PCR in excluded CAP may be that some patients with minimal or early lesions of CAP could present with a normal CT scan, as has been known for a long time for chest X-rays.
The epidemiology of CAP in adults has probably changed in recent years because of the indirect impact of pneumococcal conjugate vaccines in children [24]. However, establishing the aetiology of CAP remains difficult and the imputability of pathogens detected in the upper respiratory tract remains questionable. As in the present study, no cause is determined in half or more of the patients. Even considering the limit related to the routine non-standardized sampling for bacterial investigations in the study, Streptococcus pneumoniae was the most commonly identified bacteria associated with CAP, as is generally recognized. The rate of detection of other bacteria, and atypical agents was similar to that reported in a recent review [24]. Respiratory viruses, particularly influenza virus and respiratory syncytial virus, are commonly detected in patients with CAP as described here [14]. However, it remains challenging to differentiate viral CAP from mixed-infection or bacterial CAP. It is a hypothesis that the increasing recognition (15–54%) of viruses as a cause of CAP in adults may reflect advances in viral detection by PCR-based testing. Better ways to diagnose viral CAP would increase the use of targeted antiviral therapies. Specifically, detection of influenza virus should lead to consideration of antiviral therapy within the 48 h after onset of symptoms, which has been shown to improve prognosis [23]. It can be useful to establish the virological diagnosis for epidemiological purposes in hospitalized patients who may require infection control measures to prevent nosocomial transmission, especially in the case of a new and/or contagious virus outbreak, such as the recent Middle East respiratory syndrome coronavirus outbreak [25]. Whether the rapid viral diagnosis is likely to change individual antibiotic management and reduce antibiotic overuse has to be studied [24], [26]. Nevertheless, the relatively low sensitivity of the test and the high cost issues need to be addressed before widespread use of the multiplex PCR method.
The current study had several limitations. The multiplex PCR (RespiFinder-19) assay was not performed in real time, a limitation that may have underestimated identification of viral agents. However, this assay is generally considered to be as sensitive as monoplex PCR, which suggests that such a delay is not a major determinant of false-negative results [27]. One major issue is that the PCR test did not include S. pneumoniae, the leading organism of CAP, as a target [3], [24], [28], [29], which precludes comments on the respective roles of S. pneumoniae and viruses as CAP agents in our study. Using sputum samples in adult patients attending ED, Yang et al. found that PCR assay might be a useful diagnostic adjunct for clinicians to detect a pneumococcal pneumonia [30].
In summary, viruses in nasopharyngeal samples seem common in adults attending ED with suspected CAP. A concomitant clinical, radiological and biological analysis of the patient's chart can contribute to either confirm their role in CAP, or suggest upper respiratory tract infection or shedding. Their imputability and possible impact in early management of CAP deserve further studies.
Funding
The study was funded by the French Ministry of Health (PHRC AOM 10014) and sponsored by Assistance Publique—Hôpitaux de Paris (O. Chassany, C. Misse).
Transparency declaration
DD and CL: No potential conflicts.
Acknowledgements
We thank the patients and the investigators for their participation in the ESCAPED study. We also thank the ESCAPED Study Group and all staff members of Bichat, Cochin, Pitié-Salpêtrière and Tenon Hospitals in Paris who were involved in this study for their excellent work and dedication in executing and organizing the trial in their emergency departments. We gratefully acknowledge the invaluable contribution of the members of the adjudication committee. We are grateful to Gilles Collin for performing PCR tests and to Philippe Guerin for scientific discussions. We thank Berangere Gallet de Saint Aurin for her assistance in database construction and in data management and Melanie Huet for secretarial assistance.
Editor: L. Kaiser
The ESCAPED study group
Scientific committee:
Steering committee: Y.E. Claessens, (principal investigator), X. Duval (co-principal investigator), M.F. Carette, C. Mayaud, C. Leport.
Other members: E. Bouvard, M.P. Debray, N. Houhou, S. Tubiana.
Adjudication committee*: M. Benjoar, X. Blanc, A.L Brun, L. Epelboin, C. Ficko, A. Khalil, H. Le Floch, J.M. Naccache, B. Rammaert.
Clinical investigators*: J.C. Allo, S. Andre, C. Andreotti, A. Aubry, N. Baarir, M. Bendahou, L. Benlafia, J. Bernard, A. Berthoumieu, M.E. Billemont, J. Bokobza, A.L. Brun, E. Burggraff, P. Canavaggio, M.F. Carette, E. Casalino, S. Castro, C. Choquet, Y.E. Claessens, H. Clément, L. Colosi, A. Dabreteau, S. Damelincourt, S. Dautheville, M.P. Debray, M. Delay, S. Delerme, L. Depierre, F. Djamouri, F. Dumas, X. Duval, M.R.S. Fadel, A. Feydey, Y. Freund, L. Garcia, H. Goulet, P. Hausfater, E. Ilic-Habensus, M.O. Josse, J. Kansao, Y. Kieffer, F. Lecomte, K. Lemkarane, B. Madonna-Py, O. Meyniard, L. Mzabi, D. Pariente, J. Pernet, F. Perruche, J.M. Piquet, R. Ranerison, P. Ray, F. Renai, E. Rouff, D. Saget, K. Saïdi, G. Sauvin, E. Trabattoni, N. Trimech.
Monitoring, data management and statistical analysis: C. Auger, S. Tamazirt, F. Tubach, J.M. Treluyer, J. Wang.
*Investigators participating to the adjudication committee did not validate the cases from their own centre.
References
- 1.Ruuskanen O., Lahti E., Jennings L.C., Murdoch D.R. Viral pneumonia. Lancet. 2011;377:1264–1275. doi: 10.1016/S0140-6736(10)61459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jennings L.C., Anderson T.P., Beynon K.A., Chua A., Laing R.T., Werno A.M. Incidence and characteristics of viral community-acquired pneumonia in adults. Thorax. 2008;63:42–48. doi: 10.1136/thx.2006.075077. [DOI] [PubMed] [Google Scholar]
- 3.Johansson N., Kalin M., Tiveljung-Lindell A., Giske C.G., Hedlund J. Etiology of community-acquired pneumonia: increased microbiological yield with new diagnostic methods. Clin Infect Dis. 2010;50:202–209. doi: 10.1086/648678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Templeton K.E., Scheltinga S.A., van den Eeden W.C., Graffelman A.W., van den Broek P.J., Claas E.C. Improved diagnosis of the etiology of community-acquired pneumonia with real-time polymerase chain reaction. Clin Infect Dis. 2005;41:345–351. doi: 10.1086/431588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cesario T.C. Viruses associated with pneumonia in adults. Clin Infect Dis. 2012;55:107–113. doi: 10.1093/cid/cis297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mandell L.A., Wunderink R.G., Anzueto A., Bartlett J.G., Campbell G.D., Dean N.C. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44:S27–S72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim W.S., Macfarlane J.T., Boswell T.C., Harrison T.G., Rose D., Leinonen M. Study of community acquired pneumonia aetiology (SCAPA) in adults admitted to hospital: implications for management guidelines. Thorax. 2001;56:296–301. doi: 10.1136/thorax.56.4.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnstone J., Mandell L. Guidelines and quality measures: do they improve outcomes of patients with community-acquired pneumonia? Infect Dis Clin North Am. 2013;27:71–86. doi: 10.1016/j.idc.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Stockton J., Ellis J.S., Saville M., Clewley J.P., Zambon M.C. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J Clin Microbiol. 1998;36:2990–2995. doi: 10.1128/jcm.36.10.2990-2995.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poritz M.A., Blaschke A.J., Byington C.L., Meyers L., Nilsson K., Jones D.E. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One. 2011;6:e26047. doi: 10.1371/journal.pone.0026047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houck P.M., Bratzler D.W., Nsa W., Ma A., Bartlett J.G. Timing of antibiotic administration and outcomes for Medicare patients hospitalized with community-acquired pneumonia. Arch Intern Med. 2004;164:637–644. doi: 10.1001/archinte.164.6.637. [DOI] [PubMed] [Google Scholar]
- 12.Fine M.J., Auble T.E., Yealy D.M., Hanusa B.H., Weissfeld L.A., Singer D.E. A prediction rule to identify low-risk patients with community-acquired pneumonia. N Engl J Med. 1997;336:243–250. doi: 10.1056/NEJM199701233360402. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y.F., Gao Y., Chen M.F., Cao B., Yang X.H., Wei L. Etiological analysis and predictive diagnostic model building of community-acquired pneumonia in adult outpatients in Beijing, China. BMC Infect Dis. 2013;13:309. doi: 10.1186/1471-2334-13-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavia A.T. What is the role of respiratory viruses in community acquired pneumonia ; what is the best therapy for rinfluenza and other viral causes of CAP. Infect Dis Clin North Am. 2013;27:157–175. doi: 10.1016/j.idc.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khamis F.A., Al-Kobaisi M.F., Al-Areimi W.S., Al-Kindi H., Al-Zakwani I. Epidemiology of respiratory virus infections among infants and young children admitted to hospital in Oman. J Med Virol. 2012;84:1323–1329. doi: 10.1002/jmv.23330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi S.H., Hong S.B., Ko G.B., Lee Y., Park H.J., Park S.Y. Viral infection in patients with severe pneumonia requiring intensive care unit admission. Am J Respir Crit Care Med. 2012;186:325–332. doi: 10.1164/rccm.201112-2240OC. [DOI] [PubMed] [Google Scholar]
- 17.Falsey A.R., McElhaney J.E., Beran J., van Essen G.A., Duval X., Esen M. Respiratory syncytial virus and other respiratory viral infections in older adults with moderate to severe influenza-like illness. J Infect Dis. 2014;209:1873–1881. doi: 10.1093/infdis/jit839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodhead M. Community-acquired pneumonia guidelines–an international comparison: a view from Europe. Chest. 1998;113 doi: 10.1378/chest.113.3_supplement.183s. 183S–187S. [DOI] [PubMed] [Google Scholar]
- 19.Gerna G., Piralla A., Rovida F., Rognoni V., Marchi A., Locatelli F. Correlation of rhinovirus load in the respiratory tract and clinical symptoms in hospitalized immunocompetent and immunocompromised patients. J Med Virol. 2009;81:1498–1507. doi: 10.1002/jmv.21548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicholson K.G., Kent J., Hammersley V., Cancio E. Risk factors for lower respiratory complications of rhinovirus infections in elderly people living in the community: prospective cohort study. BMJ. 1996;313:1119–1123. doi: 10.1136/bmj.313.7065.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kusel M.M., de Klerk N.H., Holt P.G., Kebadze T., Johnston S.L., Sly P.D. Role of respiratory viruses in acute upper and lower respiratory tract illness in the first year of life: a birth cohort study. Pediatr Infect Dis J. 2006;25:680–686. doi: 10.1097/01.inf.0000226912.88900.a3. [DOI] [PubMed] [Google Scholar]
- 22.Srinivasan A., Flynn P., Gu Z., Hartford C., Lovins R., Sunkara A. Detection of respiratory viruses in asymptomatic children undergoing allogeneic hematopoietic cell transplantation. Pediatr Blood Cancer. 2013;60:149–151. doi: 10.1002/pbc.24314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ploin D., Chidiac C., Carrat F., Cohen B., Javouhey E., Mayaud C. Complications and factors associated with severity of influenza in hospitalized children and adults during the pandemic wave of A(H1N1)pdm2009 infections–the Fluco French cohort. J Clin Virol. 2013;58:114–119. doi: 10.1016/j.jcv.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 24.Musher D.M., Thorner A.R. Community-acquired pneumonia. N Engl J Med. 2014;371:1619–1628. doi: 10.1056/NEJMra1312885. [DOI] [PubMed] [Google Scholar]
- 25.Guery B., Poissy J., El Mansouf L., Sejourne C., Ettahar N., Lemaire X. Clinical features and viral diagnosis of two cases of infection with Middle East Respiratory Syndrome coronavirus: a report of nosocomial transmission. Lancet. 2013;381:2265–2272. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roson B.C.J., Fernandez-Sabe N., Tubau F., Manresa F., Gudiol F. Causes and factors associated with early failure in hospitalized patients with community-acquired pneumonia. Arch Intern Med. 2004;164:502–508. doi: 10.1001/archinte.164.5.502. [DOI] [PubMed] [Google Scholar]
- 27.Dabisch-Ruthe M., Vollmer T., Adams O., Knabbe C., Dreier J. Comparison of three multiplex PCR assays for the detection of respiratory viral infections: evaluation of xTAG respiratory virus panel fast assay, RespiFinder 19 assay and RespiFinder SMART 22 assay. BMC Infect Dis. 2012;12:163. doi: 10.1186/1471-2334-12-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruiz-Gonzalez A., Falguera M., Nogues A., Rubio-Caballero M. Is Streptococcus pneumoniae the leading cause of pneumonia of unknown etiology? A microbiologic study of lung aspirates in consecutive patients with community-acquired pneumonia. Am J Med. 1999;106:385–390. doi: 10.1016/s0002-9343(99)00050-9. [DOI] [PubMed] [Google Scholar]
- 29.Menendez R., Cordoba J., de La Cuadra P., Cremades M.J., Lopez-Hontagas J.L., Salavert M. Value of the polymerase chain reaction assay in noninvasive respiratory samples for diagnosis of community-acquired pneumonia. Am J Respir Crit Care Med. 1999;159:1868–1873. doi: 10.1164/ajrccm.159.6.9807070. [DOI] [PubMed] [Google Scholar]
- 30.Yang S., Lin S., Khalil A., Gaydos C., Nuemberger E., Juan G. Quantitative PCR assay using sputum samples for rapid diagnosis of pneumococcal pneumonia in adult emergency department patients. J Clin Microbiol. 2005;43:3221–3226. doi: 10.1128/JCM.43.7.3221-3226.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


